Introduction
The vast proliferation of human endogenous retroviruses, and their derived and dependent products, in the human genome is increasingly recognized to be important in human embryology, normal physiology and disease. I introduced this subject in a preliminary paper, in 2004,1 but progress during the ensuing five years demands a wholesale reappraisal. As with mitochondrial symbiosis, we find profound differences, conceptually, and mechanistically, between viral symbiogenesis and what we have come to expect of mutation per se. Moreover, while there are helpful commonalities with the symbiotic mechanisms of mitochondrial genetics, there are also important differences. These differences, which have evolutionary and medical implications, derive from the quintessential nature of viruses, and their evolved genetic and genomic behavioural patterns.
Retroviral symbiosis
We are familiar with exogenous retroviruses, such as the human T-cell leukaemia viruses, HTLV-I and HTLV-II, and the AIDS pandemic, caused by the human immunodeficiency viruses, HIV-1 and HIV-2. All such retroviruses enter into a state of ‘persisting infection’ with the host population, and this has symbiogenetic implications.2 For example, even in the prevailing pandemic of AIDS, there is intense co-evolution between HIV-1 and the human major histocompatibility antigens, typifying the early stage of ‘aggressive symbiosis’.3 When the virus infects a new host it first discovers its target cells, which appears to be the CD4+T lymphocyte and macrophage.4 Here, employing its retrovirus-specific enzyme reverse transcriptase, it convert its RNA-based viral genome to the homologous DNA before uniting with the host genome, subverting its normal controls to convert it into a factory for the production of virus. This means that the virus is pre-evolved to manipulate its host genome, albeit at peripheral cell level. Many retroviruses have the additional, remarkable, ability to invade the germ-lines of their hosts, a process known as ‘endogenization’, which gives rise to a new symbiogenetic evolutionary entity – the holobiontic union of virus and host.
To date, the HIV-1 virus has not been shown to endogenize. Until recently many virologists believed that the lentivirus genus of retroviruses, which includes HIV-1, were unable to do so. But lentivirus endogenization has now been confirmed in European rabbits5 and an endogenized immunodeficiency lentivirus, thought to be ancestral to all primate lentiviruses, has been found in the genome of the grey mouse lemur, a basal primate that inhabits Madagascar.6 The latter may reflect an integration event dating back at least 14 million years, suggesting that lentiviruses have been co-evolving with primates for much longer than was previously considered.
Given the structure of the human genome, it would appear that our ancestors suffered many other retroviral epidemics throughout our human and primate history, progressing to germ-line union. If we consider that each invading virus was a complete, evolutionarily-competent, entity, and that it already possessed the pre-evolved capacity to manipulate the genome it was invading, we can anticipate that such unions would have resulted in major symbiogenetic potential. We can also extrapolate that such viral symbiogenesis would differ from what we saw with mitochondria, since bacterial genes and sequences have no such capacity for host genomic manipulation.
There are a few simple terms we need to define. When an ‘exogenous’ retrovirus becomes a permanent component of the human genome it is called a ‘human endogenous retrovirus’ (HERV). Unlike mitochondrial genes, HERVs are inherited in classical Mendelian fashion, as integral components of the new holobiontic nuclear genome. In Part 1, I referred to the retroviral epidemic affecting the koala population of Australia, which is following the typical pattern of ‘aggressive symbiosis’, with endogenization well under way within 100 years or so of the beginning of the epidemic. There is also evidence of a recent endogenization in humans, where HERV-K113, found on chromosome 19 in just 29% of people of mainly African, Asian and Polynesian descent, appears to have entered the human genome after the last great migration from Africa, less than 150,000 years ago.7 We are only beginning to appreciate the evolutionary importance of such retroviral invasions. For example, the evolutionary virologist, Villarreal, has concluded that there was a major explosion of new retroviruses with the origins of jawed vertebrates, another with the origins of mammals and again with the origins of the primates.8 These are likely to have played an important role in host evolution. The vertebrate-associated explosion gave rise to large-scale endogenous colonisation with resultant genomic expansion. This was coincident with the origins of adaptive immunity, leading Villarreal to propose a complex step-by-step scenario in which retroviral-host interaction gave rise to adaptive immunity as an integral part of the evolution of a highly sophisticated system of self-identity. We shall return to Villarreal's hypothesis in considering the viral role in the auto-immune diseases in a subsequent paper. Meanwhile, we shall focus on the retroviral invasions of the vertebrate, mammalian, primate and hominid genomes that have played an important role in our pre-human and human evolution.
All retroviruses have a similar basic genomic structure ( Figure 1). The viral genome comprises three genetic domains, each of which codes for multiple potentials, for example gag codes for matrix and core shell proteins, pol codes for enzymes such as reverse transcriptase, protease, ribonuclease and integrase, and env codes for the surface and transmembrane glycoproteins. The flanking regions, labeled LTRs, or ‘long terminal repeats’, possess a range of regulatory capabilities that can manipulate the expression of neighboring genes, regardless of whether they originally derived from the host or virus. Human endogenous retroviruses comprise between 30 to 50 families, depending on definition, and these families are further subdivided into more than 200 distinct groups and subgroups.9 Each of these represents an independent evolutionary lineage, which would imply that the human genome has been subject to a large series of independent HERV colonisations. Although many of these unions took place more than 10 million years ago, a significant number took place during our human period of evolution. For example, the families, CERV 1 (PTERV1) and CERV 2, are unique to chimpanzees,10 meanwhile eight out of 10 full-length HERV-K subgroups are unique to humans.11,12
Figure 1.
Basic genome of retrovirus
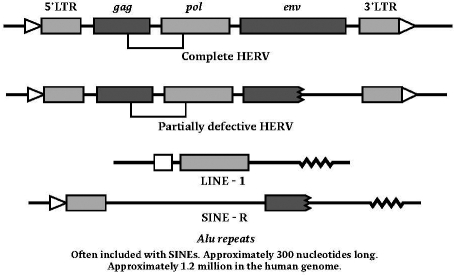
The first step in the symbiotic adaptation of an endogenized virus will be the loss of infectious behaviour. This has been achieved with the most recently endogenized human virus, HERV-K113, mentioned above, through the intervention of a single stop codon in its gag domain.13 From this point on selection working at holobiontic level will adapt both the viral and original vertebrate elements to the new partnership, so that mutation-plus-selection is now fulfilling an editorial role. Selfish elements will be silenced in the short term by epigenetic mechanisms, such as methylation, histone modification or RNAi, and in the longer term through genetic mechanisms such as mutation of their DNA sequences, or through elimination from the genome through chromosome breaks in cell division, or through deletions during homologous sexual recombination. But where viral integrity, in whole or in part, is important to the holobiont, the viral sequences will be conserved. These symbiogenetic mechanisms will apply to the elements of both host and viral genomes, and they will continue to apply long after the initial virus-host fusion, with positive selection preserving viral or host genes, viral or host translational sequences and viral or host control and developmental sequences that contribute to the evolving holobiont. This offers a comprehensive, and testable, explanation for the proliferation of whole viruses, and the so-called ‘defective HERVs’, and HERV products, such as LINEs and SINEs ( Figure 2).
Figure 2.
HERVs and products in the human genome
Some 9% of the human genome consists of complete human endogenous retroviruses, or HERVs, and their LTRs, and if we extend this to HERV genes, fragments and derivatives, such as LINEs and SINEs, the viral component amounts to roughly 43% of our DNA ( Figure 3).14,15 As with the mitochondrial symbiosis, the majority of HERV genes appear to have been silenced by the mechanisms listed above, leading an earlier generation of geneticists to dismiss all viral components as ‘junk DNA’.16,17 But now we know that many genes and sequences have remained active, and even truncated HERV genes, which would normally be assumed to be inactivated, may code for truncated proteins that may still fulfil a biologically significant function.18 Meanwhile the large percentage of the genome made up of HERVs, and their genetic legacy, beg some pertinent questions. What possible function has this vast retroviral legacy played in the evolution of the human genome? What role is it still playing in normal human embryology, physiology and biochemistry? Only when we can answer these questions is it possible to take the further step of considering the contribution of this retroviral legacy, for good or for bad, in human health and disease.
Figure 3.

DNA breakdown of human genome
The implications for human development and normal physiology
Two endogenous retroviruses, HERV-W and HERV-FRD, play an important role in the construction and physiological function of the human placenta, their env genes coding for the proteins syncytin-1 and syncytin-2, respectively, which fuse the trophoblast cells of the placenta into the confluent multinucleated syncytial layer.19–21 Each virus fulfils a specific and different role in placental construction, working in a complementary way with a third virus, ERV-3 (also known as HERV-R), in a complex developmental coordination.22 However, 1% of Caucasians may have a mutation that precludes the fusion and putative immunosuppressive functions of ERV-3, yet this does not appear to prevent pregnancy,23 suggesting that there may be some degree of redundancy, or flexibility, in the remarkable multi-HERV coordination. Only a minority of mammals has been assessed to date, but it would appear that the major mammalian groups have different patterns of placentation, involving different endogenous retroviruses, for example primates, mice and sheep,24–27 suggesting that mammalian placentation is either of polyphyletic origins or that newly incoming exogenous viruses may compete with and replace established viruses. Syncytin-2, unlike syncytin-1, has also been shown to have immunosuppressive properties that may play a role in the maternal tolerance to foetal antigens.28 Humans share their placental viruses with the great apes, and possibly old world, but not new world, monkeys.29 The exogenous retrovirus that gave rise to the HERV-W endogenous family is believed to have entered the ancestral genome almost 40 million years ago. When Bonnaud and his colleagues tracked the action of natural selection on the HERV-W locus ERVWE1 (whose env gene codes for syncytin-1) in chimpanzee, gorilla, orang-utan and gibbon, they found that the genetic signature crucial to the gene's fusogenic action had been conserved over the tens of millions of years of primate divergence.30 This is what one would anticipate from the symbiogenetic perspective, with selection operating at the level of the holobiontic genome.
There is growing evidence that many retroviruses and their products are active during human germ cell formation and embryogenesis. Based on the fact that mature spermatozoa are endowed with reverse transcriptase, Spadafora and associates have blocked its action, using the anti-retroviral drug Nevirapine, to discover that this caused an irreversible arrest of development up to the four-cell stage of mouse development.31 They further demonstrated that reverse transcriptase inhibition caused a substantial reprogramming of gene expression in arrested embryos, involving both developmental and translational genes.32 This suggests that endogenous retroviruses, or LINE type products, are playing an important role at the earliest stages of mammalian development. Larsson and colleagues have pioneered the study of HERVs in human embryogenesis, showing that ERV-3, one of the three viruses involved in placentation, is also highly expressed in many human foetal tissues, including adrenal cortex, kidney tubules, tongue, heart, liver and central nervous system.33 The same researchers have also shown that ERV-3 is highly expressed in the sebaceous glands of normal skin.34 De Parseval and colleagues searched the human genome for retroviral envelope genes with ‘open reading frames’ – genes available for transcription to proteins – to discover 16 HERV env genes, all of which were expressed in healthy tissues.35 Three of these were expressed in placenta, as seen above. One, a newly identified env gene, was expressed at a high level in the fully developed thyroid gland, and appeared to be exclusive to this organ, with a possible link to hormone secretion. Another HERV gene, envR, was expressed in the adrenal, where the association may again be linked to hormone secretion. It is significant that all 16 were expressed in the testis. This is congruent with the fact that endogenous retroviruses are a common finding in the male reproductive tract from Drosophila to mammals.36
De Parseval also found that, despite the fact that some of these genes were only transcribed at low level, all of the promoters were active, very likely in germ line expression. Others have detected HERV-W RNA in the human testis and HERV-R env mRNA appears to be expressed in the first phases of spermatogenesis but not in the Sertoli or Leydig cells.37 This expression appears to be linked to steroid hormones since, as Larsson and colleagues have shown, HERV-R contains androgen receptor sites in its 5'LTR. Proteins coded by HERV-derived L1 retrotransposons have also been found in prespermatogonia of foetal testis, in germ cells of adult testis, and in Leydig and Sertoli cells, as well as vascular endothelial cells, the latter suggesting a possible role in vasculogenesis.38
HERVs and their products appear to play an equally pervasive role in normal adult structure and physiology. Perhaps most dramatically of all, a number of researchers, including Larsson and colleagues at Uppsala, have shown expression of HERV env and gag genes in key tissues and structures of the human brain, indicating what appears to be structural or physiological function, or both.39.40 In particular, the Swedish group have demonstrated widespread and dense expression of the env proteins syncytin-1 and syncytin-2, presumably different variants of the proteins from those found in the placenta, which is undergoing further evaluation.41 Other researchers are studying the control and promotional sequences of HERVs in human physiology. For example Mager and colleagues have shown that the LTR of ERV-L controls most of the gene transcripts of the human gene β3GAL-T5 in the human colon,42 meanwhile Sverdlov and colleagues have shown two examples of HERV LTRs that are participating in the specific antisense regulation of the human genes SLC4A8 (sodium bicarbonate co-transporter) and IFT172 (intraflagellar transport protein 172).43 Previously the same researchers had shown that at least 50% of the human-specific HERV-K LTRs are active promoters for non-viral DNA transcription.44 For example, the LTR of ERV-9 has been shown to play a key role in transcriptional control of the β-globin gene cluster of humans.45,46 This key viral promoter has also been conserved throughout at least 15 million years of primate evolution, and appears to have displaced several other non-viral promoters within the locus control region. A range of virus-derived elements, including SINEs (Alus), LINEs, and LTRs, are found in a large number of human protein-coding genes, where most are inserted into introns – the non-coding regions between the coding regions, known as exons – and where they appear to influence the function of some 533 genes.47 Other LTRs have taken on the role of alternative promoters, or splice receptors, for example in the control of the endothelin B receptor and apolipoprotein C-I genes,48 and in the control of the human leptin receptor, which is involved in energy expenditure, production of sex hormones and activation of haemopoietic cells.49 Endogenous retroviruses have also been involved in the evolution, and tissue specific expression, of the enzyme amylase in humans.50,51 HERV sequences have also contributed functional genes, or parts of genes, to the human genome, including integrase52 and transaldolase.53
Viruses also have the ability to create entirely new genes (neogenes) through stitching together fragments of other genes,54 and HERVs have retained this capability. For example a novel gene, PIPSL, common to chimpanzee and human testis, appears to have been created by ‘exon-shuffling’.55 Another family of neogenes, called Mart, are located on the X chromosomes of various mammals, including humans.56 They play ubiquitous roles during embryogenesis of the mouse, with important function in the nervous system. Although largely unevaluated in humans, at least one of the Mart genes (Mart 8) is amplified in the human genome. It seems likely that, in time, more such virally-created neogenes will be discovered as part of our viral-symbiogenetic inheritance.
Virus-derived transposable elements have also been found to contribute to a substantial fraction of human regulatory sequences.57 LINEs are HERV-derived long interspersed repetitive elements that have massively reproduced themselves throughout the human genome. Although they have lost much of the original HERV genetic structures, they retain the enzymes necessary to replicate and move to new positions within the genome. This is known as ‘retrotransposition’. SINES are short interspersed repetitive elements that have also massively dispersed themselves through the genome, but they have lost the enzymes necessary for retrotransposition and so they must borrow these from other sources, most likely from LINEs.58 Recent work by Ohshima and colleagues has demonstrated evolutionary links between LINEs and SINEs, which, between them, comprise some 34% of the human genome.59 Mammalian specific LINES are known as LINE-1s (often abbreviated to L1s). Most of these have been inactivated by selection, but about 100 or so remain highly active, copying themselves and inserting elsewhere in the genome, where they play a significant role in the structure and regulation of the genome. One important role for L1s may be DNA repair.60–62 A subsection specific to humans has been specified variously as LINE-1H, or LIH, or sometimes Ta, and there is growing evidence that the rate of L1 amplification has been increasing during recent human evolution.63,64
Alu elements, also known as Alu repeats, are specific to primates and comprise a short sequence of approximately 300 nucleotides, which tend to be loosely included with SINEs. Like SINEs, they require the presence of HERV or LINE enzymes to insert themselves into a new location.65,66 They are so efficient at this that they have amplified to more than one million copies in the human genome, including 2000 or so that are specific to humans, and they continue to amplify at the rate of about one new insertion every 200 births, so that these more recent insertions have been used to track and survey human evolutionary origins.67,68 Where HERVs and LINEs tend to transpose themselves randomly within the genome, Alus appear to hone into regions that are particularly rich in vertebrate genes. This, as we shall see in the ensuing paper, results in a mutation-like propensity to disrupt normal gene expression, and thus give rise to a wide range of diseases. Nevertheless, these too have played an important role in the evolution of the human genome, through such insertions as well as recombination between elements, gene conversion and alterations in gene expression,69 and possibly in the evolution of the primate transcriptome.70
Geneticists are becoming increasingly aware of the importance of viral lineages, including genes, gene fragments, LTRs, LINES, SINES, and dependents such as Alus, on genetic and whole genomic function in humans. The observed viral roles fulfil the predictions of symbiogenetic evolution, as do the large repertoire of viral elements that code for a multiplicity of functions essential to the evolution and normal working of the genome, including the contribution of unique coding sequences, organisational roles during genomic duplication, accurate transmission to progeny cells, and a fundamental role in the cooperative molecular interactions intrinsic to nucleoprotein complexes.71,72 Indeed the viral roles in human genomic function are so widespread, yet still underestimated, that Flockerzi and colleagues have suggested the need for a specialised human endogenous retrovirus transcriptome project.73 A holistic long-term consequence of the symbiotic viral component of the genome is an increase in ‘genetic plasticity’, which has important implications for medicine as well as evolutionary biology.74 This is likely to contribute to the high level of genetic variation currently being observed between individual human genomes.75
Part 3 of this series will examine the role of HERVs and their products in miscellaneous human diseases and the autoimmune diseases.
Footnotes
DECLARATIONS —
Competing interests None declared
Funding None
Ethical approval Not applicable
Guarantor FPR
Contributorship FPR is the sole contributor
Acknowledgements
FPR would like to thank HarperCollins for permission to reproduce Figures 1, 2 and 3 from the book Virolution published in 2009
References
- 1.Ryan F. Human endogenous retroviruses in health and disease: a symbiotic perspective. J Royal Soc Med 2004;97:560–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villarreal LP. Viral persistence and symbiosis: are they related? Symbiosis 2007;43:1–9 [Google Scholar]
- 3.Kiepiela P, Leslie AJ, Honeyborne I, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 2004;432:769–74 [DOI] [PubMed] [Google Scholar]
- 4.Wahl SM, Greenwell-Wild T, Peng G, et al. Viral and host cofactors facilitate HIV-1 replication in macrophages. J Leukocyte Biology 2003;74:726–35 [DOI] [PubMed] [Google Scholar]
- 5.Katzourakis A, Tristem M, Pybus OG, Gifford RJ. Discovery and analysis of the first endogenous lentivirus. PNAS 2007;104:6261–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gifford RJ, Katzourakis A, Tristrem M, et al. A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. PNAS 2008;105:20362–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner G, Barbulescu M, Su M, et al. Insertional polymorphisms of full-length endogenous retroviruses in humans. Current Biology 2001;11:1531–5 [DOI] [PubMed] [Google Scholar]
- 8.Villarreal L. The source of self: genetic parasites and the origin of adaptive immunity. Ann New York Acad Sci (in press) [DOI] [PubMed] [Google Scholar]
- 9.Jurka J. Repbase update: a database and an electronic journal of repetitive elements. TIG 2000;16:418–20 [DOI] [PubMed] [Google Scholar]
- 10.Polaverapu N, Bowen NJ, McDonald JF. Identification, characterization and comparative genomics of chimpanzee endogenous retroviruses. Genome Biol 2006;7:R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbulescu M, Turner G, Seaman MI, et al. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr Biol 1999;9:861–8 [DOI] [PubMed] [Google Scholar]
- 12.Medstrand P, Mager DL. Human-specific integrations of the HERV-K endogenous retrovirus family. J Virol 1998;72:9782–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heslin DJ, Murcia P, Arnaud F, et al. A single amino acid substitution in a segment of the CA protein within gag that has similarity to human immunodeficiency virus type 1 blocks infectivity of a human endogenous retrovirus K provirus in the human genome. J Virol 2008;83:1105–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. PNAS 2004;101:14572–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medstrand P, van de Lagemaat LN, Mager DL. Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res 2002;12:1483–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orgel LE, Crick FHC. Selfish DNA: the ultimate parasite. Nature 1980;284:604–7 [DOI] [PubMed] [Google Scholar]
- 17.Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genomic evolution. Nature 1980;284:601–3 [DOI] [PubMed] [Google Scholar]
- 18.Schiavetti F, Thonnard J, Colau D, et al. A human retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Canc Res 2002;62:5510–16 [PubMed] [Google Scholar]
- 19.Mi S, Lee X, Li X, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 2000;403:785–9 [DOI] [PubMed] [Google Scholar]
- 20.Blond J-L, Lavillette D, Cheynet V, et al. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol 2000;74:3321–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blaise S, de Parseval N, Bénit L, et al. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. PNAS 2003;100:13013–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rote NS, Chakrabarti S, Stetzer BP. The role of human endogenous retroviruses in trophoblast differentiation and placental development. Placenta 2004;25:673–83 [DOI] [PubMed] [Google Scholar]
- 23.De Parseval N, Heidmann T. Physiological knockout of the envelope gene of the single-copy ERV-3 human endogenous retrovirus in a fraction of the Caucasian population. J Virol 1998;72:3442–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dupressoir A, Marceau G, Vernochet C, et al. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. PNAS 2005;102:725–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunlap KA, Palmarini M, Varela M, et al. Endogenous retroviruses regulate periimplantation placental growth and differentiation. PNAS 2006;103:14390–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts CT, Breed WG. Embryonic-maternal cell interactions at implantation in the fat-tailed dunnart, a dasyurid marsupial. Anat Rec 1994;240:59–76 [DOI] [PubMed] [Google Scholar]
- 27.Villarreal L. Viruses and the Evolution of Life. Washington, DC: ASM Press; 2005 [Google Scholar]
- 28.Manganey M, Renard M, Schlecht-Louf G, et al. Placental syncytins: genetic dysjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. PNAS 2007;104:20534–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HS, Yi JM, Hirohisa H, et al. Human endogenous retrovirus (HERV)-R family in primates: chromosomal location, gene expression, and evolution. Gene 2006;370:34–42 [DOI] [PubMed] [Google Scholar]
- 30.Bonnaud B, Bouton O, Oriol G, et al. Evidence of selection on the domesticated ERVWE1 env retroviral element involved in placentation. Mol Biol Evol 2004;21:1895–1901 [DOI] [PubMed] [Google Scholar]
- 31.Spadafora C. A reverse transcriptase-dependent mechanism plays central roles in fundamental biological processes. Systems Biology in Reproductive Medicine 2008;54:11–21 [DOI] [PubMed] [Google Scholar]
- 32.Pittoggi C, Sciamanna I, Mattei E, et al. Role of endogenous reverse transcriptase in murine early embryo development. Mol Reprod Dev 2003;66:225–36 [DOI] [PubMed] [Google Scholar]
- 33.Andersson A-C, Venables PJW, Tönjes RR, et al. Developmental expression of HERV-R (ERV-3) and HERV-K in human tissue. Virology 2002;297:220–5 [DOI] [PubMed] [Google Scholar]
- 34.Andersson A-C, Merza M, Venables P, et al. Elevated levels of the endogenous retrovirus ERV3 in human sebaceous glands. J Invest Dermatol 1996;106:125–8 [DOI] [PubMed] [Google Scholar]
- 35.De Parseval N, Lazar V, Casella JF, et al. Survey of human genes of retroviral origin: identification and transcriptome of the genes with coding capacity for complete envelope proteins. J Virol 2003;77:10414–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prudhomme S, Bonnaud B, Mallet F. Endogenous retroviruses and animal reproduction. Cytogenet Genome Res 2005;110:353–64 [DOI] [PubMed] [Google Scholar]
- 37.Larsson E, Andersson AC, Nilsson BO. Expression of an endogenous retrovirus (ERV3 HERV-R) in human reproductive and embryonic tissues – evidence for a function for envelope gene products. Uppsala J Med Sci 1994;99:113–20 [DOI] [PubMed] [Google Scholar]
- 38.Ergun S, Buschmann C, Heukeshoven J, et al. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J Biol Chem 2004;279:27753–63 [DOI] [PubMed] [Google Scholar]
- 39.See http://www.linnean.org/fileadmin/events2/events.php?detail=117
- 40.Perron H, Lazarini F, Ruprecht K, et al. Human endogenous retrovirus(HERV)-W ENV and GAG proteins: physiological expression in human brain and pathophysiological modulation in multiple sclerosis lesions. J Neurovirol 2005;11:23–33 [DOI] [PubMed] [Google Scholar]
- 41.Personal communication
- 42.Dunn CA, Medstrand P, Mager DL. An endogenous retroviral long terminal repeat is the dominant promoter for human β1,3-galactosyltransferase 5 in the colon. Proc Natl Acad Sci USA 2003;100:12841–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gogvadze E, Stukacheva E, Buzdin A, Sverdlov E. Human specific modulation of transcriptional activity provided by endogenous retroviral inserts. J Virol 2009; doi:10.1128/JVI.00123-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buzdin A, Kovalskaya-Alexandrova E, Gogvadze E, et al. At least 50% of human-specific HERV-K (HML-2) long terminal repeats serve in vivo as active promoters for host nonrepetitive DNA transcription. J Virol 2006;80:10752–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plant KE, Routledge SJ, Proudfoot NJ. Intergenic transcription in the human beta-globin gene cluster. Mol Cell Biol 2001;21:6507–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Routledge SJ, Proudfoot NJ. Definition of transcriptional promoters in the human beta globin locus control region. J Mol Biol 2002;323:601–11 [DOI] [PubMed] [Google Scholar]
- 47.Nekrutenko A, Li W-H. Transposable elements are found in a large number of human protein-coding genes. Trend Genet 2001;17:619–21 [DOI] [PubMed] [Google Scholar]
- 48.Medstrand P, Landry JR, Mager DL. Long terminal repeats are used as alternative promoters for the endothelin B receptor and apolipoprotein C-I genes in humans. J Biol Chem 2001;276:1896–903 [DOI] [PubMed] [Google Scholar]
- 49.Kapitonov VV, Jurka J. The long terminal repeat of an endogenous retrovirus induces alternative splicing and encodes an additional carboxy-terminal sequence in the human leptin receptor. J Mol Evol 1999;48:248–51 [DOI] [PubMed] [Google Scholar]
- 50.Samuelson LC, Wiebauer K, Snow CM, Meisler MH. Retroviral and pseudogene insertion sites reveal the lineage of human salivary and pancreatic amylase genes from a single gene during primate evolution. Mol Cell Biol 1990;10:2513–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ting C-N, Rosenberg MP, Snow CM, et al. Endogenous retroviral sequences are required for tissue-specific expression of a human salivary amylase gene. Genes Dev 1992;6:1457–65 [DOI] [PubMed] [Google Scholar]
- 52.Kitamura Y, Ayukawa T, Ishikawa T, et al. Human endogenous retrovirus K10 encodes a functional integrase. J Virol 1996;70:3302–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Banki K, Halladay D, Perl A. Cloning and expression of the human gene for transaldolase. A novel highly repetitive element constitutes an integral part of the coding sequence. J Biol Chem 1994;269:2847–51 [PubMed] [Google Scholar]
- 54.Villarreal, personal communication
- 55.Babushok DV, Ohshima K, Ostertag EM, et al. A novel testis ubiquitin-binding protein gene arose by exon shuffling in hominoids. Genome Res 2007;17:1129–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brandt J, Schrauth S, Veith AM, et al. Transposable elements as a source of genetic innovation: expression and evolution of a family of retrotransposon-derived neogenes in mammals. Gene 2005;345:101–11 [DOI] [PubMed] [Google Scholar]
- 57.Jordan K, Rogozin IB, Glazko GV, et al. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trend Genet 2003;19:68–72 [DOI] [PubMed] [Google Scholar]
- 58.Muotri AR, Marchetto MCN, Coufal NG, Gage FH. The necessary junk: new functions for transposable elements. Hum Mol Genet 2007;16:R159–R167 [DOI] [PubMed] [Google Scholar]
- 59.Ohshima K, Okada N. SINEs and LINEs: symbionts of eukaryotic genomes with a common tail. Cytogenetic Genome Res 2005;110:475–90 [DOI] [PubMed] [Google Scholar]
- 60.Morrish TA, Gilbert N, Myers JS, et al. DNA repair mediated by endonuclease-independent Line-1 retrotransposition. Nat Genet 2002;31:159–65 [DOI] [PubMed] [Google Scholar]
- 61.Sen SK, Huang CT, Kyudong H, et al. Endonuclease-independent insertion provides an alternative pathway for L1 retrotransposition in the human genome. Nucleic Acids Res 2007;[volume]:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farkash EA, Prak ETL. DNA damage and L1 retrotransposition. J Biomed Biotechnol 2006; doi 10.1155/JBB/2006/37285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boissinot S, Chevret P, Furano AV. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol 2000;17:915–28 [DOI] [PubMed] [Google Scholar]
- 64.Ovchinnikov I, Rubin A, Swergold GD. Tracing the LINES of human evolution. PNAS 2002;99:10522–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vansant G, Reynolds WF. The consensus sequence of a major Alu subfamily contains a functional retinoic acid response element. PNAS 1995;92:8229–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deininger PL, Batzer MA. Alu repeats and human disease. Mol Genet Metab 1999;67:183–93 [DOI] [PubMed] [Google Scholar]
- 67.Roy AM, Carroll ML, Kass DH, et al. Recently integrated human Alu repeats: finding needles in the haystack. Genetica 1999;107:149–61 [PubMed] [Google Scholar]
- 68.Comas D, Plaza S, Calafell F, et al. Recent insertion of an Alu element within a polymorphic human-specific Alu insertion. Mol Biol Evol 2001;18:85–8 [DOI] [PubMed] [Google Scholar]
- 69.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nature Reviews (Genetics) 2002;3:370–80 [DOI] [PubMed] [Google Scholar]
- 70.Urrutia AO, Ocaña LB, Hurst LD. Do Alu repeats drive the evolution of the primate transcriptome? Genome Biol 2008;9:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shapiro JA. A 21st century view of evolution: genome system architecture, repetitive DNA, and natural genetic engineering. Gene 2005;345:91–100 [DOI] [PubMed] [Google Scholar]
- 72.Shapiro JA, von Sternberg R. Why repetitive DNA is essential to genome function. Biol Rev 2005;80:1–24 [DOI] [PubMed] [Google Scholar]
- 73.Flockerzi A, Ruggieri A, Frank O, et al. Expression patterns of transcribed human endogenous retrovirus HERV-K(HML-2) loci in human tissues and the need for a HERV transcriptome project. BMC Genomics 2008;9:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hughes JF, Coffin JM. Evidence for genomic rearrangements mediated by human endogenous retroviruses during primate evolution. Nature Genetics 2001;29:487–9 [DOI] [PubMed] [Google Scholar]
- 75.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature 2006;444:444–54 [DOI] [PMC free article] [PubMed] [Google Scholar]