Abstract
The ability to rapidly and efficiently isolate specific viruses, bacteria, or mammalian cells from complex mixtures lies at the heart of biomedical applications ranging from in vitro diagnostics to cell transplantation therapies. Unfortunately, many current selection methods for cell separation, such as magnetic activated cell sorting (MACS), only allow the binary separation of target cells that have been labeled via a single parameter (e.g., magnetization). This limitation makes it challenging to simultaneously enrich multiple, distinct target cell types from a multi-component sample. We describe here a novel approach to specifically label multiple cell types with unique synthetic dielectrophoretic tags that modulate the complex permittivities of the labeled cells, allowing them to be sorted with high purity using the multitarget dielectrophoresis activated cell sorter (MT-DACS) chip. Here we describe the underlying physics and design of the MTDACS microfluidic device and demonstrate ~1000-fold enrichment of multiple bacterial target cell types in a single-pass separation.
The capability to sort target biological species from complex mixtures with high purity, recovery, and throughput is of paramount importance for a wide range of biotechnological applications ranging from viral diagnostics1 to cell transplantation therapies.2,3 Antibody-based magnetic selection4 is among the most useful of such separation technologies, because it allows rapid, high-throughput enrichment (positive selection) or depletion (negative selection) of specific target species including molecules, viruses, and cells. Magnetic separation has also proven invaluable as a method for the pre-enrichment of complex samples, for example, to decrease the number of nontarget cells prior to flow cytometry.5 However, magnetic selection strategies operate with only a single separation parameter (i.e., magnetization), and the development of similar methods capable of enriching multiple distinct species simultaneously has proven challenging.
We have previously demonstrated the dielectrophoresis activated cell sorter (DACS),6,7 in which target cells immunochemically labeled with synthetic dielectrophoretic tags are isolated from complex mixtures via the electrokinetic phenomenon of dielectrophoresis (DEP).8 Here, we extend this concept and demonstrate the capability to simultaneously enrich multiple, distinct target cells into independent fractions. The multitarget dielectrophoresis activated cell sorter (MT-DACS) is a two-input, multiple-output device that operates in a continuous flow manner (Figure 1A). The input consists of a running buffer and a sample mixture containing multiple types of target cells, each labeled with a distinct DEP tag. After a single pass through the device, the target and nontarget cells are separated and eluted through multiple, independent, spatially segregated outlets. Here we describe the physics, design, and fabrication of an MT-DACS chip and report on its separation performance.
Figure 1.
Multitarget bacterial cell sorting procedure using the MT-DACS device. (A) The experimental scheme. Step a: target cells (target A and target B) are labeled with DEP tags (tag A and tag B, respectively) via their respective surface markers. Step b: cells are dielectrophoretically sorted and eluted through independent outlets. Step c: collected cells from the outlets are analyzed via flow cytometry to quantify the sorting performance. (B) The physics of multitarget separation via MT-DACS. Two sets of electrodes are positioned at different glancing angles (θ1 and θ2) to select for two different targets. Target A cells labeled with tag A are selected at electrode set A (θ1 = 10°) and elute through outlet A. Target B cells labeled with tag B are sorted at electrode set B (θ2 = 8°) and elute through outlet B. The unlabeled, nontarget cells are not deflected by either electrode set, and they are eluted through the waste outlet.
EXPERIMENTAL SECTION
Samples for Bead Separation and Buffer Conditions
Polystyrene beads with a diameter of 10.0 µm (tag A) were obtained from G. Kisker GbR (Vancouver, Canada), and green and red fluorescent polystyrene beads with diameters of 5.0 µm (tag B) and 2.0 µm (nontarget) were obtained from Duke Scientific (Fremont, CA). The bead fractionation study was performed using concentrations of 0.8 × 104 (tag A), 1.3 × 104 (tag B), and 1 × 108 (nontarget) beads/mL. The bead mixture was suspended in 0.1× phosphate-buffered saline (PBS) supplemented with 1% bovine serum albumin (BSA) (Fraction V, Sigma Aldrich). To prevent settling of the beads during fractionation, the density of the solution was adjusted to that of polystyrene beads (1.06 g/mL) by addition of glycerol to a final concentration of 20% (v/v).
Cells and Reagents
All experiments were performed with MC1061 strain of E. coli [F− araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL(StrR) hsdR2 (rK− mK+) mcrA mcrB1].9 Genes coding for the surface peptides and fluorescent proteins were expressed using the plasmid pBAD33.10 The surface peptides were expressed as N-terminal fusions to eCPX.11 Cells were grown overnight at 37 °C in Luria–Bertani (LB) medium with 34 µg/ mL chloramphenicol (Sigma, St. Louis, MO) and subcultured at a 1:50 dilution for 2 h at 37 °C. Cell-surface expression of peptides and fluorescent proteins was induced by adding l-arabinose (0.02% w/v) to the culture media for 45 min at 37 °C. Approximately 104 target cells (cells expressing streptavidin-binding peptides and cells containing T7 · tag) were centrifuged (2650g, 5 min) and resuspended in 10 µL of 0.25× PBS, 0.5% BSA (PBSB). We used 56 µL (~106 beads) of SuperAvidin-coated, 9.6 µm Proactive microspheres (Bangs Laboratory, Fishers, IN) as tag A for bacterial cell sorting, which were first washed four times in BlockAid (Invitrogen, Carlsbad, CA) and then added into the PBSB containing the cells. For tag B, we used 10 µL (~106 beads) of protein A-coated, 5.5 µm Proactive microspheres (Bangs Laboratory), which were washed once in 500 µL of PBSB and then added to 100 µL of PBSB containing 20 nM mAb (anti-T7 · tag, EMD Biosciences, La Jolla, CA). Following 1 h of incubation at 4 °C on an inversion shaker, the beads were washed four times and added into the PBSB containing the cells. Approximately 108 nontarget cells expressing blue fluorescent protein (BFP) were washed in 500 µL of PBSB and resuspended in the cell/bead mixture. This mixture was incubated in an inversion shaker at 4 °C, washed once, resuspended in 500 µL of sorting buffer (0.1× PBS, 1% BSA, 20% glycerol), and finally passed through a CellTrics 20 µm mesh filter (Partec, Münster, Germany).
Cytometry Analysis
After the separation, the eluted cells from outlets A and B and the waste outlet were regrown in order to reduce the effect of the DEP tags on the flow cytometry measurements. The cells were grown overnight in LB medium with 0.2% glucose to eliminate growth bias among the cells. The cells were subcultured at a 1:50 dilution for 2 h at 37 °C. The cell-surface expression of peptides and fluorescent proteins was induced by adding l-arabinose (0.02% w/v) to the culture media for 3 h at 37 °C. Five microliters of subcultured cells from each outlet were incubated in 100 µL of 1× PBS, 0.5% BSA containing 20 nM streptavidin–phycoerythrin (SAPE) for 1 h on ice, followed by centrifugation and removal of the supernatant. Cells were then resuspended in cold PBS at ~106 cells/mL and immediately analyzed by FACS (FACSAria, BD Biosciences, San Jose, CA).
Cell Viability Measurement
Cells were grown overnight at 37 °C in LB medium with 34 µg/mL chloramphenicol (Sigma, St. Louis, MO), and subcultured at a 1:50 dilution for 2 h at 37 °C. Two 1 mL samples of the cells were centrifuged, and the supernatants were removed. One pellet of live cells was resuspended in 1 mL of 0.15 M NaCl, and the second pellet was resuspended in 70% ethanol to create a population of dead cells. The samples were incubated at room temperature for 1 h with mixing and were centrifuged and washed twice in 1 mL of 0.15 MNaCl. The two samples were mixed at a ratio of 60.2: 39.8 (live/dead) and were injected into the MT-DACS device under the same conditions as the bead and cell sorting experiment (sample flow rate = 150 µL/h, buffer flow rate = 960 µL/h). First, the electric field was turned off and eluted cells were collected. Then the electric field was turned on (voltage amplitude = 20 Vpeak to peak, frequency = 500 kHz), and the eluted cells were collected in a different tube. Both eluted fractions were stained with 5 µM SYTO-9 and 45 µM propidium iodide from LIVE/DEAD BacLight bacterial viability and counting kit (Invitrogen, Carlsbad, CA) for 15 min at room temperature and were analyzed with flow cytometry.
RESULTS AND DISCUSSION
Separation Architecture and Device Physics
In MT-DACS separation, each target cell type is first labeled with a unique DEP tag via specific receptor–ligand binding (e.g., antibody–antigen) (Figure 1A, step A). Each specific DEP tag exhibits a distinct dielectrophoretic response when placed in a nonuniform electric field. We employ polystyrene beads (PSB) as our DEP tags because, as we have previously shown,6 their low surface conductivity (σ) leads to complex permittivities that differ significantly from those of bacterial cells. Equally critically, the sizes of the DEP tags are selected such that the complex permittivity of the tag dominates that of the cell, regardless of its growth phase.
Once inside the MT-DACS device, the cells are subject to a hydrodynamic force (FHD) arising from the flow of the buffer and a dielectrophoretic force (FDEP) created by the nonuniform electric field generated by the device’s electrodes (Figure 1B). Target cells passing through the electric field are deflected if FDEP exceeds FHD in the direction perpendicular to the electrodes. Separation between two target cell types is achieved by implementing two sets of electrodes at different glancing angles (θ1 and θ2, Figure 1B). Cells bound to the larger DEP tag (tag A) are dielectrophoretically deflected by electrode set A (θ1 = 10°) and subsequently elute through outlet A, whereas cells tagged with the smaller DEP tag (tag B) and cells lacking any tag (i.e., nontarget cells) are not, because the FDEP/FHD ratio experienced by these cells is insufficient to cause deflection at this angle. The remaining cells are sorted at electrode set B (θ2 = 8°), where the tag B-bound species are efficiently deflected and eluted through outlet B. The nontarget cells remain unaffected and elute through the waste outlet.
MT-DACS utilizes an angled electrode structure (initially described by Schnelle et al.12 and subsequently used by our group and others6,7,13–15) to generate the necessary repulsive FDEP force within the microfluidic channels. Briefly, the repulsive FDEP exerted on a particle by the electrodes is given approximately16 by
| (1) |
where εm is the permittivity of the suspension medium, a is the effective radius of the cell-tag species, h is the channel depth, Re(CM) is the real component of the Clausius–Mossotti factor (which describes the competitive polarization between the buffer and the species6,16), and V is the rms magnitude of the applied voltage. Concurrently, the cell-tag species also experiences FHD due to viscous drag from the fluid flow. This force can be estimated by Stokes law to be
| (2) |
where υ and μ are the velocity and viscosity of the fluid, respectively. Cells passing through the electrode region experience a vector sum of the two forces (FTOTAL = FDEP + FHD). Since FDEP depends on the cube of the particle radius (eq 1) and FHD depends linearly on it (eq 2), the ratio FDEP/FHD increases approximately with the square of this radius, enabling precise, size-based separation.
In order to optimize the device design and predict its performance, we performed numerical simulations to calculate the concentration profiles of the DEP tags in the MT-DACS, as described previously.17 Using the relevant experimental values of permittivities and conductivities, the CM factor was calculated to be −0.5, and this value was used in the simulations. The diameters of tags A and B were set to 10 and 5 µm, respectively, and the diameter of nontarget particles was set at 2 µm. From this, the velocities of the DEP-tagged particle (υ⃗p) were calculated from the assumption that, at steady state,
| (3) |
where the drag force (F⃗DRAG) is given by the Stokes equation:
| (4) |
The fluid velocity, υ⃗ is obtained by solving the Navier–Stokes equation for a given geometry. Therefore, the DEP-modified particle velocity was expressed as
| (5) |
Once υ⃗p was calculated, the concentration profile of the bead–target complex was obtained by solving the convection–diffusion equation:
| (6) |
where C is particle concentration and D is the diffusion coefficient as calculated using the Stokes–Einstein relationship. The resulting concentration profiles for the ternary particle mixture demonstrate that particles tagged with the larger tag A should be efficiently separated as they travel through the region of electrode set A and elute through outlet A (Figure 2A). Here the smaller tag B and nontarget particles are not deflected, and their concentrations at outlet A are minimal. At electrode set B, in contrast, only particles bound to tag B are deflected and elute through outlet B (Figure 2B). The nontarget particles are not deflected by either electrode sets and, thus, elute through the waste outlet (Figure 2C).
Figure 2.
Numerical simulation of concentration profiles of tag A, tag B, and nontarget particles. Calculation of the DEP-modified particle velocities and the solution to the convection–diffusion equation during the MT-DACS operation demonstrate that (A) tag A is sorted from the sample stream at electrode set A, (B) tag B is selected at electrode set B, and (C) nontarget particles are not deflected by either electrode sets and therefore elute through the waste outlet.
The design of the MT-DACS chip benefits from many aspects of microfluidics technology. First, the microfabricated electrodes enable the precise and reproducible generation of nonuniform electric fields within the microchannel. This is important because the FDEP depends critically on the gradient of the electric fields, and the sorting mechanism relies on differentiating the amplitude of FDEP for each tag. Second, polyimide processing allows consistent fabrication of 40 µm tall microchannels that enable reproducible control of the cell velocities and, therefore, of the hydrodynamic force. Finally, the laminar nature of the flow in the microchannel (ReMT-DACS ~ 0.1) allows the sustained segregation of two streams such that the target cells are enriched with high purity by switching from the sample stream into the buffer stream.
Particle Separation Performance
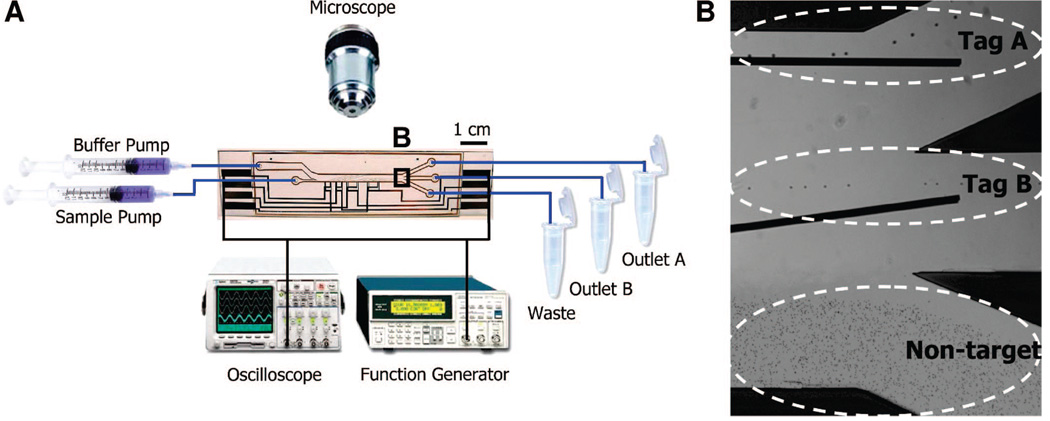
We fabricated the MT-DACS using a glass–polyimide–glass sandwich architecture via a previously described process6,17 (Supporting Information Figure S-1). The device was mounted beneath the objective of an epifluorescence microscope (DM4000B, Leica, Bannockburn, IL) in order to characterize device performance, and the electrodes were powered using a function generator (AFG320, Tektronix) through two card-edge connectors (Figure 3A). The frequency and magnitude of applied voltage were measured with a digital oscilloscope (54622A, Agilent Technologies, Palo Alto, CA).
Figure 3.
Experimental setup and optical micrographs of MT-DACS operation. (A) The MT-DACS chip is placed beneath an epifluorescence microscope with two dual-track programmable syringe pumps delivering the sample mixture and buffer fluid. The electrodes are connected to a function generator, and the frequency and magnitude of the applied voltage are measured by a digital oscilloscope. The flow pattern during the separation is monitored by a high-speed camera. (B) Overlaid optical micrographs show tag A, tag B, and nontarget beads being eluted via outlet A, outlet B, and the waste outlet, respectively. Sinusoidal voltage of 20 Vpeak to peak was applied to both sets of electrodes at 500 kHz, and the device was operated at a throughput of ~1.5 × 107 particles/h.
The separation performance of the MT-DACS chip was initially characterized with a mixture of DEP tags in order to optimize the operating conditions of the device. A ternary mixture of tag A (diameter = 10.0 µm), tag B (diameter = 5.0 µm), and nontarget beads (diameter = 2.0 µm) was suspended in the sorting buffer (0.1× PBS, 1% BSA, 20% glycerol). Two dual-track programmable syringe pumps (PhD 2000, Harvard Apparatus, Holliston, MA) were used to deliver the sample mixture and buffer into the device at flow rates of 150 (sample) and 960 (buffer) μL/h, which corresponded to a throughput of ~1.5 × 107 particles/h/microchannel. A sinusoidal voltage of 20 Vpeak to peak was applied to both sets of electrodes at 500 kHz. High-speed videos of the separation process reveal that, as expected, tags A and B were efficiently separated at their corresponding electrodes and eluted into their respective outlets (Figure 3B). The nontarget beads were not deflected and were eluted through the waste outlet. Although some particles were lost in the tubing and other fluidic interconnects, all particles that entered the device were successfully recovered and no sticking of the particles to the device was observed (data not shown).
In order to quantify the purity of the separation, the eluted fractions from outlet A, outlet B, and the waste outlet were analyzed using a flow cytometer (FACSAria, BD Biosciences, San Jose, CA). The initial mixture contained 0.008% tag A, 0.013% tag B, and 99.979% nontarget beads (Figure 4). After a single pass through the MT-DACS device, the eluted fraction at outlet A consisted mostly of tag A (98.7%) with no tag B (0%), and only 1.3% nontarget beads were present. This translates to an ~12 000-fold enrichment of tag A. Likewise, the outlet B fraction was 97.4% tag B, 0% tag A, and only 2.6% nontarget beads, corresponding to an ~7500-fold enrichment over nontarget beads. The waste fraction was comprised purely of nontarget beads, with no detectable A or B tags.
Figure 4.
Separation of DEP tags in MT-DACS measured via flow cytometry. The initial sample contained low concentrations of tag A (0.008%) and tag B (0.013%) and an excess of nontarget beads (99.979%). After a single pass through the MT-DACS device, the eluted fraction at outlet A contained mostly tag A (98.7%) and no tag B (0.0%), along with a small percentage of nontarget beads (1.3%). This corresponds to an ~12 000-fold enrichment for tag A. In contrast, the fraction eluted through outlet B consisted primarily of tag B (97.4%), with no tag A (0.0%), and some nontarget beads (2.6%); this corresponds to an ~7500-fold enrichment for tag B. No tag A (0.0%) or tag B (0.0%) was found in the fraction eluted via the waste outlet, which consisted entirely of nontarget beads.
Multitarget Bacterial Cell Separation
Finally, we characterized the performance of the MT-DACS device for sorting multiple target bacterial cells. Three distinct bacterial clones of commonly used E.coli MC1061 strain were used wherein each target cell type expressed a distinct sequence of peptides on its cell surface. Target A cells, which inducibly express a streptavidin-binding peptide sequence (SA1) on their outer membrane,18 were labeled with SuperAvidin-modified tag A beads (Bangs Laboratory, Fishers, IN). Target B cells, which express the inducible T7 · tag peptide sequence (MASMTGGQQMG) on their surface,19 were labeled using anti-T7 mAb-functionalized tag B beads. These cells also express green fluorescent protein (GFP), allowing for facile visualization. Finally, E. coli cells expressing azurite, a BFP,20 were employed as nontarget cells. During the experiment, all cells were kept on ice, and the sorting experiments were performed within ~1 h.
A sample mixture containing low concentrations of labeled target A (0.071%) and target B (0.37%) cells doped into a large excess of nontarget cells (99.559%) was suspended in the sorting buffer (0.1× PBS, 1% BSA, 20% glycerol) and pumped into the MT-DACS device. The parameters of separation (i.e., buffers, flow rates, voltage amplitude and frequency) were identical to those used in the tag separation experiment, corresponding to a throughput of ~1.5 × 107 cells/h. After the separation, the cells eluted from each outlet were cultured overnight and quantified using flow cytometry. It should be noted that the culturing was performed in the presence of glucose to repress pBAD33 gene expression which eliminated growth biases among the various cell types.
Flow cytometry analysis revealed that, after a single pass through the MT-DACS device, the population of target A cells in outlet A increased from an initial population of 0.071% (Figure 5A) to 66% (Figure 5B), corresponding to a 930-fold enrichment. Similarly, the target B cell population in outlet B was enriched 260-fold from 0.37% to 96% (Figure 5C). It is noteworthy that neither outlet A nor outlet B contained any detectable target B or target A cells, respectively, confirming that there is virtually no crossover of target cells between the outlets. The population in the waste outlet consisted of 0.062% target A cells, 0.32% target B cells, and 99.6% nontarget cells (Figure 5D), numbers similar to those of the initial sample. We suspect that this is due to the fact that a fraction of target cells become unlabeled during the separation process and thus eluted through the waste outlet.
Figure 5.
Multitarget bacterial cell sorting performance. (A) Two-color flow cytometry measurement of the initial sample show that it consisted of an excess of nontarget cells (99.559%) with low concentrations of labeled target A (0.071%) and target B (0.37%) cells suspended in sorting buffer. (B) After a single round of separation, the outlet A fraction contained target A cells (66%, a 930-fold enrichment) and no target B cells (0.0%). (C) Similarly, the outlet B fraction contained target B cells (96%, a 260-fold enrichment) and no target A cells (0.0%). (D) The fraction collected at the waste outlet consisted mostly of nontarget cells (99.6%) with small quantities of target A (0.062%) and target B (0.32%) cells.
CONCLUSIONS
Here we demonstrate, for the first time, the capability to simultaneously enrich multiple target cell types from a large background of nontarget cells using the phenomenon of dielectrophoresis. This was achieved by labeling each cell type with differing DEP tags; these cells were labeled through specific receptor–ligand binding and were therefore sorted according to distinct surface markers. In a single pass through the device, we obtained ~10 000-fold overall enrichment of DEP tags and ~1000-fold enrichment of two separate types of labeled target cells with no detectable cross-contamination within the outlet channels. From the observation that the enrichment of cell-free tags was an order of magnitude higher than that observed with labeled bacterial cells, we infer that enrichment performance is limited by the affinity and specificity of the capture reagents rather than the device itself.
The throughput of the MT-DACS in these experiments was ~1.5 × 107 cells/h. In order to increase the throughput of the device, higher DEP forces must be generated through the application of higher voltages or multiple channels must be operated in parallel. Practically, however, the operating voltage is limited by the electrolysis at the electrodes and the viability of the cells. Under the reported operating conditions, we routinely operated the MT-DACS device for 6 h continuously without electrolysis. The effect of electric fields on cell viability was measured with the LIVE/DEAD BacLight bacterial viability and counting kit (Invitrogen, Carlsbad, CA). The quantitative flow cytometry data shows that the viability of the cells was not affected by MT-DACS separation (Supporting Information Figure S-2). We attribute this to the fact that the electric field strength used in the MT-DACS device is significantly lower than the range commonly used for electroporation21 and that the MT-DACS device operates in a negative DEP mode, in which the cells are pushed away from regions of high electric fields.
We found our approach of modulating the amplitude response of the cell-tag species using differently sized DEP tags to be well suited for the separation of targets which are smaller in size with respect to the tags, including bacteria, viruses, and molecules. In order to extend our method for sorting larger, mammalian cells, we may label the target cell with a single DEP tag with distinct σ and/or ε. In this case, as we have shown for the bacterial cells, we expect the minimum size of the DEP tag to approach the size of the target cell. Alternatively, it may also prove interesting to explore the alternate approach of modulating the frequency response by labeling the target cells with multiple, smaller DEP tags.
Supplementary Material
ACKNOWLEDGMENT
We express thanks for the financial support from the ARO Institute for Collaborative Biotechnologies (DAAD1903D004), DARPA/DMEA-CNID Grant (H94003-05-2-0503), a Beckman Foundation Young Investigator Grant (8-442550-57174), and a UEPP Grant from the Livermore National Laboratories (8-482550-26752). We appreciate the helpful discussions with Dr. Paul Bessette, Gaurav Soni, and Karen Dane, and we thank Professor Kevin Plaxco and Mr. Michael Eisenstein for their careful reading of the manuscript. Microfabrication was carried out in the Nanofabrication Facility at UC Santa Barbara.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Patterson BK, Till M, Otto P, Goolsby C, Furtado MR, McBride LJ, Wolinsky SM. Science. 1993;260:976–979. doi: 10.1126/science.8493534. [DOI] [PubMed] [Google Scholar]
- 2.Thomas ED, Storb R, Clift RA, Fefer A, Johnson FL, Neiman PE, Lerner KG, Glucksberg H, Buckner CD. N. Engl. J. Med. 1975;292:832–843. doi: 10.1056/NEJM197504172921605. [DOI] [PubMed] [Google Scholar]
- 3.Thomas ED, Storb R, Clift RA, Fefer A, Johnson FL, Neiman PE, Lerner KG, Glucksberg H, Buckner CD. N. Engl. J. Med. 1975;292:895–902. doi: 10.1056/NEJM197504242921706. [DOI] [PubMed] [Google Scholar]
- 4.Miltenyi S, Muller W, Weichel W, Radbruch A. Cytometry. 1990;11:231–238. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- 5.Herzenberg LA, Parks D, Sahaf B, Perez O, Roederer M. Clin. Chem. 2002;48:1819–1827. [PubMed] [Google Scholar]
- 6.Hu XY, Bessette PH, Qian JR, Meinhart CD, Daugherty PS, Soh HT. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15757–15761. doi: 10.1073/pnas.0507719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bessette PH, Hu XY, Soh HT, Daugherty PS. Anal. Chem. 2007;79:2174–2178. doi: 10.1021/ac0616916. [DOI] [PubMed] [Google Scholar]
- 8.Pohl HA. Dielectrophoresis: The Behavior of Neutral Matter in Nonuniform Electric Fields. Cambridge; New York: Cambridge University Press; 1978. [Google Scholar]
- 9.Casadaban MJ, Cohen SN. J. Mol. Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 10.Guzman LM, Belin D, Carson MJ, Beckwith J. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rice JJ, Daugherty PS. Protein Eng. Des. Sel. 2008;21:435–442. doi: 10.1093/protein/gzn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schnelle T, Hagedorn R, Fuhr G, Fiedler S, Muller T. Biochim. Biophys. Acta. 1993;1157:127–140. doi: 10.1016/0304-4165(93)90056-e. [DOI] [PubMed] [Google Scholar]
- 13.Fiedler S, Shirley SG, Schnelle T, Fuhr G. Anal. Chem. 1998;70:1909–1915. doi: 10.1021/ac971063b. [DOI] [PubMed] [Google Scholar]
- 14.Kralj JG, Lis MTW, Schmidt MA, Jensen KF. Anal. Chem. 2006;78:5019–5025. doi: 10.1021/ac0601314. [DOI] [PubMed] [Google Scholar]
- 15.Seger U, Gawad S, Johann R, Bertsch A, Renaud P. Lab Chip. 2004;4:148–151. doi: 10.1039/b311210a. [DOI] [PubMed] [Google Scholar]
- 16.Gascoyne PRC, Vykoukal J. Electrophoresis. 2002;23:1973–1983. doi: 10.1002/1522-2683(200207)23:13<1973::AID-ELPS1973>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim U, Shu CW, Dane KY, Daugherty PS, Wang JYJ, Soh HT. Proc. Natl. Acad. Sci. U.S.A. 2007;104:20708–20712. doi: 10.1073/pnas.0708760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bessette PH, Rice JJ, Daugherty PS. Protein Eng. Des. Sel. 2004;17:731–739. doi: 10.1093/protein/gzh084. [DOI] [PubMed] [Google Scholar]
- 19.Rice JJ, Schohn A, Bessette PH, Boulware KT, Daugherty PS. Protein Sci. 2006;15:825–836. doi: 10.1110/ps.051897806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mena MA, Treynor TP, Mayo SL, Daugherty PS. Nat. Biotechnol. 2006;24:1569–1571. doi: 10.1038/nbt1264. [DOI] [PubMed] [Google Scholar]
- 21.Fox MB, Esveld DC, Valero A, Luttge R, Mastwijk HC, Bartels PV, van den Berg A, Boom RM. Anal. Bioanal. Chem. 2006;385:474–485. doi: 10.1007/s00216-006-0327-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.