Abstract
The cuticle of Caenorhabditis elegans, a complex, multi-layered extracellular matrix, is a major interface between the animal and its environment. Biofilms produced by the bacterial genus Yersinia attach to the cuticle of the worm, providing an assay for surface characteristics. A C. elegans gene required for biofilm attachment, bah-1, encodes a protein containing the domain of unknown function DUF23. The DUF23 domain is found in 61 predicted proteins in C. elegans, which can be divided into three distinct phylogenetic clades. bah-1 is expressed in seam cells, which are among the hypodermal cells that synthesize the cuticle, and is regulated by a TGF-β signaling pathway.
Introduction
The cuticle of the nematode Caenorhabditis elegans is vital for proper morphology, locomotion, and protection from pathogens and environmental stresses [1]. After the nematode hatches, this multi-layered, dynamic extracellular matrix is synthesized four times, with new cuticle replacing old at each molt [2]. The cuticle basal layer is attached to the hypodermal cells [1], and distal to it are the medial and cortical layers. These three layers, composed primarily of cross-linked collagens and cuticlins, provide structure, integrity and flexibility [2], [3]. The next layer, the epicuticle, contains lipids and perhaps proteins, but these have not been analyzed in detail [4], [5]. The most distal layer, the surface coat, is also poorly characterized. It can be visualized in transmission electron microscopy with cationized ferritin particles, suggesting that it is anionic [2], and indirect evidence suggests that the surface coat contains O-linked glycoproteins [6]. In the parasitic nematode Toxocara canis the surface coat is labile, requiring only ethanol for extraction [7], but this has not been examined for C. elegans.
Several phenotypes have been used in genetic studies of C. elegans surface determination. Antibody- and lectin-binding aberrations were used to isolate srf mutants [6], [8], [9], [10], [11]. Resistance to the nematode-specific pathogen Microbacterium nematophilum yielded numerous novel bus (bacterially unswollen) genes as well as alleles of srf genes. The bus study also described an assay for cuticle integrity using sensitivity to alkaline-hypochlorite treatment [12]. The causative agent of bubonic plague, Yersinia pestis, and its close relative, Yersinia pseudotuberculosis, form biofilms on the surface of the C. elegans head [13], [14], [15]. A screen for mutants resistant to biofilm attachment produced bus and srf alleles and also identified three novel bah (biofilm absent on head) genes: bah-1, bah-2 and bah-3 [16].
We now describe the cloning and initial characterization of bah-1, which encodes a member of a previously undescribed protein family found in nematodes, insects, plants and some non-mammalian vertebrates.
Materials and Methods
C. elegans strains
The wild-type strain N2 Bristol and JR667 (wIs51) SCM::GFP were used as were the following mutant genotypes of N2 Bristol; chromosomes are indicated by Roman numerals: (I) dpy-5(e61), daf-16(mgDf47), unc-101(m1), daf-8(e1393), (II) daf-5(e1386), rrf-3(pk1426), (III) daf-2(e1370), daf-4(m63), daf-7(e1372), (IV) daf-1(m40), daf-14(m77), (V) him-5(e1467), (X) daf-3(e1376), daf-12(m25), lin-15(n765).
Biofilm formation on C. elegans
Biofilm assays were performed as previously described [14]. Briefly, adult worms were placed on lawns of Y. pestis or Y. pseudotuberculosis at room temperature and the presence of biofilm scored after 4 hours. For the analysis of the TGF-β pathway, all nematodes were grown at 15° C and assayed as adults at room temperature.
Screen for additional bah-1 alleles
Strain N2 was mutagenized with ethyl methanesulfonate (EMS) as described [17]. F2 animals resistant to biofilm accumulation were isolated as previously described [16]. New alleles were confirmed by complementation test against bah-1(br1). In addition, a non-complementation screen was performed. Strain CB1467 him-5(e1467) was mutagenized with EMS and F1 male progeny mated to C. elegans strain DC1065 dpy-5(e61) bah-1(br1) unc-101(m1). F1 progeny were treated with alkaline hypochlorite to release their eggs, which were washed in M9 buffer and placed onto lawns of Y. pseudotuberculosis strain YPIII. Bah non-Dpy non-Unc worms were selfed for two generations to recover a strain homozygous for a new bah-1 allele. From approximately 9000 mutagenized genomes, one additional allele of bah-1 was isolated, br12.
Transgenic lines
Transgenic strains were produced by injecting yeast artificial chromosome (YAC), cosmid, or plasmid DNA into the gonad of young adult hermaphrodites as described [17]; for YAC experiments, we used total yeast DNA from strains carrying a YAC. Two transformation markers, lin-15 and rol-6, were used. A wild-type lin-15 (pbLH98) construct was injected, at approximately 60 ng/µl marker DNA and approximately 100–150 ng/µl test DNA, to transform strain DC1077 bah-1(br1) I; lin-15(n765ts) X. Stable F2 Lin+ lines were obtained and tested for rescue of the Bah phenotype on Yersinia pseudotuberculosis lawns. For the GFP expression construct, 100 ng/µl of rol-6(su1006) (pRF4), a dominant marker, was injected into wild-type C. elegans along with approximately 100 ng/µl pCBD156 and lines established from Rol progeny. For the epitope-tagged localization construct, plasmid pCBD219 was injected into DC1077 along with the lin-15 marker as described above.
Transgenic rescue
bah-1 was previously mapped to a region deleted by the deficiency hDf17 [16]. YAC clones Y53A2, Y47H9, Y53C10, and Y47H10 span this region. After complementation with Y53A2, cosmids spanning this YAC were obtained: T02A8, M04D5, C27D12, C27C7, ZK1025, K08C9, F32B4, F15D3, and C17D12. After cosmid ZK1025 complemented the Bah phenotype, single gene rescue was tested with a PCR-derived clone specific to the genes ZK1025.3, ZK1025.4, and ZK1025.7. All cosmid and YAC clones were obtained from the Sanger Institute (Cambridge, UK). To sequence mutant alleles, genomic DNA was harvested and PCR products corresponding to ZK1025.7 were analyzed for mutations.
Plasmid construction
For rescue experiments with full length bah-1, a PCR fragment including bah-1 and approximately 2 kb upstream, amplified with primers 5′-CGAACCTTAACTGGGGAGTAC and 5′-GACGATGATACGTGTACC, was cloned into pCR2.1-TOPO vector (Invitrogen, San Diego, CA), making the plasmid pCBD154. For expression analysis 2 kb upstream of bah-1 was PCR amplified with primers 5′-CTGCAGGCGAACCTTAACTGGGGAGTAC and 5′-GGATCCTGTAATCAATATATGCCTTG that added restriction sites PstI and BamHI, respectively. The PCR product was ligated into vector pPD95.81 (Addgene plasmid 1497) to make pCBD156. For the 3×hemagglutinin (HA) tagged bah-1 construct, full-length bah-1 along with the 2 kb upstream was obtained with PCR primers 5′-GCCACTAGTCGAACCTTAACTGGGGAG and 5′-GCTCTGCAGGACGATGATACGTGTAC that introduced the restriction sites SpeI and PstI, respectively. The product was ligated into the vector pKH3 [18] to make pCBD219.
Phylogenetic analysis of C. elegans DUF23
Phylogenetic analyses and construction was accomplished using MEGA4 [19]. Cladistic analysis used the maximum parsimony method [20] and the bootstrap consensus tree was inferred from 10,000 replicates. The Close-Neighbor-Interchange algorithm [21] was used to obtain the initial trees with the random addition of 10 replicates sequences.
RNA interference (RNAi)
RNAi was performed using constructs from the Ahringer RNAi feeding library [22]. Briefly, an overnight culture was diluted and grown to exponential phase before the addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) to induce double-stranded RNA production for approximately 2.5 hours at 37° C. Cultures were spiked with additional IPTG and used to seed Nematode Growth Medium (NGM) plates [17]. The next day L1/L2 stage rrf-3(pk1426) animals (hypersensitive to RNAi) were placed on these plates and incubated at 20° C for two days. Gravid adults from these plates were transferred to new NGM plates freshly seeded with the same RNAi construct induced as before. On the third day the F1 progeny were tested for cuticle defects.
Immunofluorescence
C. elegans strain DC1154 carrying the construct pCBD219 was grown at 25° and transgenic worms identified by a Lin+ phenotype. L4 and early adult stage worms were fixed in 1% formaldehyde as described [17]. Incubation with anti-HA mouse monoclonal antibody 16B12 conjugated to Alexa Fluor 488 (Molecular Probes, Eugene OR) was carried out overnight at 4° at a 1∶25 dilution. Immunolocalization was carried as described using phosphate buffered saline (pH 8.0) containing 0.1% Tween 20 and 5% bovine serum albumin as a blocking agent [17]. Fluorescence was visualized using a Zeiss Axioplan 2 microscope.
Results
bah-1 animals are resistant to biofilm attachment
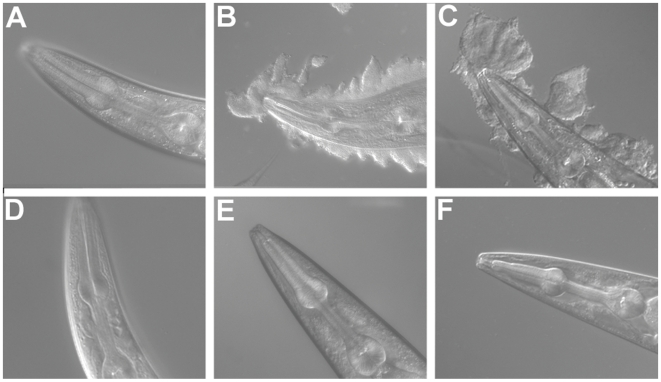
Biofilms produced by the bacterial species Y. pestis and Y. pseudotuberculosis attach to and accumulate on the head of C. elegans (Figure 1B and 1C). The biofilms physically block feeding, which for larval worms results in developmental arrest [23]. A screen for C. elegans mutants with altered biofilm binding identified three novel genes, including bah-1 [16]. No biofilm attaches to bah-1 animals in adult (Figure 1) or larval (not shown) stages, and their development is identical whether fed E. coli or Yersinia.
Figure 1. bah-1 worms are resistant to biofilm attachment.
(A–C) Wild-type C. elegans exposed to (A) E. coli, (B) Y. pestis, and (C) Y. pseudotuberculosis. (D–F) bah-1 worms exposed to (D) E. coli, (E) Y. pestis, and (F) Y. pseudotuberculosis.
Molecular identification of bah-1
The genetic deficiency hDf17 failed to complement bah-1 [16], narrowing the interval containing bah-1 to approximately 4 map units on the right arm of chromosome I. YACs spanning this interval were injected into bah-1 animals and transgenic lines established for each clone. YAC Y53A2 complemented the Bah phenotype, further narrowing the interval to approximately 350 kb. Next, overlapping cosmid clones spanning the Y53A2 region were injected, and rescue obtained with ZK1025, which contains nine genes. Finally, rescue was achieved with a single full-length gene on this cosmid, ZK1025.7 (Figure S1). Sequencing of seven bah-1 mutant alleles confirmed that ZK1025.7 is bah-1 (Table 1). An allele generated by the C. elegans Gene Knockout Consortium, ok2197, is a deletion that removes most of the coding region and is therefore presumptively null.
Table 1. Location and consequence of bah-1 mutations.
| Allele | Locationa | Mutation | Consequence |
| br1 | 1332 | a to t | E268V |
| br12 | 965 | g to a | premature stop |
| br21 | 965 | g to a | premature stop |
| br22 | 1657 | g to a | aberrant splice junction |
| br29 | 1388 | a to t | H279L |
| 1486 | g to a | G312E | |
| br42 | 1284 | c to t | H245Y |
| ok2197 | 319–2060 | deletion | incomplete transcript |
Distance from start ATG in base pairs.
bah-1 belongs to a large C. elegans gene family
The predicted bah-1 mRNA (GenBank CAA18365) encodes a 461-amino acid, 53 kDa predicted protein. Amino acid residues 1–215 and 232–461 are confirmed by six expressed sequence tags (ESTs) in the Nematode Expression Pattern Database, National Institute of Genetics, Japan (clones yk841d05, yk1713e02, yk759d11, yk838c01, yk1505d06, yk842d05). No splice variants are predicted by these ESTs. We sequenced the cDNA clone yk1713e02 to confirm that residues 216–231 are expressed (data not shown). The SignalP and PSORT algorithms both predict a cleavable signal peptide, suggesting extracellular localization [24], [25]. The protein contains eight N-glycosylation motifs (Asn-Xaa-Ser/Thr, where Xaa is any amino acid except proline).
BAH-1 contains the domain of unknown function DUF23, comprising 267 amino acids, and has no other domains in the PFAM database [26]. DUF23 is found in 60 additional predicted C. elegans proteins. No mutations in any of the corresponding genes have been reported, although a genome-wide RNA interference (RNAi) screen found a molting defect for a single DUF23 gene, Y47D3B.1 [27]. There are no observable molting defects produced by any bah-1 mutations, including the ok2197 large deletion. Phylogenetic analysis (see below) indicates that Y47D3B.1 is an outlier within the DUF23 family, and unlike BAH-1 and most other C. elegans DUF23 proteins, it lacks a predicted signal sequence.
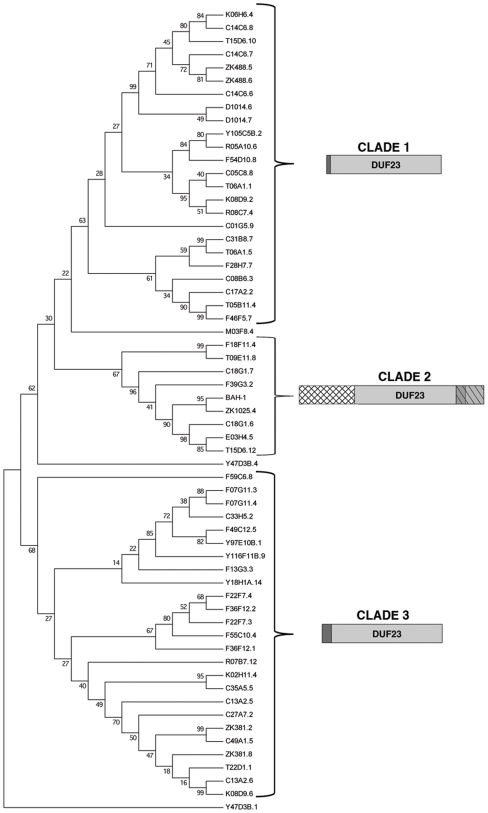
We analyzed the C. elegans DUF23 family using a maximum parsimony phylogenetic method. Most proteins fell into one of three distinct clades, each of which has unique features (Figure 2). Clade 1 proteins, which cluster away from other DUF23 proteins with a 63% bootstrap value, have a common motif consisting of approximately 10 amino acids directly N-terminal to the DUF23 domain. Clade 2, of which BAH-1 is a member, is characterized by two motifs. The first, approximately 150 residues, lies N-terminal to DUF23, while the second begins within the C-terminal region of DUF23 and continues beyond it for approximately 75 residues. The portion of this second motif that lies within DUF23 is more highly conserved within clade 2 than in the other two clades. Lastly, clade 3 proteins (68% bootstrap) have a common motif of approximately 25 amino acids N-terminal to DUF23. A number of proteins do not cluster with any of these clades, including the one implicated in molting, Y47D3B.1 (Figure 2).
Figure 2. Phylogenetic relationships of 61 C. elegans DUF23 proteins determined by maximum parsimony.
The bootstrap consensus tree shown is inferred from 10,000 replicates. Bootstrap values shown next to the branches were used to divide the family into three clades based on characteristic motif arrangements diagramed to the right of each clade. The DUF23 domain is shaded in light gray while conserved motifs found within each clade are distinguished by darker gray or a crosshatching pattern. The previously annotated DUF23 genes C13A2.1, C13A2.11, C13A2.2, D1014.5, K08D9.5, and T13H10.2 were eliminated from consideration because they lack the conserved cysteines and acidic amino acid motif, while C27C7.2 and ZK1025.5 were eliminated due to a high probability of being pseudogenes.
The DUF23 domain is found in insects, plants, and some bacteria. It is also in two vertebrates, the frog Xenopus laevis and the fish Tetraodon nigroviridis, but absent from all sequenced mammalian genomes. Genes encoding DUF23 proteins are far more abundant in free-living nematode genomes than in the genomes of other taxa. The fruit fly Drosophila melanogaster has five predicted DUF23 proteins while the mosquitoes Anopheles gambia and Aedes aegypti each have three. Figure 3 shows the alignment of DUF23 domains of BAH-1, insects and X. laevis. DUF23 domains of other species are more similar to BAH-1 and other members of clade 2 than they are to the C. elegans DUF23s in the other clades.
Figure 3. CLUSTAL alignment of the DUF23 domains of BAH-1 and its closest relatives in three insect species and X. laevis.
Boxed regions are the most highly conserved within DUF23 proteins. Arrows show cysteine residues with conserved spacing. Bars designate conserved acidic motif.
Across the entire family, the most highly conserved DUF23 features are a small acidic motif and two cysteines with conserved spacing (Figure 3). The br1 mutation results in a glutamic acid to valine substitution in the acidic motif. The Bah phenotype of br1 animals is identical to that of worms carrying the presumptively null ok2197 deletion, suggesting that br1 is also null and therefore that the acidic motif is essential to protein function. All other bah-1 mutations analyzed also disrupt the DUF23 domain.
Six C. elegans proteins annotated as DUF23 in databases (13, 29) lack the conserved cysteines, lack the acidic domain or have low overall similarity to the majority of family members, and we consider these to be misannotated (Figure 2).
bah-1 is expressed in seam cells
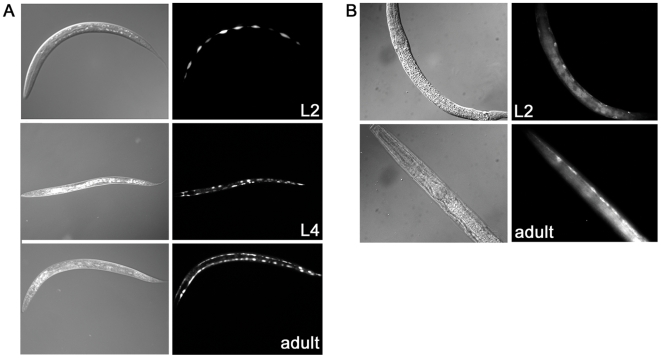
We fused the putative bah-1 promoter to a green fluorescent protein (GFP) gene and observed expression in the lateral seam cells (Figure 4A), a subset of the hypodermis. We obtained a strain with an integrated seam-cell specific promoter fused to GFP, JR667 (wIs51) SCM::GFP, and compared the signals to our bah-1 promoter-GFP signals; at all stages the expression patterns were the same (data not shown). Expression was detectable beginning in the early larval stages (L1/L2) and continuing into adulthood, consistent with the temporal expression reported in a functional genomics study [28]. To determine protein localization we generated transgenic worms expressing full-length BAH-1 with an added C-terminal HA tag. Expression of this construct rescued a bah-1 mutation, indicating that the modification did not disrupt function. In fixed, permeabilized worms, BAH-1 was detected in association with hypodermal cells, including the seam cells, consistent with the promoter fusion data (Figure 4B). We were unable to detect BAH-1 on the surface of unpermeabilized worms with antibody against the HA tag.
Figure 4. bah-1 expression in seam cells.
(A) Expression of promoter-GFP fusion (Experimental Methods) in L2, L4 and early adult. (B) Immunofluorescence detection of epitope-tagged BAH-1 in two representative worms.
bah-1 is regulated by the TGF-β signaling pathway
A TGF-β pathway regulates C. elegans entry into the alternative developmental stage known as the dauer larva, which the worm uses to survive under certain harsh conditions. TGF-β signaling inhibits the transcriptional regulators DAF-3 and DAF-5 that are required for dauer formation. Mutants with defects in the TGF-β ligand gene daf-7, the receptor genes daf-1 and daf-4, and the downstream SMAD signaling genes daf-8 and daf-14 constitutively make dauers at 25°C under non-inducing conditions (Daf-c phenotype). Conversely, daf-3 and daf-5 mutants are dauer defective under inducing conditions (Daf-d) [29].
A global transcriptional analysis showed that bah-1 expression was down-regulated an average of 19-fold in the absence of daf-7, daf-8 or daf-14 [30]. We confirmed that daf-7 animals have decreased bah-1 expression using semiquantitative RT-PCR (data not shown). The down-regulation prompted the hypothesis that mutants with TGF-β signaling defects would be Bah, and indeed this was previously reported for an unspecified daf-1 allele [15].
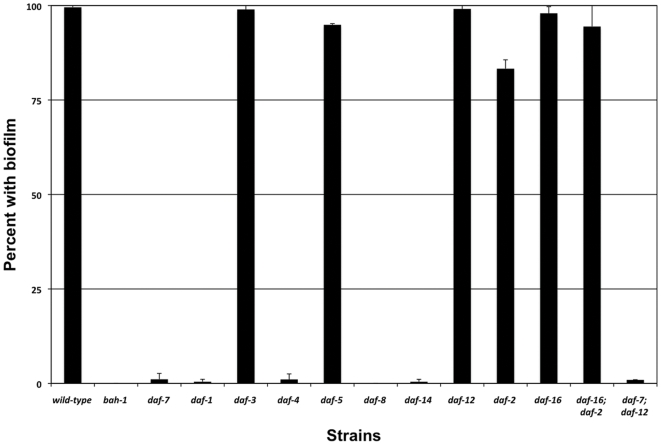
We previously showed that biofilms do not form on dauers [16]. To examine the biofilm phenotypes of non-dauer animals with defects in TGF-β signaling, mutants were grown at 15°C, because at this temperature even worms with null mutations in Daf-c genes do not become dauers [31]. We tested daf-7(e1372), daf-1(m40), daf-4(m63), daf-8(e1393) and daf-14(m77), each of which has a strong Daf-c phenotype [32], [33] and one of which, daf-14(m77), is probably null [31]. Each mutation produced a complete or nearly complete Bah phenotype, with no more than 2% of animals acquiring biofilms (Figure 5). The results are consistent with the bah-1 down-regulation observed in daf-7, daf-8 or daf-14 mutants [30], but do not exclude the possibility that TGF-β regulates other genes required for biofilm attachment. Mutants with defects in the Daf-d genes daf-3 and daf-5 were not Bah (Figure 5), indicating that TGF-β positive regulation of bah-1 is not mediated through DAF-3 or DAF-5.
Figure 5. Biofilm attachment phenotypes of bah-1 null, dauer defective, and dauer constitutive mutants.
Biofilm attached after 4 hours of exposure to Y. pseudotuberculosis. Data are mean±S.D. of two independent trials, with a minimum of 82 animals from each genotype per trial.
Dauer formation is also regulated by insulin/IGF-1 signaling, and mutations in the receptor gene of this pathway, daf-2, produce a Daf-c phenotype at 25°C. DAF-2 represses dauer formation by inhibiting the FOXO transcription factor DAF-16. Both daf-16 null animals and daf-16; daf-2 double mutants are Daf-d.
When grown at 15°C to avoid dauer formation, about 85% of daf-2 adults acquired biofilms (Figure 5). This weak Bah phenotype is in contrast to the nearly complete absence of biofilms on TGF-β pathway mutants. daf-16 null and daf-16; daf-2 animals had the wild-type biofilm phenotype (Figure 5).
A steroid hormone receptor required for dauer formation, DAF-12, is downstream of both TGF-β and insulin-like pathways [32], [34]. daf-12 mutants are not Bah, while a daf-7; daf-12 mutant was biofilm defective, indicating that TGF-β positive regulation is not mediated through DAF-12.
Disruption of other DUF23 genes does not produce the Bah phenotype
Using RNAi we tested 21 other C. elegans DUF23 proteins for aberrant biofilm attachment (Table S1). Three criteria were used to choose those tested: they are in the same clade as BAH-1 (Figure 2), the developmental timing of their expression is similar to that of BAH-1 [28], or they are regulated by the TGF-β pathway [30]. Only RNAi on bah-1 and ZK1025.4 resulted in a Bah phenotype (Table S1). ZK1025.4 is the C. elegans DUF23 most similar to bah-1, and there is sufficient DNA sequence identity to predict cross-reaction of RNAi constructs. RT-PCR analysis showed that ZK1025.4 RNAi indeed reduced bah-1 expression, suggesting that the Bah phenotype observed was an off-target effect (data not shown). Attempts to reduce ZK1025.4 expression by RNAi without affecting bah-1 expression were unsuccessful.
Discussion
Biofilm attachment reveals a role for a DUF23 protein in determining C. elegans surface characteristics
The composition, structure and function of the C. elegans exterior surface are poorly understood. Biochemical analysis, using sequential extractions of increasing harshness, provided evidence for a heterodimeric protein complex distal to the collagen layers, probably in the epicuticle [4]. The identities of the two proteins were not determined. Genetic analysis, using antibodies and lectins to probe the surface, identified genes srf-2, srf-3 and srf-5, which appear to have primary defects in surface properties [9], [10]. Here, too, the surface defects have not been determined at the molecular level. srf-3 encodes a nucleotide sugar transporter, a protein that mediates intracellular trafficking of the glycosylation substrate UDP-galactose [35]. A biochemical analysis confirmed that srf-3 mutants are globally deficient in galactosylation [36], but the specific glycoconjugates that are absent or aberrant are not known. Infection with M. nematophilum identified additional glycosylation genes that affect the surface: bus-17 [37], bus-2, bus-4 and bus-12 (J. Hodgkin, personal communication). Mutations in any of these four genes produce the Bah phenotype [16].
Yersinia sp. biofilms that bind the C. elegans exterior provide an additional assay for the nematode surface. Ultrastructural analysis of biofilm binding does not show any physical perturbation of the cuticle's subsurface collagen layers [13], and biofilm binding is normal when collagen genes, e.g. dpy-5, dpy-9, dpy-17 and rol-6, are mutated (data not shown). Even bli-6 mutants, whose cuticle becomes heavily blistered, bind biofilm normally (data not shown). In contrast, mutations in any of the glycosylation genes found to affect the surface (bus-2, bus-4, bus-12, bus-17, srf-3) produce the Bah phenotype, as do mutations in the uncloned surface-determining genes srf-2 and srf-5 [16]. Together, these observations indicate that biofilm attachment is to distal cuticle components, i.e. the surface coat and/or epicuticle. The Bah phenotypes of multiple glycosylation mutants suggest that the biofilm receptor either is itself a glycoconjugate or that its function requires at least one glycoconjugate.
Using the absence of biofilm phenotype (Bah) in a mutant screen, we identified three genes, bah-1, bah-2 and bah-3, without other strong phenotypes [16]. In particular, they behave normally in response to M. nematophilum infection, suggesting that they are specific for the surface characteristics that mediate biofilm attachment. bah-1 and bah-2 cuticles are mildly sensitive to alkaline hypochlorite treatment, while bah-3 mutants have normal integrity [16].
bah-1 encodes one of the many C. elegans proteins containing a DUF23 domain. A fully penetrant Bah phenotype is produced by each of seven alleles, four of which (br12, br21, br22 and ok2197) are likely null and three of which (br1, br29 and br42) are missense mutations (Table 1). The br1 mutation could be especially informative about DUF23 function because it disrupts the most highly conserved motif in the family (Figure 3).
No DUF23 genes have been identified in previous C. elegans mutant screens. A genome-wide RNAi screen found a molting defect for Y47D3B.1, but not for any other DUF23 protein [27]. Although Y47D3B.1 has the common cysteines and acidic motif of DUF23 domains, it is an outlier in the C. elegans family. It does not fall within one of the three clades in our phylogenetic analysis, and unlike BAH-1 and all other members of clade 2, it has no predicted signal sequence. There is no molting defect in bah-1 mutants, including the presumptively null deletion allele ok2197, nor is there evidence that DUF23 proteins other than Y47D3B.1 function in molting. We conclude that bah-1 does not participate in molting.
BAH-1 expression in seam cells
The C. elegans cuticle is secreted by hypodermal cells. Seam cells, a hypodermal subset in bilateral rows along the length of the animal, participate in this process [27], [38]. BAH-1 is expressed in seam cells (Figure 4), and its temporal regulation correlates with that of collagen and cuticlin genes that encode the major cuticle components [1], [39]. One function of seam cells is to synthesize alae, which are lateral cuticle ridges that run the length of the animal directly over the seam cells in L1 larvae, dauer larvae, and adults [38]. Alae are not present in L2, L3 and L4 stage worms, but Yersinia sp. biofilms bind these stages [14], indicating that alae are not required for biofilm binding and that BAH-1 function is not alae-specific. Mutations in the gene srf-3 also lead to aberrant surface phenotypes including resistance to biofilm attachment [35], [40]. Like bah-1, srf-3 is expressed in lateral seam cells, further implicating these cells in the production of surface determinants and suggesting a role for these proteins in cuticle development.
The presence of a signal sequence, conserved cysteine spacing and N-glycosylation motifs together suggest that BAH-1 is extracellular, consistent with the possibility that it is part of the epicuticle or surface coat. However, we did not detect epitope-tagged BAH-1 on the surface of live, undisrupted worms. This suggests that BAH-1 remains cell-associated, but we cannot exclude the possibilities that it is within the extracellular matrix but not detected because of low abundance, covalent modification before or during secretion, masking of the epitope by other surface components, or loss during processing for staining.
Regulation by TGF-β
C. elegans is capable of sensing its environment and adapting gene expression accordingly [41]. A well-studied example is sensing of environmental stress (scarce food, overcrowding and high temperature) and subsequent entry into the non-feeding, non-reproducing dauer stage [4], [38]. This developmental decision, made during the L1 stage, is regulated by both TGF-β and insulin/IGF-1 signaling pathways [29]. Under non-stress conditions, receptors in both pathways are activated, dauer formation is repressed, and reproductive development proceeds. When signaling is down-regulated or absent, the dauer pathway is activated.
Dauers differ from the corresponding normal larval stage (L3) in morphology, physiology and behavior. Of particular relevance for the current study, the dauer cuticle has a markedly different ultrastructure and, unlike any other stage, is resistant to 1% sodium dodecylsulfate [2]. Differences in cuticle collagen composition between dauers and non-dauers have been examined [42], but the surface components that are distal to collagen layers have not been determined for either dauers or non-dauers.
Yersinia biofilms do not bind dauers, but bind the surface of all non-dauer stages in a bah-1-dependent manner. A global transcriptional analysis showed that bah-1 expression requires the TGF-β ligand gene daf-7 and the SMAD genes daf-8 and daf-14 [30]. We confirmed that mutations in these genes, as well as the daf-1 and daf-4 receptor genes, produce a strong Bah phenotype (Figure 5). Because loss of bah-1 is sufficient for Bah, our data do not address whether TGF-β has other targets affecting surface characteristics.
Evidence from another assay indicates that it does. A monoclonal antibody, M37, binds the surface of wild-type C. elegans first stage (L1) larvae but no other developmental stages. However, M37 aberrantly binds all non-dauer larval stages of some mutants, a phenotype known as constitutive larval display (Cld) [11]. Mutations in daf-1, daf-4, daf-7, daf-8 and daf-14 all produced a strong Cld phenotype when worms were grown under non-dauer-inducing conditions [43]. The antigen recognized by M37 is unknown, but it is present in bah-1 mutants and therefore cannot be BAH-1 [16]. The Bah and Cld phenotypes of TGF-β mutants indicate that the surface properties of Daf-c mutants are aberrant, even when the worms are grown at 15° C and do not actually form dauers. The defect cannot be explained by the simple hypothesis that the non-dauer Daf-c mutants have surfaces identical to those of dauers, because M37 does not bind dauer larvae [11].
Like multiple TGF-β signaling mutants, daf-2 mutants are strongly Daf-c. Unlike the TGF-β mutants, however, daf-2 animals have only a weak Bah phenotype. Most daf-2 adults acquired biofilms; only about 15% did not, and the defect was suppressed by a daf-16 mutation (Figure 5). Also unlike TGF-β Daf-c mutants, daf-2 animals do not have the Cld phenotype [11]. We could not determine the Bah phenotype of daf-2 null mutations, because such mutations are lethal [44]. However, the daf-2(e1370) allele we tested has a fully penetrant Daf-c phenotype at 25° C [32] and a strong longevity phenotype [45], indicating that even strong daf-2 mutations produce only a weak Bah phenotype.
These results suggest that during dauer formation, DAF-16 represses surface-determining genes, perhaps including bah-1, that are only required in non-dauer stages. In wild-type animals under reproductive growth conditions, repression would not occur because DAF-2 inhibits DAF-16. In daf-2 mutants under these conditions, DAF-16 is aberrantly activated, leading to repression of surface-determining genes. This is plausible in light of findings that in daf-2 mutants, dauer-stage genes are inappropriately expressed in non-dauers [46]; presumably inappropriate repression of non-dauer-stage genes also occurs.
Although the role of TGF-β in regulating dauer formation has been extensively investigated, the pathway's regulation of non-dauer characteristics has been little examined [47]. Taken as a whole, our analysis of dauer pathway regulatory mutants suggests that the TGF-β pathway is a major positive regulator of surface-determining genes such as bah-1 that are expressed during reproductive growth. Unlike TGF-β activity in the dauer pathway, the surface gene regulation is not mediated by DAF-3, DAF-5 or DAF-12. Our results also indicate a possible subsidiary role for the insulin/IGF-1 pathway in repressing surface-determining genes during dauer formation.
The DUF23 family in C. elegans and other organisms
C. elegans has 61 proteins containing DUF23, and the family is also large in the related species Caenorhabditis remanei (90 representatives) and Caenorhabditis briggsae (60 representatives). Accordingly, these proteins have representatives that fall into the same three phylogenetic clades as C. elegans [48], WormBase Release WS170). Interestingly, searches of the incomplete genomes of the human parasitic nematodes Brugia malayi and Trichinella spiralis and plant-parasitic nematodes Meloidogyne hapla and Heterodera glycines did not find DUF23 domains.
In our phylogenetic analysis we divided the C. elegans family into three clades (Figure 2). We found six C. elegans proteins that appeared to be misclassified, because they did not have the conserved acidic motif that is present in all others DUF23 proteins. Nevertheless, the number of DUF23 proteins in C. elegans and in related nematodes greatly exceeds that in any other sequenced organism. In the Pfam database (http://pfam.sanger.ac.uk) the most DUF23 proteins outside of nematodes is 13 in the mustard Arabidopsis thaliana. No more than five are in the genome of any animal that is not a nematode [26].
At least one DUF23 is present in every insect genome available in GenBank. This is significant because the biofilm system used to identify and characterize bah-1 is a model for Y. pestis colonization of its vector, the flea [13], [23]. Formation of a biofilm in the flea digestive tract is essential for transmission of this pathogen [49]. The BAH-1 requirement for C. elegans biofilm attachment, coupled with the existence of DUF23 proteins in several sequenced insect genomes, suggests that it will be worthwhile to determine whether fleas express this domain at the site of biofilm binding and, if so, whether it has a role in biofilm adherence.
Bioinformatic analysis has grouped protein families such as DUF23 in larger, multi-family “clans” that are believed to be of common evolutionary origin [50]. DUF23 is assigned to clan GT-A, which contains multiple glycosyltransferase families as well as other types of carbohydrate-interacting proteins [51]. BAH-1 may directly interact with carbohydrates by binding to specific sugar residues or indirectly by participating in a pathway for the specific glycoconjugate production required for biofilm attachment to the surface of C. elegans. Regardless, a BAH-1 interaction with carbohydrates, is consistent with the finding that multiple genes known to function in glycosylation are required for biofilm adherence to C. elegans.
Concluding remarks
The nature of the C. elegans surface and the manner in which it interacts with the environment are largely unknown. Using attachment of Yersinia sp. biofilms as a probe for the surface, we have found that bah-1 plays a role in determining the nematode's surface characteristics. Neither bah-1 nor any member of the large DUF23 family to which it belongs was identified by other mutant phenotypes, which demonstrates the utility of the biofilm phenotype in analyzing C. elegans surface characteristics. We have shown that BAH-1, possibly a secreted protein, is expressed in seam cells, a subset of the cuticle-synthesizing hypodermis. Obviously the evolved purpose of BAH-1 is not to mediate adherence of a deleterious microbial product. The normal function is not known, nor is the reason why C. elegans and related free-living nematodes have many more DUF23 proteins than organisms in other taxa. As a motile soil organism, C. elegans undoubtedly encounters a wide variety of environmental threats. Mutation in bah-1 affect the worm's interaction with a microbial exopolysaccharide, and also cause a slight increase in the cuticle's sensitivity to harsh reagents. These findings prompt the hypothesis that DUF23 proteins function in maintaining cuticle integrity against environmental assaults.
Supporting Information
bah-1 cloning. (A) Chromosome I, the deficiency hfDf17, Yeast artificial chromosomes (YACs) and cosmid clones in approximate relative positions. Numbers indicate ratios of rescuing lines to total transgenic lines for each clone. “Genes” shows rescue results for PCR-derived single gene constructs. (B) Exon-intron structure and locations of mutations were obtained from full length cDNA sequences from the National Institute of Genetics, Mishima, Japan.
(9.68 MB TIF)
(0.03 MB XLS)
Acknowledgments
We thank Jonathan Stoltzfus for assistance with RNAi experiments, Michael Miller and Kaveh Ashrafi for RNAi clones and worm strains, Bradley Yoder for plasmid pPD95.81, Yuji Kohara for cDNA clones, Karen Yook for assistance with mutagenesis, and the C. elegans Gene Knockout Consortium for ok2197. Some strains were provided by the Caenorhabditis Genetics Center, which is supported by the NIH National Center for Research Resources.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the National Institutes of Health (NIH) research grant AI057512 (C.D.), NIH training grant HL007553 (K.D.), and the Sandler Family Supporting Foundation (C.D.). Some strains were provided by the Caenorhabditis Genetics Center, which is supported by the NIH National Center for Research Resources. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Page AP, Johnstone IL. The cuticle. WormBook. 2007:1–15. doi: 10.1895/wormbook.1.138.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox GN, Kusch M, Edgar RS. Cuticle of Caenorhabditis elegans: its isolation and partial characterization. J Cell Biol. 1981;90:7–17. doi: 10.1083/jcb.90.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lassandro F, Sebastiano M, Zei F, Bazzicalupo P. The role of dityrosine formation in the crosslinking of CUT-2, the product of a second cuticlin gene of Caenorhabditis elegans. Mol Biochem Parasitol. 1994;65:147–159. doi: 10.1016/0166-6851(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 4.Blaxter ML. Cuticle surface proteins of wild type and mutant Caenorhabditis elegans. J Biol Chem. 1993;268:6600–6609. [PubMed] [Google Scholar]
- 5.Proudfoot L, Kusel JR, Smith HV, Harnett W, Worms MJ, et al. Rapid changes in the surface of parasitic nematodes during transition from pre- to post-parasitic forms. Parasitology. 1993;107 (Pt 1):107–117. doi: 10.1017/s0031182000079464. [DOI] [PubMed] [Google Scholar]
- 6.Hemmer RM, Donkin SG, Chin KJ, Grenache DG, Bhatt H, et al. Altered expression of an L1-specific, O-linked cuticle surface glycoprotein in mutants of the nematode Caenorhabditis elegans. J Cell Biol. 1991;115:1237–1247. doi: 10.1083/jcb.115.5.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Page AP, Rudin W, Fluri E, Blaxter ML, Maizels RM. Toxocara canis: a labile antigenic surface coat overlying the epicuticle of infective larvae. Exp Parasitol. 1992;75:72–86. doi: 10.1016/0014-4894(92)90123-r. [DOI] [PubMed] [Google Scholar]
- 8.Politz SM, Chin KJ, Herman DL. Genetic analysis of adult-specific surface antigenic differences between varieties of the nematode Caenorhabditis elegans. Genetics. 1987;117:467–476. doi: 10.1093/genetics/117.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Politz SM, Philipp M, Estevez M, O'Brien PJ, Chin KJ. Genes that can be mutated to unmask hidden antigenic determinants in the cuticle of the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1990;87:2901–2905. doi: 10.1073/pnas.87.8.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Link CD, Silverman MA, Breen M, Watt KE, Dames SA. Characterization of Caenorhabditis elegans lectin-binding mutants. Genetics. 1992;131:867–881. doi: 10.1093/genetics/131.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grenache DG, Caldicott I, Albert PS, Riddle DL, Politz SM. Environmental induction and genetic control of surface antigen switching in the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1996;93:12388–12393. doi: 10.1073/pnas.93.22.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gravato-Nobre MJ, Nicholas HR, Nijland R, O'Rourke D, Whittington DE, et al. Multiple genes affect sensitivity of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics. 2005;171:1033–1045. doi: 10.1534/genetics.105.045716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darby C, Hsu JW, Ghori N, Falkow S. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature. 2002;417:243–244. doi: 10.1038/417243a. [DOI] [PubMed] [Google Scholar]
- 14.Tan L, Darby C. A movable surface: formation of Yersinia sp. biofilms on motile Caenorhabditis elegans. J Bacteriol. 2004;186:5087–5092. doi: 10.1128/JB.186.15.5087-5092.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joshua GW, Karlyshev AV, Smith MP, Isherwood KE, Titball RW, et al. A Caenorhabditis elegans model of Yersinia infection: biofilm formation on a biotic surface. Microbiology. 2003;149:3221–3229. doi: 10.1099/mic.0.26475-0. [DOI] [PubMed] [Google Scholar]
- 16.Darby C, Chakraborti A, Politz SM, Daniels CC, Tan L, et al. Caenorhabditis elegans mutants resistant to attachment of Yersinia biofilms. Genetics. 2007;176:221–230. doi: 10.1534/genetics.106.067496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epstein HF, Shakes DC. San Diego: Academic Press; 1995. Caenorhabditis elegans: modern biological analysis of an organism.xxi659 [Google Scholar]
- 18.Mattingly RR, Macara IG. Phosphorylation-dependent activation of the Ras-GRF/CDC25Mm exchange factor by muscarinic receptors and G-protein beta gamma subunits. Nature. 1996;382:268–272. doi: 10.1038/382268a0. [DOI] [PubMed] [Google Scholar]
- 19.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 20.Eck R, Dayhoff M. Silver Spring, Md. Washington, D.C.: National Biomedical Research Foundation; Georgetown University Medical Center; 1966. Atlas of Protein Sequence and Structure. [Google Scholar]
- 21.Nei M, Kumar S. Oxford; New York: Oxford University Press; 2000. Molecular evolution and phylogenetics.xiv333 [Google Scholar]
- 22.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 23.Darby C, Ananth SL, Tan L, Hinnebusch BJ. Identification of gmhA, a Yersinia pestis gene required for flea blockage, by using a Caenorhabditis elegans biofilm system. Infect Immun. 2005;73:7236–7242. doi: 10.1128/IAI.73.11.7236-7242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 25.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, et al. The Pfam protein families database. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frand AR, Russel S, Ruvkun G. Functional genomic analysis of C. elegans molting. PLoS Biol. 2005;3:e312. doi: 10.1371/journal.pbio.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lund J, Tedesco P, Duke K, Wang J, Kim SK, et al. Transcriptional profile of aging in C. elegans. Curr Biol. 2002;12:1566–1573. doi: 10.1016/s0960-9822(02)01146-6. [DOI] [PubMed] [Google Scholar]
- 29.Hu PJ. Dauer. WormBook. 2007:1–19. doi: 10.1895/wormbook.1.144.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu T, Zimmerman KK, Patterson GI. Regulation of signaling genes by TGF-beta during entry into dauer diapause in C. elegans. BMC Dev Biol. 2004;4:11. doi: 10.1186/1471-213X-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue T, Thomas JH. Targets of TGF-beta signaling in Caenorhabditis elegans dauer formation. Dev Biol. 2000;217:192–204. doi: 10.1006/dbio.1999.9545. [DOI] [PubMed] [Google Scholar]
- 32.Vowels JJ, Thomas JH. Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics. 1992;130:105–123. doi: 10.1093/genetics/130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Levy DE. C. elegans STAT cooperates with DAF-7/TGF-beta signaling to repress dauer formation. Curr Biol. 2006;16:89–94. doi: 10.1016/j.cub.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 34.Thomas JH, Birnby DA, Vowels JJ. Evidence for parallel processing of sensory information controlling dauer formation in Caenorhabditis elegans. Genetics. 1993;134:1105–1117. doi: 10.1093/genetics/134.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoflich J, Berninsone P, Gobel C, Gravato-Nobre MJ, Libby BJ, et al. Loss of srf-3-encoded nucleotide sugar transporter activity in Caenorhabditis elegans alters surface antigenicity and prevents bacterial adherence. J Biol Chem. 2004;279:30440–30448. doi: 10.1074/jbc.M402429200. [DOI] [PubMed] [Google Scholar]
- 36.Cipollo JF, Awad AM, Costello CE, Hirschberg CB. srf-3, a mutant of Caenorhabditis elegans, resistant to bacterial infection and to biofilm binding, is deficient in glycoconjugates. J Biol Chem. 2004;279:52893–52903. doi: 10.1074/jbc.M409557200. [DOI] [PubMed] [Google Scholar]
- 37.Yook K, Hodgkin J. Mos1 mutagenesis reveals a diversity of mechanisms affecting response of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics. 2007;175:681–697. doi: 10.1534/genetics.106.060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sapio MR, Hilliard MA, Cermola M, Favre R, Bazzicalupo P. The Zona Pellucida domain containing proteins, CUT-1, CUT-3 and CUT-5, play essential roles in the development of the larval alae in Caenorhabditis elegans. Dev Biol. 2005;282:231–245. doi: 10.1016/j.ydbio.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 39.Kim SK, Lund J, Kiraly M, Duke K, Jiang M, et al. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- 40.Drace K, Darby C. The hmsHFRS operon of Xenorhabdus nematophila is required for biofilm attachment to Caenorhabditis elegans. Appl Environ Microbiol. 2008;74:4509–4515. doi: 10.1128/AEM.00336-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bargmann CI. Chemosensation in C. elegans. WormBook. 2006:1–29. doi: 10.1895/wormbook.1.123.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cox GN, Hirsh D. Stage-specific patterns of collagen gene expression during development of Caenorhabditis elegans. Mol Cell Biol. 1985;5:363–372. doi: 10.1128/mcb.5.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olsen DP, Phu D, Libby LJ, Cormier JA, Montez KM, et al. Chemosensory control of surface antigen switching in the nematode Caenorhabditis elegans. Genes Brain Behav. 2007;6:240–252. doi: 10.1111/j.1601-183X.2006.00252.x. [DOI] [PubMed] [Google Scholar]
- 44.Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, et al. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 46.McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 47.Savage-Dunn C. TGF-beta signaling. WormBook. 2005:1–12. doi: 10.1895/wormbook.1.22.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stein L, Sternberg P, Durbin R, Thierry-Mieg J, Spieth J. WormBase: network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res. 2001;29:82–86. doi: 10.1093/nar/29.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jarrett CO, Deak E, Isherwood KE, Oyston PC, Fischer ER, et al. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J Infect Dis. 2004;190:783–792. doi: 10.1086/422695. [DOI] [PubMed] [Google Scholar]
- 50.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu J, Mushegian A. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 2003;12:1418–1431. doi: 10.1110/ps.0302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bah-1 cloning. (A) Chromosome I, the deficiency hfDf17, Yeast artificial chromosomes (YACs) and cosmid clones in approximate relative positions. Numbers indicate ratios of rescuing lines to total transgenic lines for each clone. “Genes” shows rescue results for PCR-derived single gene constructs. (B) Exon-intron structure and locations of mutations were obtained from full length cDNA sequences from the National Institute of Genetics, Mishima, Japan.
(9.68 MB TIF)
(0.03 MB XLS)