Abstract
During the last 20 years, the identification of triadin function in cardiac and skeletal muscle has been the focus of numerous studies. First thought of as the missing link between the ryanodine receptor and the dihydropyridine receptor and responsible of skeletal type excitation–contraction coupling, the current hypothesis on triadin function has slowly evolved, and triadin is envisaged now as a regulator of calcium release, both in cardiac and skeletal muscle. Nevertheless, none of the experiments performed up to now has given a clear cut view of what triadin really does in muscle. The problem became more complex with the identification of multiple triadin isoforms, having possibly multiple functions. Using a different approach from what has been done previously, we have obtained new clues about the function of triadin. Our data point to a possible involvement of triadin in reticulum structure, in relation with the microtubule network.
Excitation–contraction (EC) coupling, the process which results in muscle contraction after electrical stimulation, is performed by a macromolecular protein complex. The core of this complex is composed of two calcium channels, the dihydropyridines receptor (DHPR) and the ryanodine receptor (RyR). Around these two channels are organized a number of other proteins (calsequestrin, triadin, junctin, …), and an efficient muscle contraction relies on the synchronous work of all these. One peculiar feature of this complex is that it is anchored in two different membranes: the T-tubule membrane, an invagination of the plasma membrane into the cytoplasm, which contains the DHPR, and the sarcoplasmic reticulum terminal cisternae, which contains the RyR. In the skeletal muscle, it has been demonstrated that both channels, even anchored in two different membranes, are physically associated (Marty et al. 1994). The function of the calcium release complex is therefore based upon the precise organization of the triad (close apposition of two sarcoplasmic reticulum terminal cisternae with one T-tubule) and the implantation of the complex in this membrane structure. This extremely precise organization has to be maintained in a cell, the skeletal muscle fibre, which undergoes large deformations during each contraction. Triadin was first identified as a 95 kDa protein, specifically localized in the triad of the skeletal muscle, thought to connect DHPR to RyR (Kim et al. 1990). Since its identification in 1990, the specific function of triadin has been the matter of numerous studies, in cardiac as well as in skeletal muscle, the two muscles in which triadin expression has been demonstrated.
Triadin, a multiprotein family
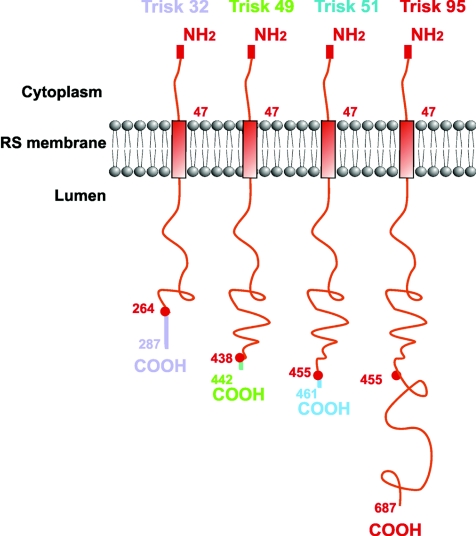
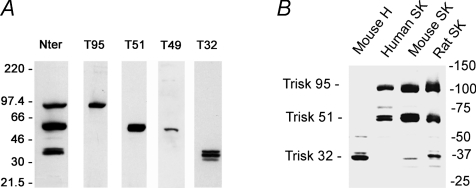
Triadin was first believed to be a skeletal muscle specific protein, responsible of skeletal muscle EC coupling (Caswell et al. 1991). In 1994, the presence of triadin in cardiac muscle was also demonstrated (Peng et al. 1994), as a 32 kDa protein (theoretical molecular mass). Nowadays, it is clear that the triadin gene can undergo multiple splicing, leading to a number of proteins. At least four isoforms are expressed in rodent skeletal muscles, named Trisk 95 (the 95 kDa triadin first identified), Trisk 51, a 51 kDa protein, Trisk 49 and Trisk 32, respectively 49 kDa and 32 kDa (Marty et al. 2000; Vassilopoulos et al. 2005). None of these isoforms is specific to a skeletal muscle type, and their pattern of expression is the same in fast or slow twitch muscles. Trisk 51 and Trisk 49 show an apparent molecular mass of 65 kDa in SDS-PAGE, and Trisk 32 migrates as a triplet centred on 37 kDa. All the skeletal isoforms are truncated versions of Trisk 95, with a specific C-terminal end of variable length (Fig. 1). We have developed a number of antibodies, and depending on their epitope localization, in the common (N-terminal end) or in specific regions (C-terminal ends), these antibodies are either reactive on every triadin isoform, or specific for one isoform (Fig. 2A). The antibody directed against the N-terminal end recognizes all triadins in all tissues and all species (mammalian) tested up to now, as this part is extremely conserved between all the spliced isoforms. Therefore this antibody allows the quantification of the relative expression level of the different isoforms in a given tissue. On the contrary, the antibodies specific for the C-terminal end of each isoform are only specific for a single isoform, and often specific for one species. Using antibodies against the common N-terminal part, we have observed that in rat skeletal muscle Trisk 95 and Trisk 51 are expressed at similar levels, each representing about 40% of the total triadin amount, Trisk 32 accounting for the remaining 20%. The situation is almost identical in mouse muscle. In adult human skeletal muscle, the major isoform is Trisk 51 (about 60% of all triadins), and Trisk 95 represents the remaining 40%. Trisk 32 is undetectable (Fig. 2B).
Figure 1.
The rat skeletal muscle triadin isoforms
Figure 2. Western blot analysis of triadin isoforms.
A, the rat skeletal muscle isoforms, detected by an antibody in common region (Nter) or in specific C-terminal region of each isoform. B, the triadin isoforms expressed in different tissues (mouse heart, human skeletal muscle, mouse skeletal muscle, rat skeletal muscle) detected with an antibody against the common N-terminal part of all triadins, and therefore able to react with all the triadins and used to quantify their relative amount. H: heart; SK: skeletal muscle.
The major cardiac triadin is CT1 (Kobayashi & Jones, 1999), but isoforms of higher molecular weight (CT2 and CT3) can be detected in cardiac muscle (Guo et al. 1996; Kobayashi & Jones, 1999; Hong et al. 2001). Trisk 32 is unambiguously identical to CT1, as demonstrated by cDNA cloning and by the reactivity of Trisk 32 specific antibodies with both skeletal and cardiac muscle. It is the only triadin isoform expressed in both muscles. The minor cardiac triadins detected with the anti-N-terminal antibody in heart homogenate are not recognized by the skeletal muscle specific antibodies of the same species, and are therefore cardiac muscle specific. As Trisk 32 is the only triadin isoform expressed in both muscles it could be renamed Trisk32/CT1, in order to clarify the nomenclature of the proteins, whereas the other tissue specific triadins should still be named Trisk (TRIadin Skeletal) or CT (Cardiac Triadin) respectively for the skeletal and cardiac specific triadins.
Localization of the triadin isoforms
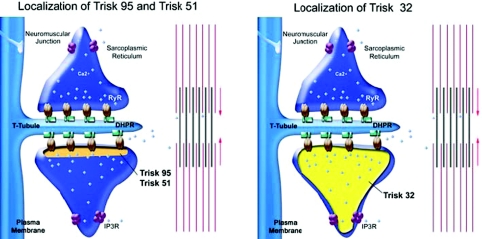
The question of the localization of triadin seems trivial, as this protein has been named ‘triadin’ according to the specific triad localization of the first identified isoform, Trisk 95. Nevertheless, considering the identification of multiple isoforms, this question has to be carefully re-examined. Trisk 95 and Trisk 51 localization is restricted to the triad of skeletal muscle where they are associated with RyR. Therefore, these two triadin isoforms are full members of the calcium release complex (Marty et al. 2000; Vassilopoulos et al. 2005). The situation is much more complex for Trisk 32, which behaves differently in heart and in skeletal muscle. In heart, it is both colocalized and associated with the cardiac RyR. In skeletal muscle, the protein is mainly in the longitudinal sarcoplasmic reticulum (Vassilopoulos et al. 2005), and therefore not associated with RyR. Interestingly, we have shown that in the longitudinal sarcoplasmic reticulum, Trisk 32 is associated with the IP3 receptor. Nevertheless, a closer examination showed that in fact Trisk 32 is localized in the whole sarcoplamic reticulum, both in the terminal cisternae which are part of the triad and in the longitudinal reticulum which connects two adjacent triads. Immunoprecipitation experiments confirmed that in fact a low amount of RyR1 is associated with Trisk 32. This means that even though the majority of Trisk 32 is not at the triad, the low amount present in the triad is associated with RyR. The localization of the main triadin isoforms in skeletal muscle is schematized in Fig. 3.
Figure 3. Schematic representation of the localization and possible partners of each triadin isoform.
Trisk 95 and Trisk 51 are only at the terminal cisternae of sarcoplasmic reticulum, where they are associated with the RyR. Trisk 32 is located in the whole sarcoplasmic reticulum, associated with the RyR in the terminal cisternae and with the IP3 receptor (IP3R) in the longitudinal sarcoplasmic reticulum.
Function of triadin: overexpression or knock out/knock down experiments
In order to identify the function of triadin, a number of experiments were performed, based on the modification of triadin expression level, followed by the evaluation of the resulting calcium release by RyR.
First, triadin (Trisk 95) was overexpressed in primary culture of rat skeletal myotubes, using adenovirus gene transfer, which resulted in a blocking of depolarization induced calcium release (Smida Rezgui et al. 2005).
On the other hand, experiments were performed on a calcium release complex completely or partially depleted of triadin. Mutant forms of RyR1 containing deletions in the triadin interacting domains (identified by Lee et al. 2004) were expressed in dyspedic myotubes, hence leading to suppression of triadin from the calcium release complex. In these myotubes, electrical depolarization was unable to induce calcium release from sarcoplasmic reticulum (Goonasekera et al. 2007), probably because of a reduction in the amplitude and kinetics of the calcium release. Similar results were obtained by reducing triadin expression with siRNA in C2C12 myotubes (Wang et al. 2009). Lowering the amount of triadin resulted in a partial inhibition of the depolarization induced calcium release. The total deletion of triadin from the calcium release complex was performed by the production of a triadin KO mouse (Shen et al. 2007). These experiments are more difficult to analyse, as compensatory reduction in calsequestrin expression is observed in the muscles of these mice. Nevertheless, in addition to the lower amount of stored calcium, a reduction in the depolarization induced calcium release was observed in myotubes which presented no reduction in calsequestrin.
Therefore, the conclusion of all these experiments is that any modification in triadin expression levels, either by overexpression or by knock-out/knock-down, results in a common effect: reduction in depolarization-induced calcium release. Such apparent conflicting results underline the necessity of developing new approaches to understand triadin function.
Triadin expression in non-muscle cells
To get insight into the function of triadin, we decided to use a different approach in order to reveal the intrinsic properties of triadin in a system devoid of its usual muscle partners. We expressed triadin in a non-myogenic cell line, the COS-7 cells, and analysed both the endoplasmic reticulum (ER) structure and the microtubule network, using specific antibodies (Fauréet al. 2009). We observed that triadin induced drastic modifications of the ER structure, forming rope-like ER structures, associated with a massive reorganization of the microtubule network, the microtubules being bundled at the periphery of the cell. Such modifications were never observed when other ER-homing proteins, RyR for example, were overexpressed. In addition, these bundled microtubules are stable, resisting a depolymerization induced by nocodazole. Similar observations have already been made after overexpression of proteins involved in anchoring the ER to microtubules (Vedrenne & Hauri, 2006). These different experiments thus point to a possible new function of triadin. Triadin could act as a protein connecting the reticulum (or the calcium release complex) to the microtubule network, and could also be involved in the stabilization of microtubules in muscle cells. This function would be a major one, since microtubules are known to depolymerize under high Ca2+ concentration. If microtubules are indeed involved in triad organization, their depolymerization during each contraction would probably be dramatic. If triadin is involved in this structural function, it is probably not the only protein responsible for this anchoring/stabilization role, because triadin knock-out mice muscles do not exhibit massive triad disorganization. Nevertheless, 30% of the triads show an abnormal orientation in the muscle of these triadin KO animals (Shen et al. 2007), and this proportion is confirmed in a triadin null mouse line we have developed in our team (unpublished data). This amount of misoriented triads was reproducibly observed in two different mouse lines, leading to a strong confirmation of the possible involvement of triadin in maintaining the triad structure. Therefore, these studies in a non-muscle cell line have allowed us to unmask a specific intrinsic property of triadin, which is more difficult to visualize in muscle cells, perhaps because of other muscle proteins partially sharing similar function with triadin.
Conclusion
We have observed that triadin expression in COS-7 cells induces drastic modification in the organisation of the endoplasmic reticulum and microtubules. Therefore our current hypothesis is that triadin could be involved in the sarcoplamic reticulum structure in skeletal muscle, and could perform a regulatory function on RyR through this structural function. The overexpression as well as a down-regulation of triadin in skeletal muscle cells would result in local sarcoplasmic reticulum disorganization, thereby inducing an uncoupling between RyR and the facing DHPR. Nevertheless, additional studies need to be performed to confirm this hypothesis in skeletal muscle.
Acknowledgments
The work of our team was supported by grants from Association Française contre les Myopathies (AFM) and GIS-Maladies Rares.
References
- Caswell AH, Brandt NR, Brunschwig JP, Purkerson S. Localization and partial characterization of the oligomeric disulfide-linked molecular weight 95,000 protein (triadin) which binds the ryanodine and dihydropyridine receptors in skeletal muscle triadic vesicles. Biochemistry. 1991;30:7507–7513. doi: 10.1021/bi00244a020. [DOI] [PubMed] [Google Scholar]
- Fauré J, Fourest-Lieuvin A, Oddoux S, Pernet-Gallay K, Brocard J, Marty I. Triadin function in sarcoplasmic reticulum structure? Abstract at the 53th meeting of the Biophysical Society
- Goonasekera SA, Beard NA, Groom L, Kimura T, Lyfenko AD, Rosenfeld A, Marty I, Dulhunty AF, Dirksen RT. Triadin binding to the C-Terminal luminal loop of the ryanodine receptor is important for skeletal muscle excitation–contraction coupling. J Gen Physiol. 2007;130:365–378. doi: 10.1085/jgp.200709790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Jorgensen AO, Jones LR, Campbell KP. Biochemical characterization and molecular cloning of cardiac triadin. J Biol Chem. 1996;271:458–465. doi: 10.1074/jbc.271.1.458. [DOI] [PubMed] [Google Scholar]
- Hong CS, Ji JH, Kim JP, Jung DH, Kim DH. Molecular cloning and characterization of mouse cardiac triadin isoforms. Gene. 2001;278:193–199. doi: 10.1016/s0378-1119(01)00718-1. [DOI] [PubMed] [Google Scholar]
- Kim KC, Caswell AH, Talvenheimo JA, Brandt NR. Isolation of a terminal cisterna protein which may link the dihydropyridine receptor to the junctional foot protein in skeletal muscle. Biochemistry. 1990;29:9281–9289. doi: 10.1021/bi00491a025. [DOI] [PubMed] [Google Scholar]
- Kobayashi YM, Jones LR. Identification of triadin 1 as the predominant triadin isoform expressed in mammalian myocardium. J Biol Chem. 1999;274:28660–28668. doi: 10.1074/jbc.274.40.28660. [DOI] [PubMed] [Google Scholar]
- Lee JM, Rho SH, Shin DW, Cho C, Park WJ, Eom SH, Ma J, Kim DH. Negatively charged amino acids within the intraluminal loop of ryanodine receptor are involved in the interaction with triadin. J Biol Chem. 2004;279:6994–7000. doi: 10.1074/jbc.M312446200. [DOI] [PubMed] [Google Scholar]
- Marty I, Robert M, Villaz M, Lai Y, De Jongh KS, Catterall WA, Ronjat M. Biochemical evidence for a complex involving dihydropyridine receptor and ryanodine receptor in triad junctions of skeletal muscle. Proc Natl Acad Sci U S A. 1994;91:2270–2274. doi: 10.1073/pnas.91.6.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty I, Thevenon D, Scotto C, Groh S, Sainnier S, Robert M, Grunwald D, Villaz M. Cloning and characterization of a new isoform of skeletal muscle triadin. J Biol Chem. 2000;275:8206–8212. doi: 10.1074/jbc.275.11.8206. [DOI] [PubMed] [Google Scholar]
- Peng M, Fan H, Kirley TL, Caswell AH, Schwartz A. Structural diversity of triadin in skeletal muscle and evidence of its existence in heart. FEBS Lett. 1994;348:17–20. doi: 10.1016/0014-5793(94)00556-7. [DOI] [PubMed] [Google Scholar]
- Shen X, Franzini-Armstrong C, Lopez JR, Jones LR, Kobayashi YM, Wang Y, Kerrick WG, Caswell AH, Potter JD, Miller T, Allen PD, Perez CF. Triadins modulate intracellular Ca2+ homeostasis but are not essential for excitation–contraction coupling in skeletal muscle. J Biol Chem. 2007;282:37864–37874. doi: 10.1074/jbc.M705702200. [DOI] [PubMed] [Google Scholar]
- Smida Rezgui S, Vassilopoulos S, Brocard J, Platel JC, Bouron A, Arnoult C, Oddoux S, Garcia L, De Waard M, Marty I. Triadin (TRISK 95) over-expression blocks excitation-contraction coupling in rat skeletal myotubes. J Biol Chem. 2005;280:39302–39308. doi: 10.1074/jbc.M506566200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilopoulos S, Thevenon D, Smida Rezgui S, Brocard J, Chapel A, Lacampagne A, Lunardi J, DeWaard M, Marty I. Triadins are not triad-specific proteins: two new skeletal muscle triadins possibly involved in the architecture of sarcoplasmic reticulum. J Biol Chem. 2005;280:28601–28609. doi: 10.1074/jbc.M501484200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedrenne C, Hauri HP. Morphogenesis of the endoplasmic reticulum: beyond active membrane expansion. Traffic. 2006;7:639–646. doi: 10.1111/j.1600-0854.2006.00419.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li X, Duan H, Fulton TR, Eu JP, Meissner G. Altered stored calcium release in skeletal myotubes deficient of triadin and junctin. Cell Calcium. 2009;45:29–37. doi: 10.1016/j.ceca.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]