Abstract
The transcriptional repressor E2F4 is important for cell cycle exit and terminal differentiation in epithelial cells, neuronal cells and adipocytes but its role in T lymphocytes proliferation and memory formation is not known. Herein, we investigated the function of E2F4 protein for the formation of functional murine memory T cells. Murine transgenic CD8+ T cells were infected in vitro with lentivirus vector expressing a shRNA targeted against E2F4 followed by in vitro stimulation with SIINFEKL antigenic peptide. For in vivo assays, transduced cells were injected into congenic mice which were then infected with HSV-OVA. The primary response, memory formation and secondary stimulation were determined for CD8+ lentivirus transduced cells. In the absence of E2F4 cell cycle repressor, activated CD8+ T cells underwent intensive proliferation in vitro and in vivo. These cells had the ability to differentiate into memory cells in vivo, but they were defective in recall proliferation. We show that transient suppression of E2F4 during CD8+ T cell priming enhances primary proliferation and has a negative effect on secondary stimulation. These findings demonstrate that the cell cycle repressor E2F4 is essential for the formation of functional memory T cells. A decrease in CD8+ T-lymphocyte compartment would diminish our capacity to control viral infections.
Keywords: cell cycle, E2F4, CD8+ T cells, lentivirus
Introduction
Activated CD8+ T cells normally undergo many cycles of proliferation before differentiation into cytotoxic T lymphocytes (CTL). CTL deliver host protection through both cytokine synthesis and cytotoxic killing of antigen-expressing targets. At the end of the primary response, the majority of these CTL undergo apoptosis but a subset persists as memory cells. Memory cells appear to be derived mostly from cycling blast cells, which has given rise to the so-called “linear model” of CTL differentiation [1]. It follows from this model that more primary proliferation is always good for memory CD8+ T cell formation.
The link between antigen-driven proliferation and maturation to a memory phenotype has recently been questioned. First, long-lived CD8+ T cells that have undergone spontaneous proliferation in antigen-free hosts acquire some of the markers and properties of memory cells [2]. Second, during the immune response to the bacterium Listeria monocytogenes, H3M2- (non-classical MHC class I)-restricted CTL can form memory T cells without proliferation [3]. Memory-like activity may also be induced by the brief engagement of the T cell receptor, insufficient to cause clonal proliferation [4]. This argues that proliferation may not always be strictly necessary for the development of memory T cells.
The progression of cells through the G1 phase and into the S phase of the cycle operates through the phosphorylation of the pocket proteins (p130, p107 and pRb). In the resting state, two inhibitory transcription factors, E2F4 and E2F5, are complexed with a DNA-binding protein (DP) and a hypo-phosphorylated pocket protein, either p130 or p107. The activity of cyclin-dependent kinases (cdk) phosphorylates the pocket proteins, releasing the E2F4 or E2F5 and the DP. These molecules harbor nuclear export sequences, and are thus expelled from the nucleus, to be replaced by the activating factors E2F1, E2F2 and E2F3 [5]. The decision of an individual cell to enter S phase emerges from a titration of E2F4/5-containing complexes against E2F1/2/3 containing complexes. Quiescent cells exhibit increased levels of E2F4 [6] while actively cycling cells show low levels of E2F4 protein [7]. Overexpression of E2F4 induces growth arrest and apoptosis in mammalian cells [8].
Besides cell cycle, E2F4 is also involved in cell differentiation. We have recently reported that E2F4 is required for early lymphoid lineage cell differentiation [9]. However, the role of E2F4 in naïve and mature T cell proliferation and function is unknown. Herein, we have investigated the effect of E2F4 absence on naïve CD8+ T cells division, memory formation and secondary proliferation.
Our findings reveal that E2F4 protein is essential for cell cycle exit of primary CD8+ T cell blasts and for the formation of normal, functional memory CD8+ T cells. Thus, in T cells E2F4 has a dual role, acting both in the regulation of proliferation and in differentiation.
Materials and methods
Mice and cell culture
Mice of C57BL/6J strain (CD45.2) and the congenic B6.SJL-Ptprca Pep3b/BoyJ (termed B6.CD45.1) mice were obtained from The Jackson Laboratory (Bar Harbor, ME). OT-1 mice express a transgenic TCR that recognizes the 8-mer SIINFEKL peptide derived from residues 257–264 of OVA. A colony of OT-1 mice was maintained on the B6.CD45.1-expressing background and was bred in house. Mice were maintained in the specific pathogen-free environment in compliance with institutional guidelines for animal care. OT1 T cells were purified using a CD8+ T cell isolation kit (Miltenyi Biotec, Auburn, CA), from spleens and lymph nodes of 6~8 week old mice, after the depletion of red blood cells using Lympholyte® -M (Cedarlane Laboratories, Ontario, Canada) centrifugation. The purity of OT1 cells was > 95% (as determined by CD8 and CD45.1 surface staining). For in vitro experiments, OT1 cells were cultured in RPMI1640 cell growth medium (Invitrogen, Carlsbad, CA) containing 10% FBS (Invitrogen), 2 mM L-Glutamine (Invitrogen), 50 μM β-Mercaptoethanol (Sigma-Aldrich, St. Louis, MO), 2 μg/ml penicillin, 2 μg/ml streptomycin. To transduce naïve T cell by Lentiviral vectors, cells were cultured in the above medium with 15 ng/ml murine recombinant IL-7 (Endogen).
Construction of lentivirus vectors for expression of RNA short hairpin
Block-it™ U6 RNAi entry vector kit was purchased from Invitrogen. Single stranded oligonucleotides against E2F4 or Lac-Z (LAZ) mRNA were designed using Block-it ™ RNAi software and synthesized by Invitrogen. Briefly, the single-stranded oligos were annealed to create double strand oligos in vitro in annealing buffer according to the manufacturer’s protocol. The double strand oligos were ligated into pENTR™/U6 vector to generate the short hairpin-expressing entry vectors, which were used to screen for effective short hairpin RNA by transient transfection of NIH 3T3 cells. To build a cellular marker for tracking of T cells, DsRed cDNA were amplified from pCMV-DsRed-Express vector (BD Biosciences) using PCR. The primer sequences were:
Forward primer: GTCCCCGCGGGACCGTATTACCGCCATGCATTAG.
Reverse primer: TGGGGTACCCCGCTACAGGAACAGGTGGTGG.
A 952bp PCR product was digested by SacII and KpnI and ligated into pLenti6/BLOCK-iT™ –DEST vector, which was digested using the same two enzyme to generate the destination Lentivirus vector for SH-RNA targeting either E2F4 or LAZ. Finally, the tested U6 entry vectors were recombined with the destination vector to generate the Lentivirus vector expressing short hairpin RNA, by LR clonase™ reaction performed according to the manufacturer’s direction (Invitrogen). The sequences of single-stranded oligonucleotides for E2F4 were:
Top strand: 5′-CACCGCCAGAAACGGCGGATCTACGTTCAAGAG ACGTAGATCCGCCGTTTCTGGC.
Bottom strand: 5′-AAAAGCCAGAAACGGCGGATCTACGTCTCTTGAACGTAGATCCGCCGTTTCTGGC.
Oligos for LAZ
Top strand: 5′-CACCGCTACACAAATCAGCGATTTCGAAAAATCGCTGATTTGTGTAG.
Bottom strand: 5′-AAACTACACAAATCAGCGATTTTTCGAAATCGCTGATTTGTGTAGC.
Generation of Lentivirus
293FT cells (Invitrogen) were transfected with DNA mixture containing E2F4-SH or LAZ-SH-expressing Lentivirus vector and the virus packaging vectors (Invitrogen) using lipofectamine 2000 (Invitrogen). After 48 hours, the virus supernatant was harvested and filtered through a 0.48 μM filter. Virus was concentrated by ultracentrifugation at 22,000 rpm for 2 hours using Sorvall SW28 rotor, and re-suspended in 200 μl culture medium. Virus stock was frozen at −80°C. Virus titer was determined by transduction of NIH 3T3 cells.
Lentivirus transduction of T cells
OT1 T cells were cultured in RPM1640 medium with 15 ng/ml IL-7 for 4 days, washed once and 2 ×106 cells were re-plated into a 12 well plate in 1.8 ml culture medium containing 10 mg/ml polybrene (Sigma). 200 μl Lentivirus (MOI 20) was added and the plate was centrifuged at 1400 rpm for one hour. Cells were incubated at 37°C with 5% CO2 for 24–48 hours.
Generation of and use of HSV-1 amplicon expressing OVA
The HSV-OVA amplicon vector [10] and methods to package helper virus-free HSV amplicon stocks were previously described [11]. Viral pellets were resuspended in PBS and stored at −80°C until use. Vectors were titered as described previously [12]. OT1 (CD45.1) cells were injected into congenic C57BL/6 (CD45.2) mice by iv. Twenty-four hours later, animals were inoculated with 1 ×106 IFU HSV amplicon expressing OVA protein by intra-muscular (im) injection into the quadriceps femoris muscle.
RT-PCR for E2F4 mRNA
T cells were maintained in culture medium for four days and infected overnight using Lentivirus expressing SH-RNA targeting E2F4 or LAZ at MOI 20. Cells were washed, then 2 × 106 cells were re-suspended in fresh media in a 12-well plate containing splenocytes pulsed with 1 μM of SIINFEKL peptide. After 48 hours, both DsRed positive and negative populations were sorted using a FACS Aria (Becton-Dickinson). RNA from sorted cells was extracted using Trizol reagent (Invitrogen) and quantified. Reverse transcription was performed using SuperScript™ First-Strand Synthesis System (Invitrogen) for RT-PCR.
PCR amplification was carried out using the following conditions: 94 °C denaturation for 3 minutes for 1 cycle, 94°C 1 min, 58°C 1 min, 72°C 1 min, for 35 cycles. PCR products were confirmed on 1.5% agarose gel and detected with ethidium bromide staining. Primers for E2F4 cDNA: Forward: 5′-ACTCAAGTCTCAGAGGTGGC Reverse: 5′-GCAGCTCGGAGCTCATGCA. Primers for β-actin PCR amplification were obtained from Promega.
Western Blotting
Lentivirus vector-transduced NIH 3T3 cells were lysed in buffer containing 0.5% NP40, 100 mM NaCl, 50 mM Tris-HCl (pH 7.4) and protease inhibitor cocktail (Calbiochem Inc, San Diego, CA) by 3 cycles of free-thaw. Cell lysates were resolved on a 10% SDS-PAGE gel, transferred to Hybond™ ECL™ (Amersham Pharmacia Biotech, Piscataway, NJ) nitrocellulose membranes and blocked with TBS containing 5% dry milk. Immunoblots were probed overnight with primary antibodies in TBS with 0.05% Tween 20, containing 5% dry milk, and washed 3 times, then incubated with peroxidase conjugated goat anti-rabbit for 1 hour. The blots were washed 5 times and developed using ECL-PLUS reagent (Amersham Pharmacia Biotech) following the manufacturer’s protocol. Rabbit anti-E2F4 was purchased from Santacruz (Santa Cruz, Ca) and anti-β-actin antibody was obtained from Sigma-Aldrich.
Flow cytometry
For cell surface staining, 1 × 106 cells were incubated with antibodies in 500 μl PBS containing 5% FBS for 30 min on ice in the dark. Fluorescein isothiocyanate (FITC) conjugated anti-Mouse CD127, CD62L and CD44 and Peridinin chlorophyll- α protein (PerCp) conjugated anti-CD8 (53-6.7) were all obtained from Pharmingen. Anti-CD45.1-allophycocyanin (APC) was from E Biosciences (San Diego, CA).
For determining cell proliferation, T cells were labeled using 5 μM carboxyfluorescein diacetate, succinimidyl ester (CFSE) for 10 minutes at 37°C, followed by 2 washes in Hank’s balanced salt solution.
Intracellular staining of IFN-γ was detected as follows: lymphoid and spleen cells were cultured in media containing 1 μM GolgiPlug (BD Biosciences), in the presence or absence of antigen (1 μM SIINFEKL). After 6 hours, cells were washed and stained for CD45.1 surface marker, then fixed, permeabilized using the Cytofix/Cytoperm kit (BD Pharmingen) and subjected to intracellular staining for IFN-γ using a rat anti-mouse antibody (BD Biosciences).
Fluorescence was detected using a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA).
Statistical analysis
At least 3 independent experiments were performed for each figure panel. Error bars represent the SD of the mean (n=3). The significance of differences in the frequency of OT1 T cells between different groups of animals was evaluated using a two-tailed unpaired t-test.
Results
Lentivirus vector expressing shRNA effectively suppressed E2F4
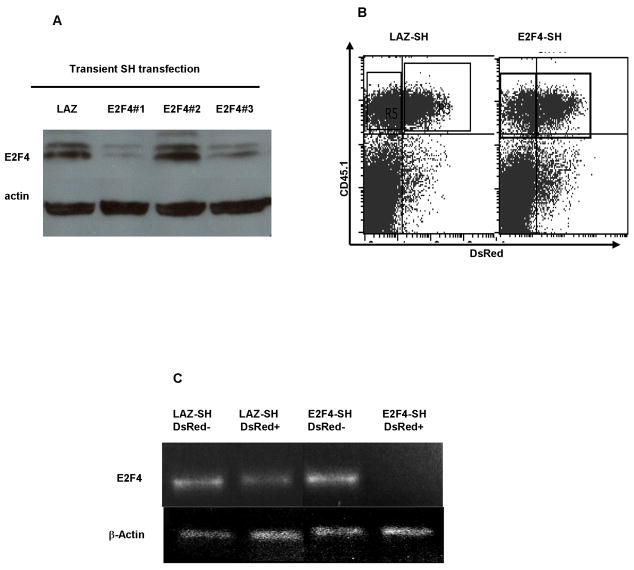
In order to investigate the function of E2F4 in T cell activation, proliferation and effector function, we employed short hairpin RNA to interfere with the stability of murine E2F4 mRNA. To validate this approach, the effect of these short hairpins was first tested in NIH-3T3 cells using transient transfection. Among three such constructs, two (E2F4-SH1 and E2F4-SH3) significantly reduced the E2F4 protein level, as detected by Western blotting in NIH-3T3 cells (Figure 1A).
Figure 1. Depletion of E2F-4 using short-hairpin (SH) siRNA.
(A) Three siRNA SH constructs, versus a control SH targeting β˜-galactosidase (LAZ), were transiently transfected into NIH-3T3 cells. 24h after transfection cells were harvested and cell lysates were prepared. Lanes were loaded with equal quantities of total protein, and E2F4 was detected by Western blotting. The E2F4-SH#1 and E2F4-SH#3 constructs depleted E2F4. (B) Activated CD8+ T cells were infected with Lentivirus vector expressing E2F4-SH1 and also DsRed as a marker of infection. 48h after SIINFEKL activation, cells were surface stained for CD45.1 and analysed by Flow cytometry. (C) 48h after lentivirus infection DsRed-expressing cells were FACS-sorted and examined by RT-PCR for the presence of E2F4 mRNA. The E2F4-SH transduced cells had lost E2F4 RNA, while control LAZ-SH transduced cells retained E2F4 RNA. This shows that SH-mediated siRNA is an effective tool to test the function of E2F4. The E2F4-SH1 was used in the subsequent experiments.
To test the efficiency of the short hairpin in blocking the expression of E2F4 mRNA in primary T cells, we generated Lentivirus vectors expressing the E2F4-SH1 construct. This vector also expressed a DsRed marker, which we used to compare the titer of batches of virus, and to positively identify transduced T cells. T cell receptor transgenic OT1 T cells on a CD45.1 background were purified and cultured in the presence of IL-7 for 4 days, and then the cells were washed and transduced with Lentivirus vector overnight. The OT1 cells were then washed and re-suspended in culture medium containing splenocytes from B6 mice pulsed with the ovalbumin-derived SIINFEKL peptide, for which OT1 cells are specific. After a further 48 hours, OT1 cells were sorted for CD45.1 and DsRed expression (Figure 1B). Analysis using RT-PCR showed that in DsRed positive, Lentivirus-infected cells, the E2F4 mRNA level decreased dramatically compared with non-transduced cells. In contrast, the DsRed-positive T cells infected with a Lentivirus expressing a short hairpin targeting β-galactosidase (LAZ) retained normal E2F4 mRNA levels. These data showed that Lentiviral vector-mediated RNA interference targeting E2F4 mRNA was an effective tool for studying the function of this molecule in primary T cells.
Increased proliferation of OT1 cells lacking E2F4 in vitro
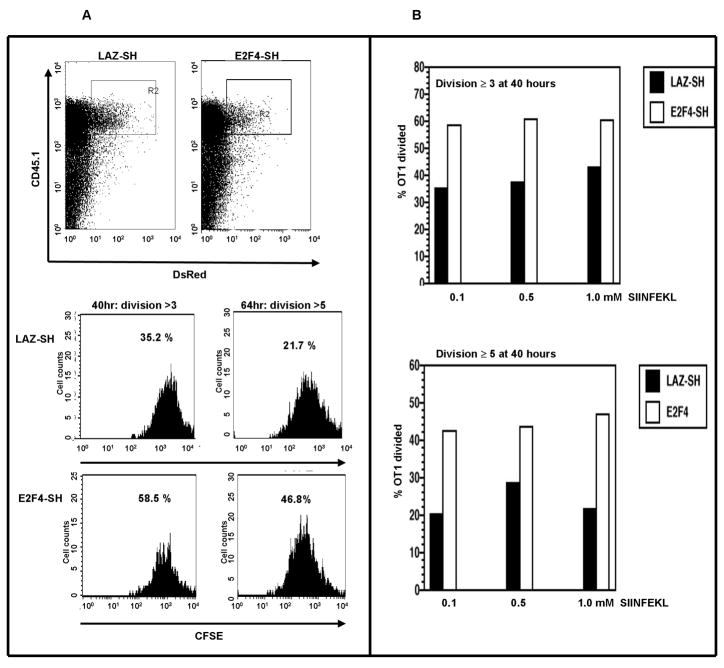
To test whether E2F4 plays a role in T cell proliferation, OT1 cells were labeled with the cytophilic dye CFSE, and transduced with Lentivirus vector expressing either short hairpin targeted against RNA encoding E2F4 (E2F4-SH) or against LAZ (LAZ-SH) as a control. Cells were activated with splenocytes from B6 mice in the presence of SIINFEKL antigenic peptide, and harvested at various time points. Cell division of OT1 cells was analyzed by gating on the CD45.1+, DsRed+ cells (Figure 2A). OT1 cells transduced with vector expressing E2F4-SH had undergone more cell divisions, compared with LAZ-SH transduced control cells. At 40 hours post-activation, 58.5% of E2F4-SH transduced OT1 cells had divided more than 3 times, while only 35.2% of the LAZ-SH transduced control cells have gone through 3 cell divisions over the same time interval. A similar difference was evident in cells that had divided 5 or more times by 64 hours. The data from multiple experiments with different concentrations of SIINFEKL peptide support this conclusion (Figure 2B), indicating the role of E2F4 in restricting the proliferation of OT1 T cells.
Figure 2. The effect of E2F4 depletion on T cell proliferation in vitro.
(A) Proliferating T cells, labeled with CFSE, were infected with Lentivirus vectors encoding either LAZ-SH or E2F4-SH. At 40 and 64 hours, the frequency of T cells that had divided more than 3 and 5 times respectively was increased in the E2F4-SH transduced cells. (B) These effects were independent of the concentration of antigenic peptide.
Enhanced proliferation of OT1 T cells in primary responses in vivo
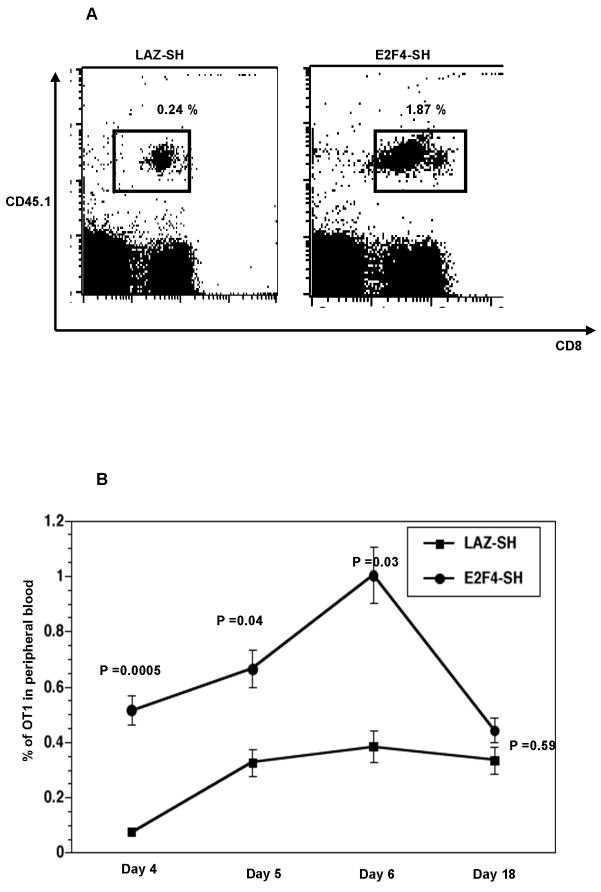
In order to investigate the consequence of loss of E2F4 in T cells in vivo, 2×106purified OT1 CD8+ T cells (CD45.1) were transduced with either Lentivirus vector expressing E2F4-SH or the LAZ-SH control for 24h and injected into congenic C57BL/6 (CD45.2) mice. Twenty-four hours later, these mice were immunized by IM injection with 1 × 106 IFU of a Herpes simplex virus type 1 (HSV-1) based amplicon vector expressing the entire ovalbumin protein (HSV-OVA). Clonal expansion of the OT1 cells was observed in peripheral blood from immunized animals based on staining for CD8 and CD45.1 (Figure 3A). In our experiments DsRed intensity was decreasing during the culture period in vitro and was not evident during or after an immune response in vivo; thus we were unable to use DsRed expression post-hoc to identify the transduced cells. We therefore identified the OT1 T cells based on their CD45.1+ expression. Even if only a subset of OT1 cells were transduced, these cells had a dramatically increased proliferation that accounted for the increased abundance in the blood. As expected, OT1 cells infected with the LAZ-SH control Lentivirus responded to the antigen, with a peak in the frequency of OT-1 T cells at day 6. The OT-1 cells transduced with E2F4-SH vector showed a greatly enhanced expansion compared to the control cells (Figure 3B), with the greatest effect (around 5 fold) at the earliest time-points (days 4), but sustained and statistically significant (ie P<0.05) through the peak of the response at day 6. Importantly, we did not observe any significant difference in the longer-term survival of T cells from these two groups; the percentage of OT1 cells in the blood dropped to around 0.4% after 18 days, suggesting that E2F4 does not influence the ability of T cells to survive the acute death of activated cells at the end of primary responses.
Figure 3.
In vivo clonal expansion of control LAZ-SH-transduced versus E2F4-SH-transduced T cells. (A) T cell receptor transgenic OT1 T cells on a CD45.1 background were transduced in vitro using Lentivirus vectors for 48h then transferred to C57BL/6 (CD45.2) mice. 24h later these mice were injected with HSV-based amplicon encoding ovalbumin. On day 6 OT1 T cells were identified among peripheral blood lymphocytes by CD45.1 and CD8 staining. (B) Those OT1 T cells that were transduced with the E2F4-SH vector exhibited increased clonal expansion in vivo. The increase was most profound early in the response; by day 18 there was no significant effect on circulating OT1 T cell frequency.
E2F4-SH lentivirus transduced OT1 T cells E2F4 become memory cells
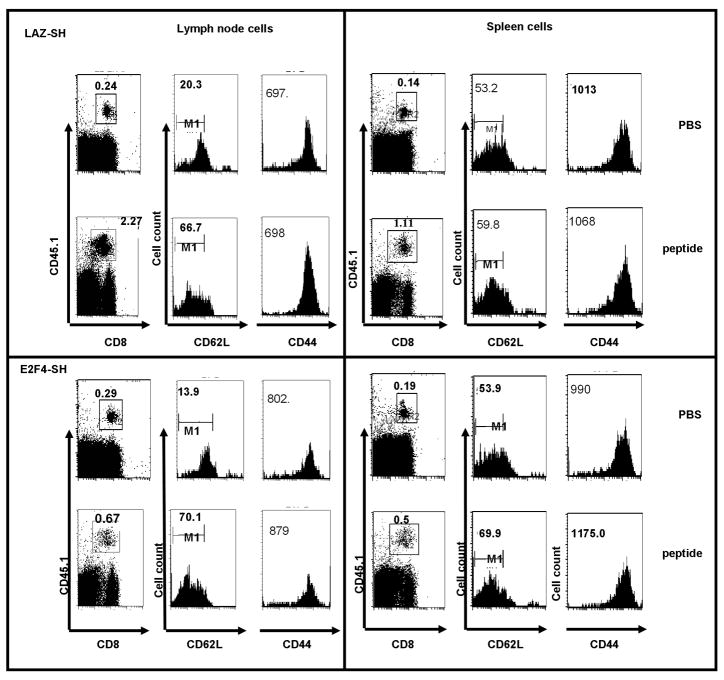
We tested whether the increased expansion of OT1 T cells in the primary response would yield any consequences during memory formation and the secondary response. Thus, lentivirus transduced OT1 cells (CD45.1) were adoptively transferred into congenic C57BL/6 (CD45.2) mice and these animals were immunized with HSV-OVA as above. After 10 weeks, animals were re-challenged once with the specific antigenic peptide, SIINFEKL, or with PBS as a control. For secondary stimulation we used OVA soluble peptide instead of HSV-OVA amplicon since we could expect this antigenic challenge to sample all memory OT1 T cells in the organism. Lymph nodes and spleens were harvested on day 5 following SIINFEKL challenge. HSV-OVA primed T cells, transduced with either E2F4-SH Lentivirus or control LAZ-SH, formed long-lived memory cells that were present at 10 weeks. These cells expressed CD62L and CD44 (Figure 4, second and third column of each panel). In lymph nodes, OT1 cells infected with either E2F4-SH or LAZ-SH primarily expressed a high level of CD62L and of CD44, the typical “central memory” phenotype. Cells in the spleen included more CD62L-low but CD44-high cells. Cells were also stained for CD127, the IL-7Rα chain. Naïve T cells express this receptor at a high level, but lose this marker when they are activated during the primary response; the receptor returns on memory T cells [13]. Figure 5 (first and third row of panels) illustrates that OT1 memory T cells from both the E2F4-SH and the LAZ-SH groups expressed high CD127.
Figure 4. Cell surface markers on long-lived OT1 T cells following in vivo priming with HSV-OVA amplicons.
Lymph node OT1 T cells were mostly CD62L-high and CD44-high, and down-regulated CD62L upon in vivo challenge with the SIINFEKL peptide (5 days after challenge). Spleen OT1 cells were CD44-high but showed a broad spectrum of CD62L expression, and this did not change substantially after challenge. There was no difference between E2F4-SH transduced and control LAZ-SH transduced OT1 cells in terms of these markers.
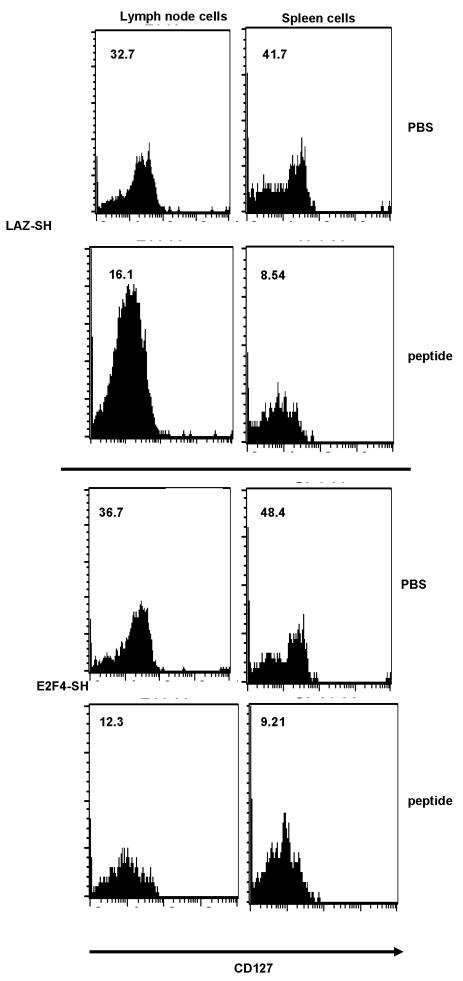
Figure 5. Expression of CD127 on long-lived OT1 cells after priming in vivo with HSV-OVA amplicons.
Both lymph node and spleen OT1 cells expressed CD127, and down-regulated its expression upon challenge (5 days after challenge). Numbers are the MFI for each fluorescence histogram. There was no difference between E2F4-SH transduced and control LAZ-SH transduced OT1 cells in terms of CD127 expression.
Upon re-challenge with OVA peptide, lymph node and spleen OT1 T cells from both the E2F4-SH and the LAZ-SH groups down-regulated both CD62L and CD127 (Figures 4 second column of each panel and figure 5, second and fourth row of panels), indicating a normal recall response. There was no significant difference between the groups in terms of the changes in expression of activation markers, indicating that memory T cells resulting from naive cells expressing normal E2F4, or lacking E2F4, were both capable of reacting to the secondary challenge. We conclude that the lack of E2F4 during priming did not impair the differentiation of the CD8+ T cells into long-lived, antigen-responsive cells.
Defect in clonal expansion of memory T cells that lacked E2F4 during priming
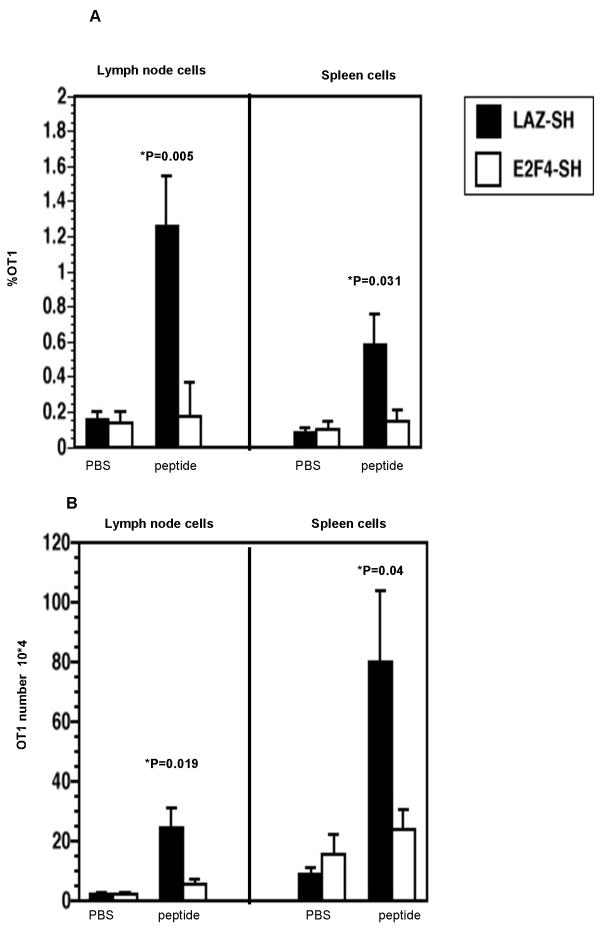
A normal memory CD8+ T cell response involves secondary proliferation, and to test whether this aspect of memory cell function was intact, we recorded the secondary expansion of these cells in vivo. While memory OT1 cells transduced with LAZ-SH control vector proliferated in response to the re-challenge (Figure 6, filled bars), T cells infected with E2F4-SH virus showed a severe defect in clonal expansion (Figure 6, open bars). In lymph node, the percentage of control LAZ-SH OT1 cells expanded from a mean of 0.2% to 1.2% of total cells (Figure 6A, filled bars) while the cell number increased from less than 4 ×104 to 2.5 × 105 (Figure 6B, filled bars). In contrast, the E2F4-SH transduced OT1 cells did not show a significant increase in either OT1 cell number (Figure 6A, open bars) or percentage (Figure 6B, open bars), compared with the PBS-challenged group. The same phenomenon was observed in spleen (Figure 6A and 6B). These data show that OT1 cells lacking E2F4 during priming could not undergo an effective secondary expansion, indicating an indispensable role of this molecule in the formation of proliferation-competent memory T cells.
Figure 6.
E2F4-SH transduced, long-lived, in vivo primed OT1 cells showed a specific defect in secondary clonal expansion. Control LAZ-SH transduced OT1 cells (solid bars) showed significant clonal expansion in response to SIINFEKL peptide challenge, evident as an increase in cell frequency (A) and cell number (B). In contrast, E2F4-SH transduced OT1 cells (open bars) failed to undergo clonal expansion.
Memory T cells primed without E2F4 are functional
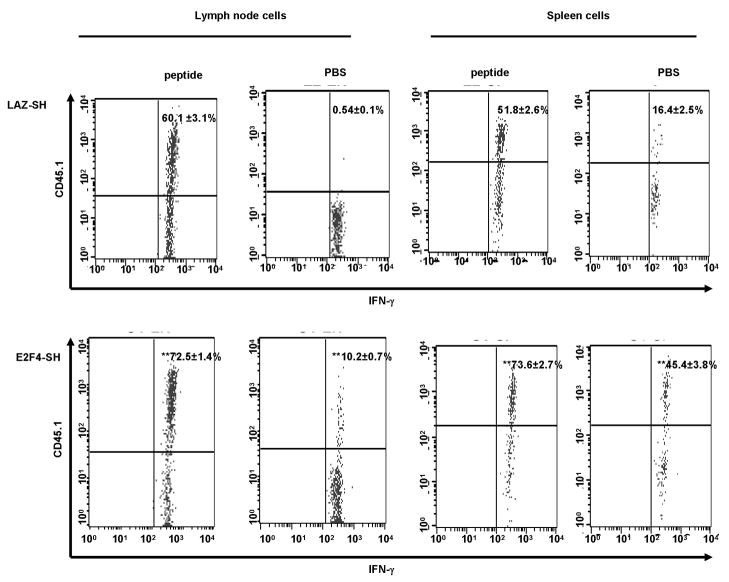
To test whether memory T cells derived from OT1 cells that lacked E2F4 during the priming phase could produce cytokine, we stained these cells for intracellular IFN-γ. Lymph node and spleen cells, taken 10 weeks after HSV-OVA immunization, were washed and plated with or without SIINFEKL peptide for 6 hours. Data in Figure 7 show that OT1 memory cells resulting from naïve cells transduced with either LAZ-SH control vector or the E2F4-SH vector were both capable of producing IFN-γ. We found that there is a significant difference in IFN-γ production when comparing peptide stimulated OT1 memory cells transduced with E2F4-SH (~72% produce IFN-γ; Figure 7, lower panel, first column) to OT1 memory cells transduced with LAZ-SH (~60% produce IFN-γ; Figure 7, upper panel, first column). The same pattern is found in OT1 memory cells in spleen (Figure 7, third column). The increase in the number of IFN-γ producing OT1 cells (E2F4-SH transduced) might compensate for the defect in the secondary clonal expansion, however our results show that there is an increase in the percentage of OT1 cells (E2F4-SH transduced) producing IFN-γ even in the absence of the peptide. Taken together, our data indicated that the E2F4 molecule plays a crucial role in controlling normal T cell proliferation during an immune reaction, and that the de-regulated primary expansion due to the lack of E2F4 generated memory cells which had lost the capacity for proliferation during a secondary response. However, E2F4 did not appear to play a role in priming these T cells for survival or for effector function.
Figure 7.
Long-lived, in vivo primed OT1 T cells from lymph nodes and spleen synthesized IFN-γ in response to peptide re-stimulation in vitro. 10 weeks after HSV-OVA immunization, LN and splenocytes were collected, stimulated with SIINFEKL peptide for 6 h followed by intracellular staining for IFN-γ. There is a significant difference in IFN-γ production when comparing OT1 memory cells transduced with E2F4 to those transduced with LAZ-SH in both LN (first and second column, compare upper to lower panel) and spleens (third and fourth column, compare upper to lower panel). The percentage of IFN-γ producing cells are plotted for each group with the corresponding SD (n=3 mice/group). Statistical analysis was performed using one-way ANOVA followed by post-hoc Tukey test. **, p < 0.001 (OT1 cells transduced with E2F4-SH compared to those transduced with LAZ-SH).
Discussion
It is very important to understand the relationship between T cell proliferation and memory T cell formation. If the linear model were strictly correct, measurements of CD8+ T cell proliferation in response to a pathogen or a vaccine would always predict the formation of protective memory T cells. In contrast, if excessive proliferation sometimes compromises the formation of T cell memory, we need to understand exactly how and why this occurs, so that analyses of experimental vaccines can aim for optimum proliferation, and so that we can develop better surrogate markers that predict the formation of protective memory.
In this study, we manipulated the cell proliferation machinery by suppressing the expression of E2F4. The approach was to “knock down” expression of E2F4 using a short-hairpin RNA, delivered using a Lentivirus vector in vitro, followed by adoptive transfer of the T cells to mice immunized with an HSV-based amplicon vector expressing our test antigen, the SIINFEKL peptide of ovalbumin. We prefer this approach rather than the use of gene-targeted E2F4-deficient mice because these mice harbor defects in several hematopoietic lineages, including T cells [14], and also have reduced viability. Employing the short-hairpin siRNA carried in a replication-defective Lentivirus vector, we limited the effect of E2F4 deficiency to the mature CD8+ T cells of interest, and were able to extinguish E2F4 in these cells. This method provides the advantage of the introduction and stable integration of the shRNA cassette into the host genome [15] thus the transgene cassettes introduced by the Lentivirus are inherited by the daughter cells during cell division. Although RNAi provides a rapid means for assessing the effects of the loss of gene function, this approach provokes in mammalian cells an antiviral/interferon response which results in the global shutdown of protein synthesis [16]. In order to avoid the activation of the interferon pathway the shRNA molecules targeting E2F4 and the LAZ control were composed of 19–22 nucleotides, which can induce gene silencing evading interferon response [17].
E2F4 loss resulted in the hyper-proliferation of CD8+ T cells at the time of initial activation, followed by the formation of long-lived memory T cells. These memory cells were able to recognize antigen upon secondary encounter, and were fully competent to synthesize IFN-γ. However, the cells were unable to undergo secondary proliferation.
Our results are not in agreement with previous data obtained in different cell types. Humbert et al [18] showed that E2F4-deficient MEFs (mouse embryonic fibroblasts) proliferated at the same rate as wild type controls in different culture conditions. They conclude that E2F4 is fully dispensable for the regulation of cell cycle arrest and proliferation. The different results might be the consequence of using different cell types and stimulation conditions. The MEFs were cultured in vitro and entered cell cycle in response to the reintroduction of serum, whereas in our experiment E2F4-deficient OT1 cells are activated via the T cell receptor, using the OVA peptide in vitro and in vivo. Another key difference is that our approach resulted in transient loss of E2F4 at the time of priming. As seen in our experiments, long-term culture of transformed cells leads to a significant reduction in the reporter gene intensity in both E2F4-deficient and control cells. The loss of DsRed expression signified loss of the co-expressed E2F4-SH. We would therefore conclude that we were testing memory responses in T cells that had been E2F4-deficient during priming, but not during the secondary response. This allowed us to draw conclusions about the role of E2F4 in programming for memory, without a superimposed effect on the function of the memory T cells.
Since E2F4 is ubiquitously expressed in cycling as well as in differentiated cells in the form of complexes with the pocket proteins pRb, p130 and p107, one would like to know whether the absence of E2F4 has any consequences for the expression of other members of the E2F family or on pocket proteins. In our adoptive transfer model, very low cell numbers made it difficult to assess the abundance of these proteins, since they are regulated at the protein level by protein-protein interactions and post-translational modifications, best revealed by immunoprecipitation and Western blotting. However, we are reassured that in E2F4-deficient ES cells, there were no changes in the expression level of other E2F family members or in the composition of E2F-pocket protein complexes [18]. Thus, we think our experiment is best interpreted as a consequence of an isolated, transient lack of E2F4 during CD8+ T cell priming, and as showing an effect of E2F-4 on programming of memory T cells.
Activation signals from an immunological synapse drive naïve cells from a resting G0 state into G1. This involves the synthesis of cyclins D, cdk4 and cdk6. Memory CD8+ T cells start from a different point, since they already express cyclins D, as well as cdk4 and cdk6 [19]. However, these cdk may be held in check by cdk inhibitors (CDKI) of the INK family, p16 and p18 [20]. In parallel with these events, synapse-associated signaling activates transcription factors that drive the IL-2 gene; the IL-2 acts as an autocrine/paracrine growth factor and causes the phosphorylation, ubiquitinization and breakdown of the CDKI, p27Kip1 [21]. This releases cdk activity, resulting in phosphorylation of pocket proteins and a shift from E2F4/5 mediated inhibition to E2F1/2/3 mediated transcriptional activation. Through a feed-forward mechanism, the E2F-responsive genes include cyclins E and A, which are active later in the cycle, complexed with cdk2. Extracellular signals can either promote or inhibit this process. For example, the anti-proliferative cytokine, Transforming Growth Factor β-1 (TGF-β1) promotes synthesis of the CDKI, p15INK4B [22, 23], which inhibit primarily cdk4 and cdk6.
The actions of E2F4 during cell differentiation are two-fold. First there is the effect on proliferation, and second a distinct effect on differentiation. Thus, in keratinocytes the E2F1 factor is found in the germinative layer of the skin, but E2F4 appears only in the differentiating layer [7]. The question arises as to whether the effects of E2F4 in promoting differentiation are simply an effect of its action in inhibiting cell cycle progression. In PC12 neuronal precursors cells, E2F4 promotes neurite outgrowth, and in this model, the effect is not accompanied by any change in cell proliferation [24]. Similarly, in adipocytes, E2F4 controls differentiation through pathways that do not control the cycle, and do not involve pocket proteins [25]. Thus, it appears that the actions of E2F4 during proliferation and differentiation may be separate.
We have shown that the suppression of E2F4 during T cell priming results in functionally impaired memory cells, but which of the two mechanisms of E2F4 action is likely to be important in memory T cell formation? The cells survive as long-lived cells expressing a memory-like phenotype and they are competent to synthesize IFN-γ, supporting the notion that their differentiation is intact. We therefore argue that E2F4 is most important in limiting the proliferation of CD8+ T cells during the primary response, and it is through this mechanism that E2F4 controls the function of CD8+ memory T cells.
The regulation of cell cycle progression during the priming event is likely to have evolved to optimize clonal expansion, and at the same time to place some limits on future expansion potential. By disabling one of the molecules that promotes cell cycle arrest, we revealed the importance of this mechanism in preserving the potential of CD8+ T cells to proliferate during a secondary response.
Acknowledgments
This work was supported by NIH grant AI050195 (to I.N.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Opferman JT, Ober BT, Ashton-Rickardt PG. Linear differentiation of cytotoxic effectors into memory T lymphocytes. Science. 1999:283. 1745–1748. doi: 10.1126/science.283.5408.1745. [DOI] [PubMed] [Google Scholar]
- 2.Kieper WC, Jameson SC. Homeostatic expansion and phenotypic conversion of naive T cells in response to self peptide/MHC ligands. Proc Natl Acad Sci. 1999:96. 13306–13311. doi: 10.1073/pnas.96.23.13306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerksiek KM, Ploss A, Leiner I, Busch DH, Pamer EG. H2-M3-restricted memory T cells: persistence and activation without expansion. J Immunol. 2003:170. 1862–1869. doi: 10.4049/jimmunol.170.4.1862. [DOI] [PubMed] [Google Scholar]
- 4.Munitic I, Ryan PE, Ashwell JD. T cells in G1 provide a memory-like response to secondary stimulation. J Immunol. 2005:174. 4010–4018. doi: 10.4049/jimmunol.174.7.4010. [DOI] [PubMed] [Google Scholar]
- 5.Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005:24. 2796–2809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- 6.Gaubatz S, Lees JA, Lindeman GJ, Livingston DM. E2F4 Is Exported from the Nucleus in a CRM1-Dependent Manner. Mol and Cel Biol. 2001:21. 1384–1392. doi: 10.1128/MCB.21.4.1384-1392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paramio JM, Segrelles C, Casanova ML, Jorcano JL. Opposite Functions for E2F1 and E2F4 in Human Epidermal Keratinocyte Differentiation. J Biol Chem. 2000:275. 41219–41226. doi: 10.1074/jbc.M004973200. [DOI] [PubMed] [Google Scholar]
- 8.Chang YC, Nakajima H, Illenye S, Lee YS, Honjo N, Makiyama T, Fujiwara I, Mizuta N, Sawai K, Saida K, Mitsui Y, Heintz NH, Magae J. Caspase-dependent apoptosis by ectopic expression of E2F-4. Oncogene. 2000:19. 4713–4720. doi: 10.1038/sj.onc.1203833. [DOI] [PubMed] [Google Scholar]
- 9.Enos ME, Bancos S, Bushnel T, Crispe IN. E2F4 Modulates Differentiation and Gene Expression in Hematopoietic Progenitor Cells during Commitment to the Lymphoid Lineage. J Immunol. 2008:180. 3699–3707. doi: 10.4049/jimmunol.180.6.3699. [DOI] [PubMed] [Google Scholar]
- 10.Willis RA, Bowers WJ, Turner MJ, Fisher TL, Abdul-Alim CS, Howard DF, Federoff HJ, Lord EM, Frelinger JG. Dendritic cells transduced with HSV-1 amplicons expressing prostate-specific antigen generate antitumor immunity in mice. Human Gene Ther. 2001:12. 1867–1879. doi: 10.1089/104303401753153929. [DOI] [PubMed] [Google Scholar]
- 11.Bowers WJ, Howard DF, Brooks AI, Halterman MW, Federoff HJ. Expression of vhs and VP16 during HSV-1 helper virus-free amplicon packaging enhances titers. Gene Ther. 2001:8. 111–120. doi: 10.1038/sj.gt.3301340. [DOI] [PubMed] [Google Scholar]
- 12.Bowers WJ, Howard DF, Federoff HJ. Discordance between expression genome transfer titering of HSV amplicon vectors: recommendation for standardized enumeration. Mol Ther. 2000:1. 294–299. doi: 10.1006/mthe.2000.0039. [DOI] [PubMed] [Google Scholar]
- 13.Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003:4. 1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 14.Rempel RE, Saenz-Robles MT, Storms R, Morham S, Ishida S, Engel A, Jakoi L, Melhem MF, Pipas JM, Smith C, Nevins JR. Loss of E2F4 activity leads to abnormal development of multiple cellular lineages. Mol Cell. 2000:6. 293–306. doi: 10.1016/s1097-2765(00)00030-7. [DOI] [PubMed] [Google Scholar]
- 15.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Zhang M, McManus MT, Gertler FB, Scott ML, Van Parijs L. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice. Nature Genet. 2003:33. 401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 16.McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nature Rev Genet. 2002:3. 737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 17.Reynolds A, Anderson EM, Vermeulen A, Fedorov Y, Robinson K, Leake D, Karpilow J, Marshall WS, Khvorova A. Induction of the interferon response by siRNA is cell type– and duplex length–dependent. RNA. 2006:12. 988–993. doi: 10.1261/rna.2340906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humbert PO, Rogers C, Ganiatsas S, Landsberg RL, Trimarchi JM, Dandapani S, Brugnara B, Erdman S, Schrenzel M, Bronson RT, Lees JA. E2F4 is essential for normal erythrocyte maturation and neonatal viability. Molecular Cell. 2000:6. 281–291. doi: 10.1016/s1097-2765(00)00029-0. [DOI] [PubMed] [Google Scholar]
- 19.Veiga-Fernandes H, Rocha B. High expression of active CDK6 in the cytoplasm of CD8 memory cells favors rapid division. Nat Immunol. 2004:5. 31–37. doi: 10.1038/ni1015. [DOI] [PubMed] [Google Scholar]
- 20.Latner DR, Kaech SM, Ahmed R. Enhanced expression of cell cycle regulatory genes in virus-specific memory CD8+ T cells. J Virol. 2004:78. 10953–10959. doi: 10.1128/JVI.78.20.10953-10959.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nourse J, Firpo JE, Flanagan WM, Coats S, Polyak K, Lee MH, Massague J, Crabtree GR, Roberts JM. Interleukin-2 mediated elimination of p27-kip-1 cyclin dependent kinase inhibitor prevented by rapamycin. Nature. 1994:372. 570–573. doi: 10.1038/372570a0. [DOI] [PubMed] [Google Scholar]
- 22.Li JM, Nichols MA, Chandrasekharan S, Xiong Y, Wang XF. Transforming growth factor beta activates the promoter of cyclin-dependent kinase inhibitor p15INK4B through an Sp1 consensus site. J Biol Chem. 1995:270. 26750–26753. doi: 10.1074/jbc.270.45.26750. [DOI] [PubMed] [Google Scholar]
- 23.Sandhu C, Garbe J, Bhattacharya N, Daksis J, Pan CH, Yaswen P, Koh J, Slingerland JM, Stampfer MR. Transforming growth factor beta stabilizes p15INK4B protein, increases p15INK4B-cdk4 complexes, and inhibits cyclin D1-cdk4 association in human mammary epithelial cells. Mol Cell Biol. 1997:17. 2458–2467. doi: 10.1128/mcb.17.5.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Persengiev SP, Kondova II, Kilpatrick DL. E2F4 actively promotes the initiation and maintenance of nerve growth factor-induced cell differentiation. Mol Cell Biol. 1999:19. 6048–6056. doi: 10.1128/mcb.19.9.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landsberg RL, Sero JE, Danielian PS, Yuan TL, Lee EY, Lees JA. The role of E2F4 in adipogenesis is independent of its cell cycle regulatory activity. Proc Natl Acad Sci. 2003:100. 2456–2461. doi: 10.1073/pnas.0138064100. [DOI] [PMC free article] [PubMed] [Google Scholar]