Abstract
Transcriptional regulation of the galactose metabolizing genes in Saccharomyces cerevisiae depends on three core proteins - Gal4p, the transcriptional activator that binds to upstream activating DNA sequences (UASGAL), Gal80p, a repressor that binds to the C-terminus of Gal4p and inhibits transcription, and Gal3p, a cytoplasmic transducer which upon binding galactose and ATP, relieves Gal80p repression. The current model of induction relies on Gal3p sequestering Gal80p in the cytoplasm. However, the rapid induction of this system implies that there is a missing factor. Our structure of Gal80p in complex with a peptide from the C-terminal activation domain of Gal4p reveals the existence of a dinucleotide that mediates the interaction between the two. Biochemical and in vivo experiments suggests that NADP plays a key role in the initial induction event.
Saccharomyces cerevisiae senses galactose/melibiose in the surrounding medium and shuttles it into the cytoplasm. Galactose is enzymatically converted by the GAL enzymes, Gal1p, Gal5p, Gal7p and Gal10p to glucose-1-phosphate (1). The regulatory control of this pathway is governed by ‘the galactose regulon’ (Fig S1). The very short induction time for GAL genes presents a quandary because Gal3p is localized in the cytoplasm and does not appear to enter the nucleus to physically disrupt Gal80p binding to Gal4p (2). Gal80p, localized to the nucleus and the cytoplasm (2), might therefore be sequestered in the cytoplasm upon induction but this would require rapid shuttling of the repressor out of the nucleus, or rapid turnover of the Gal4p/Gal80p complex. It therefore appears that there is a missing link to initiate rapid induction and switch the system on. In order to understand the molecular mechanism of the GAL regulatory system, we determined the structure of S. cerevisiae Gal80p (ScGal80p) with the activation domain of ScGal4p.
Gal4p has a C-terminal (768–881) acidic activation domain (AD), a region that is also required to bind its repressor, Gal80p (3–5). We determined the structures of Gal80pS2:P20 and Gal80S0:P21 (Gal80pS2 and Gal80pS0 are two super-repressor mutants of ScGal80p). P21 is a 21 amino acid peptide that contains the conserved region of the C-terminal AD of Gal4p (aa 854–874). P20 is a peptide that was identified from a phage-display screen selected for Gal80p binding and was also shown to activate transcription (6).
The crystal structures of ScGal80p reveal a three-domain architecture with an N-terminal domain consisting of a Rossmann fold, normally associated with binding of NAD(P) co-factors. ScGal80p does not possess a classic NAD(P) binding sequence motif, GXGXX(G/A), but a slightly altered motif, GFVGLNAA, spanning amino acids 21–28. The C-terminal domain consists of a large β-sheet that forms an extensive dimer interface with another monomer (Fig. 1A, Fig. S2). A large cleft is apparent between these two domains. A smaller third domain, located between the N and C-terminal domains, consists of three smalḷ β -strands and a helix that resemble a set of fingers at the entrance of the cleft. ScGal80p dimers form tetramers in both crystal forms (see SOM text, Fig S3). The structure is similar to that of Gal80p from K. lactis (KlGal80) (7) and closely resembles structures of several oxidoreductases.
Figure 1.
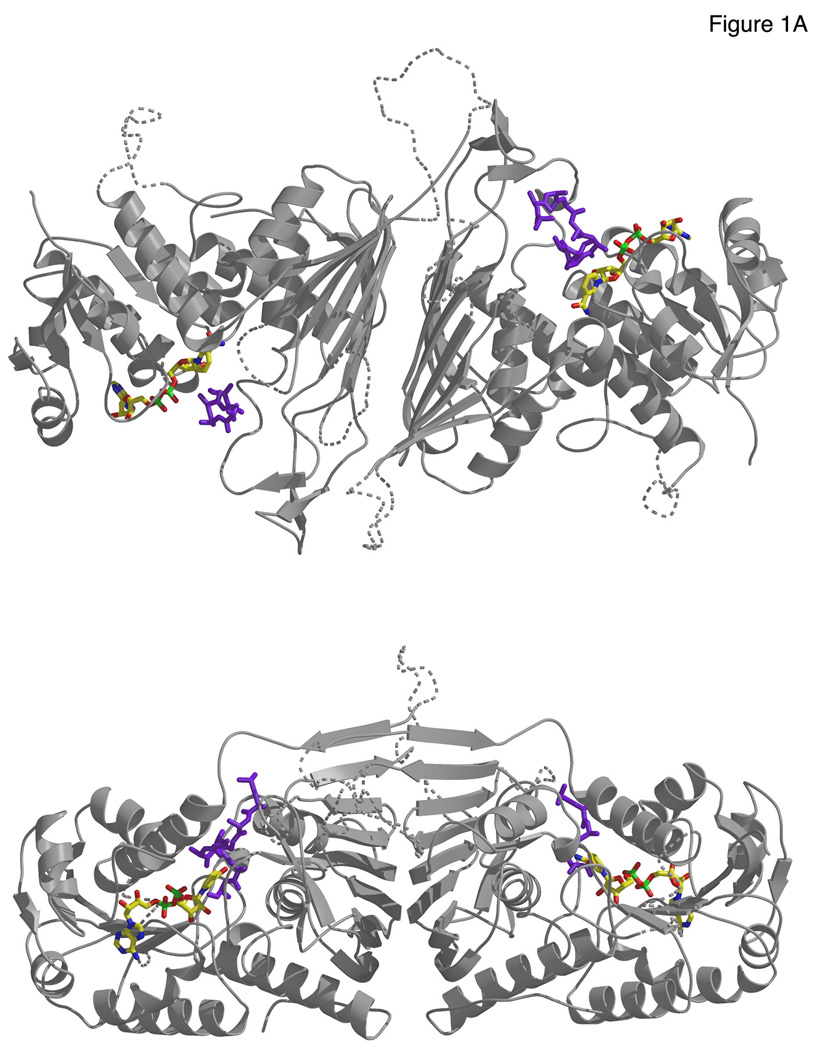
(A) Two views of the ScGal80pS0-ScGal4AD-NAD dimer. ScGal80p is depicted as gray ribbons, the Gal4p AD peptide in purple sticks and the NAD in atom colors in stick (carbon in yellow, oxygen in red, nitrogen in blue and phosphorus in green). Disordered regions are shown as a dashed coil. The β-sheet regions of the C-terminal domains form an extensive dimeric interface.
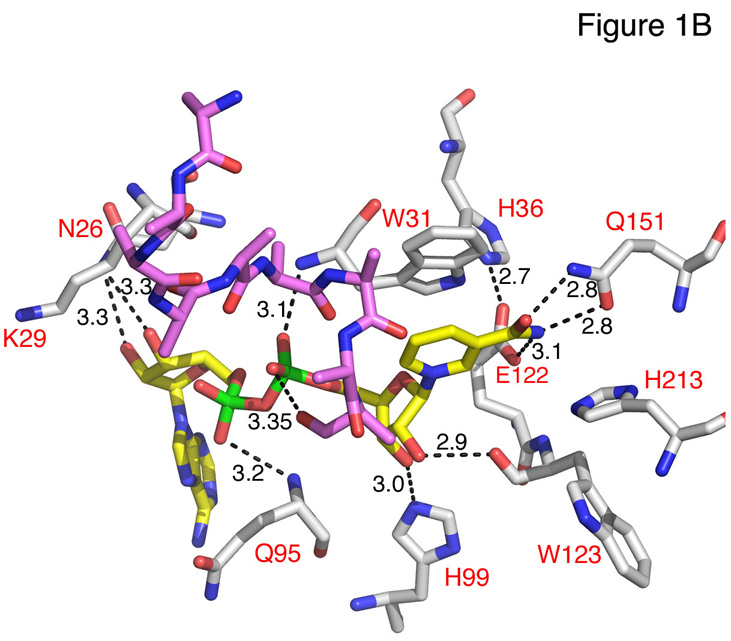
(B) NAD binding. Hydrogen-bonding interactions involving the NAD dinucleotide are shown as dashed lines with the corresponding distances indicated (Å). The side chain of W31 stacks on the NAD nicotinamide ring. Atoms are shown in atom colors as in A with Gal80p carbons in gray, NAD carbons in yellow and the Gal4p-AD peptide carbons in purple.
In the ScGal80pS2:P20 structure, we identified electron density indicating an NAD dinucleotide bound to the Rossmann fold. We therefore soaked the ScGal80pS0:P21 crystals, which diffracted to higher resolution, with NAD. Not only did the density of this dinucleotide become even more apparent than in the unsoaked crystals (Fig. S4), but we were then able to locate a portion of the Gal4p AD peptide, which we were unable to observe previously, bound to the cleft in each monomer of Gal80p (Fig. S5). We have modeled a segment of the peptide consisting of 9 residues for one monomer and 5 residues for the other. Although the backbone electron density is clear, most side chains seem to be somewhat disordered and could not be unequivocally assigned. Nevertheless the peptide appears to interact with the nicotinamide portion of the dinucleotide (Fig. 1). NAD nestles between Gal80p and P21 making several key interactions with ScGal80p (Fig. 1B). The crystal structure of KlGal80p did not show any bound dinucleotide (7).
To test the role of NAD as a possible cofactor of the ScGal80p-ScGal4p interaction, we performed pull-down assays (8) of Gal80p with purified recombinant GST-Gal4p(768–881), containing the acidic AD, in the presence of NAD and NADH (Fig. 2A). As Gal80p also interacts with the transducer, Gal3p, we used GST-Gal3p as a control. There was no change in binding for either of these two dinucleotides. We then tested binding in the presence of increasing concentrations of NADP and NADPH (Fig. 2A). A clear reduction in binding is observed at 5 µM NADP, more substantial at 500 µM. NADPH also shows some reduction in Gal80p binding at 500 µM, though this could be due to the presence of NADP impurities, which are on the order of 1–3%. There is no effect of Gal80p binding to GST-Gal3p with any of the four dinucleotides (Fig. 3A), indicating that this dinucleotide-regulated binding is specific to the Gal80p-Gal4p interaction. Modeling of NADP in the place of NAD in the structure does not show any clashes with either the Gal80p protein or with the Gal4p peptide.
Figure 2. NADP disrupts the GAL4pAD-Gal80p interaction.
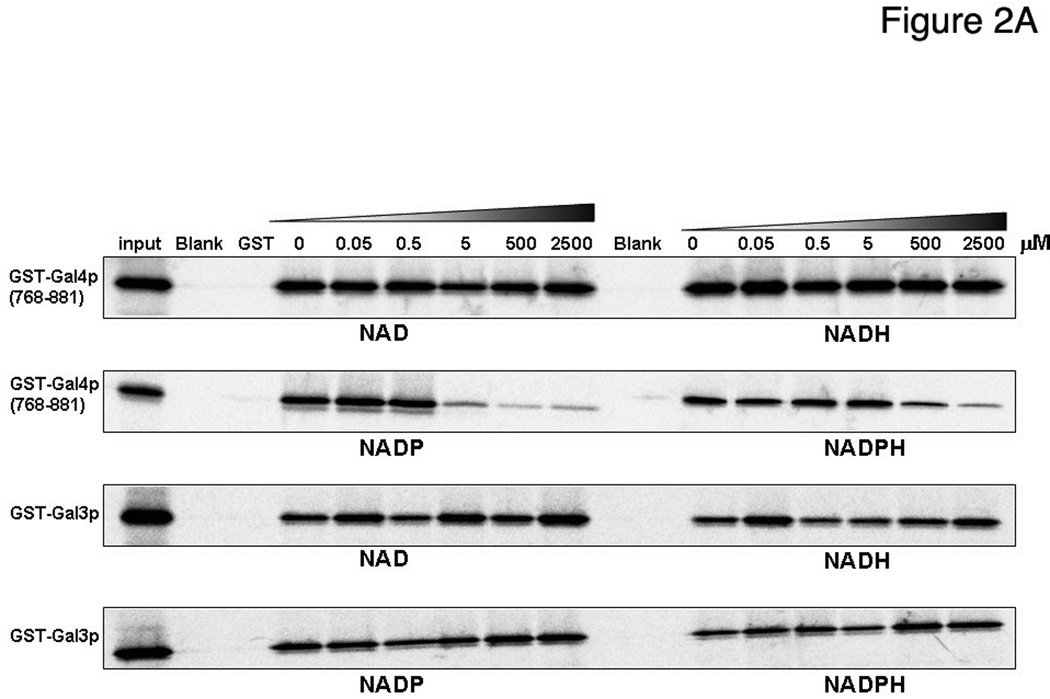
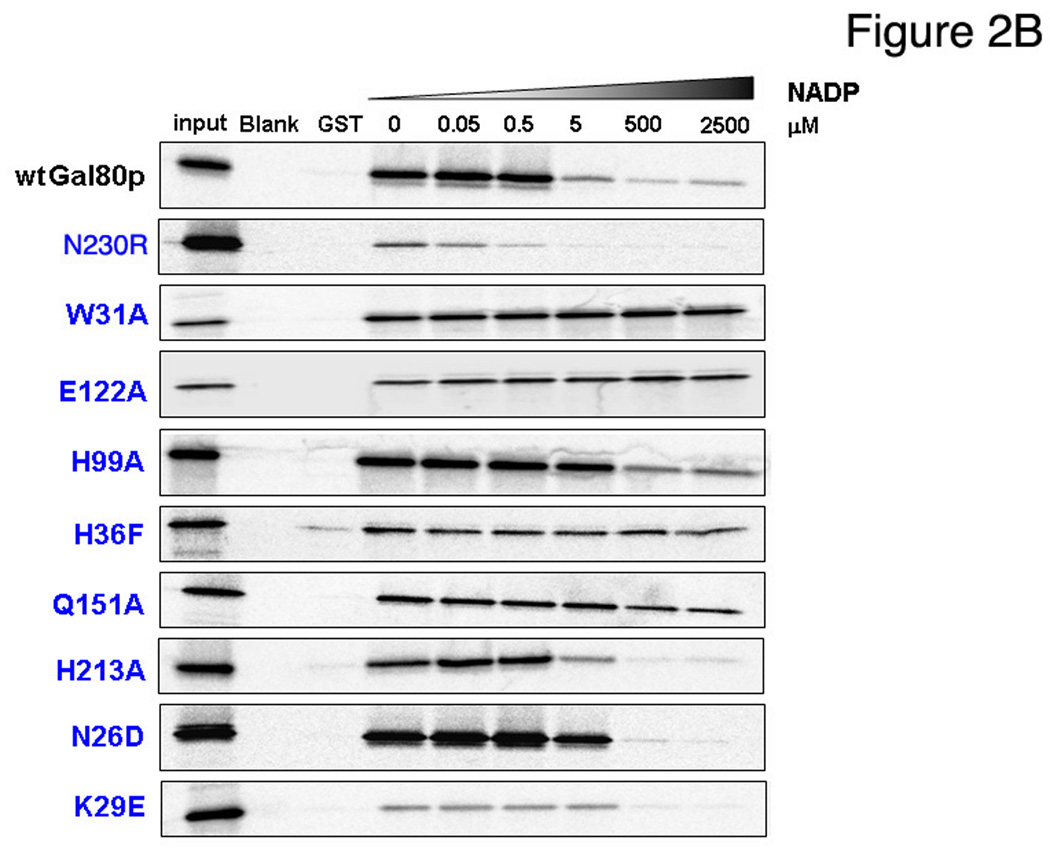
(A) GST, GST-Gal4pAD or GST-Gal3p were used in GST pull-down experiments with 35S-methionine-labeled Gal80p expressed in rabbit reticulocyte in the presence of increasing concentrations of NAD, NADH, NADP of NADPH (see Materials and Methods). 1% of input was loaded in the lanes marked “input”.
(B) Representative GST pull-down experiments of GST-Gal4pAD with Gal80p mutants in the presence of increasing concentrations of NADP (see Material and Methods).
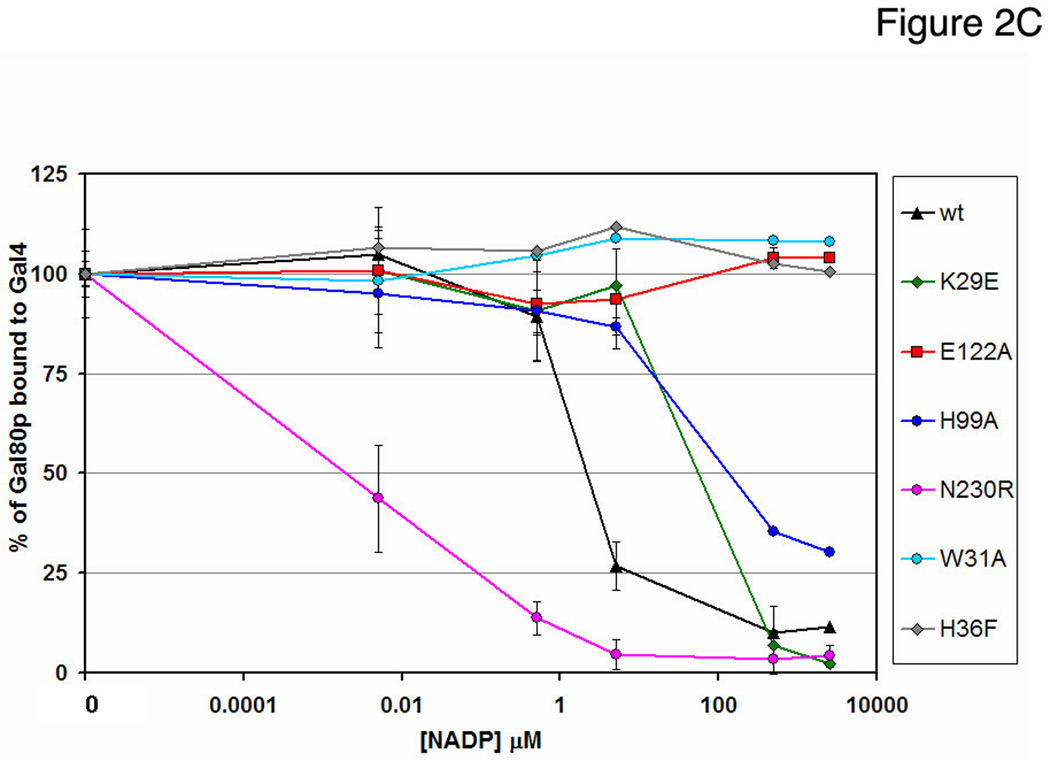
(C) Bands from the pull-down experiments (each carried out at least in triplicate) were quantified and plotted as a function of NADP concentration. Data are shown as mean and error bars indicate +/− standard deviations.
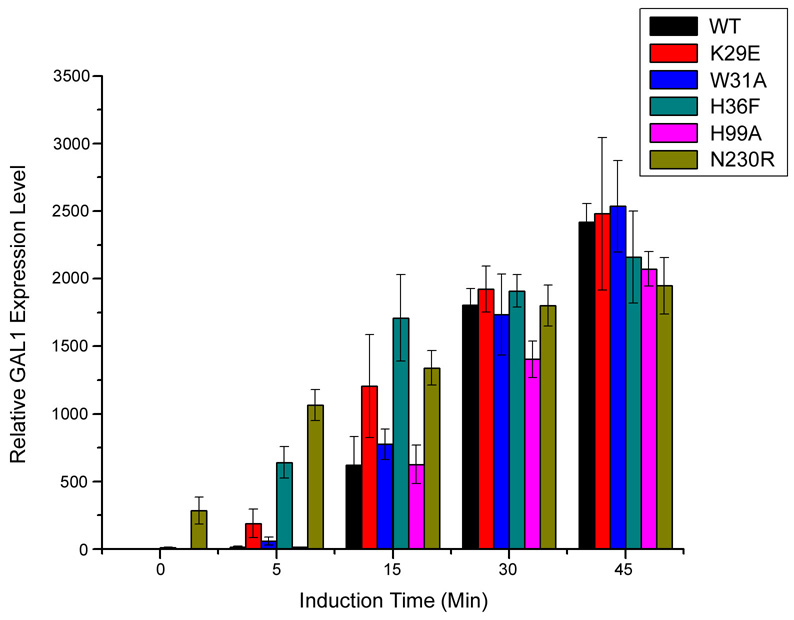
Figure 3. Alterations in the NADP binding site changes the rate of induction in vivo.
GAL1 mRNA expression as a function of time after galactose induction. Data are shown for wild-type Gal80p and for Gal80p point mutants. All data were normalized to RNA levels measured for a control gene, PMA1. A gal80Δ mutant has a high expression level even when uninduced, as high as seen for wild-type Gal80p when fully induced. The dimer mutant, N230R, also shows expression in the uninduced state (see SOM).
In order to further test the effect of NADP on Gal80p binding to Gal4p-AD, we generated a panel of Gal80p mutants and Gal4p binding was tested by GST pull-down experiments at varying NADP concentrations (Fig. 2B, Fig. S6). Disruption of the Gal80p dimer interface (N230R) by disruption of hydrogen bonding interactions between the monomers, caused substantial decrease in overall binding to Gal4p-AD and this low-level binding was almost completely abolished even in the presence of low concentrations of NADP. Several mutants that should alter NAD(P) binding had lost sensitivity to NADP compared to the wild-type protein. W31A, designed to disrupt stacking with the nicotinamide ring, exhibits no sensitivity to NADP over the range of concentrations tested (0–2.5 mM). E122, which forms a hydrogen bond with the nicotinamide N7, was changed to an alanine and showed a decrease in overall binding but no sensitivity to NADP. H36F, which should alter positioning of E122, showed a slight decrease in binding at 2.5 mM NADP. H99A, which likely disrupts interaction of the histidine with the ribose of the nicotinamide group, behaves similarly to the wild-type protein. From modeling studies, N26 and K29 might interact with the additional 2’ phosphate of NADP. N26D showed very similar behavior to the wild-type protein and K29E showed overall weaker binding.
In order to test the biological effect of altering the NADP binding site we prepared Gal80p wild type and mutants with a C-terminal FLAG tag and monitored each mutant’s effect on GAL1 expression in yeast at different time points following induction with galactose (Fig. 3, Fig. S7). All mutants predicted to affect dinucleotide binding still repressed normally in the uninduced state. Several mutants - H36F, K29E, W31A, and E122A showed higher levels of expression at earlier time points after induction with galactose relative to the wild-type proteins. H99A was more similar to wild type. All mutants leveled off to wild-type expression levels at 30–45 minutes post induction. This suggests that alterations in the NAD(P) binding site affects the initial rate of induction, but not overall final expression levels. It appears that NAD might facilitate Gal80p binding to Gal4p, since we could only identify Gal4p-AD with NAD bound, and NADP destabilizes this interaction. The mutations, affecting both NAD and NADP binding, would therefore disrupt both the stabilizing effect of NAD and destabilizing effect of NADP with a net result of faster induction for the mutants compared to wild type.
The involvement of dinucleotides and metabolic factors in transcriptional regulation is seen in a few other systems. The coactivator of Oct-1, OCA-S, contains two glycolytic enzymes – glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and lactate dehydrogenase (9). The binding of the transcriptional corepressor complex, CtBP, is enhanced by the reduced dinucleotide NADH compared to the oxidized form (10) and it possesses a NAD-dependant dehydrogenase activity (11). The DNA-binding activity of the transcription factor neuronal PAS domain protein 2 (NPAS2) is sensitive to the oxidation state of NAD, with DNA binding enhanced by the reduced form of the dinucleotide (12). While we do not understand precisely how this trigger for GAL regulation functions, nor the involvement of NADP versus NAD, we speculate that switching the cell to a fermentable galactose medium causes a change in NADP/NADPH or NADP/NAD ratios in the cell, and Gal80p effectively senses the metabolic state of the cell. NADP might be acting as a “second messenger” in triggering the system. Alternatively, Gal80p may function as an oxidoreductase, actively converting NADPH to NADP in the presence of a substrate causing it to disassociate from Gal4p.
Supplementary Material
References
- 1.Bhat PJ, Murthy TVS. Molec. Microbiol. 2001;40:1059. doi: 10.1046/j.1365-2958.2001.02421.x. [DOI] [PubMed] [Google Scholar]
- 2.Peng G, Hopper JE. Mol. Cell. Biol. 2000;20:5140. doi: 10.1128/mcb.20.14.5140-5148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma J, Ptashne M. Cell. 1987;50:137. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- 4.Keegan L, Gill G, Ptashne M. Science. 1986;231:699. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- 5.Johnston SA, Salmeron JM, Jr, Dincher SS. Cell. 1987;50:143. doi: 10.1016/0092-8674(87)90671-4. [DOI] [PubMed] [Google Scholar]
- 6.Han Y, Kodadek T. J. Biol. Chem. 2000;275:14979. doi: 10.1074/jbc.275.20.14979. [DOI] [PubMed] [Google Scholar]
- 7.Thoden JB, Sellick CA, Reece RJ, Holden HM. J. Biol. Chem. 2007;282:1534. doi: 10.1074/jbc.C600285200. [DOI] [PubMed] [Google Scholar]
- 8.Einarson MB. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press; 2001. pp. 18.55–18.59. [Google Scholar]
- 9.Zheng L, Roeder RG, Luo Y. Cell. 2003;114:255. doi: 10.1016/s0092-8674(03)00552-x. [DOI] [PubMed] [Google Scholar]
- 10.Fjeld CC, Birdsong WT, Goodman RH. Proc. Natl. Acad. Sci., USA. 2003;100:9202. doi: 10.1073/pnas.1633591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar V, et al. Mol. Cell. 2002;10:857. doi: 10.1016/s1097-2765(02)00650-0. [DOI] [PubMed] [Google Scholar]
- 12.Rutter J, Reick M, Wu LC, McKnight SL. Science. 2001;293:510. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 13.We thank Troy Messick, Khalid Siddiqui, Marlies Rossman, Jim Hicks, Alex Gann, Stephen Harrison, and members of the Joshua-Tor lab for discussions and advice, Annie Heroux, Michael Becker and Howard Robinson for support at the National Synchrotron Light Source (NSLS) and Stephan Ginell and Norma Duke for support at the Advanced Photon Source (APS). The NSLS and the APS are supported by the US Department of Energy, Office of Basic Energy Sciences. Coordinates and structure factors have been submitted to the Protein Data Bank. Accession numbers: Gal80pS0-Gal4AD-NAD: 3BTS; Gal80pS2: 3BTU; Gal80pS0: 3BTV. This work was supported by NIH grants GM074075 (to LJ) and GM55641 (to RS).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.