Abstract
Hemes are often incorporated into designed proteins. The importance of the heme ligand type and its orientation is still a matter of debate. Here, heme ligands and ligand orientation were investigated using a nonredundant (87 structures) and a redundant (1503 structures) set of structures to compare and contrast design features of natural b- and c-type heme-binding proteins. Histidine is the most common ligand. Marked differences in ligation motifs between b- and c-type hemes are higher occurrence of His-Met in c-type heme binding motifs (16.4% vs. 1.4%) and higher occurrence of exchangeable, small molecules in b-type heme binding motifs (67.6% vs. 9.9%). Histidine ligands that are part of the c-type CXXCH heme-binding motif show a distinct asymmetric distribution of orientation. They tend to point between either the heme propionates or between the NA and NB heme nitrogens. Molecular mechanics calculations show that this asymmetry is due to the bonded constraints of the covalent attachment between the heme and the protein. In contrast, the orientations of b-type hemes histidine ligands are found evenly distributed with no preference. Observed histidine heme ligand orientations show no dominating influence of electrostatic interactions between the heme propionates and the ligands. Furthermore, ligands in bis-His hemes are found more frequently perpendicular rather than parallel to each other. These correlations support energetic constraints on ligands that can be used in designing proteins.
Keywords: heme, pdb survey, ligand orientation, continuum electrostatics, molecular mechanics
INTRODUCTION
The heme cofactor is ubiquitous and essential for life.1 It is used as an electron transporter,2 plays major roles in molecule transport, storage, and detection,3–8 is often used to facilitate redox chemistry (e.g., for detoxification)6,9,10 and even takes part in the control of gene expression.6,8,11 The heme iron is coordinated by the four heme nitrogens, leaving two positions open for additional ligands. Hemes that are involved in electron transfer reactions generally have two axial ligands donated by the protein, whereas those involved in transport or catalysis have one ligand from the protein leaving one site free to be filled by substrate or water. Although hemes bound to proteins are essential for the function of a cell, free hemes can be toxic because of their ability to transfer absorbed light energy to oxygen, transforming it into singlet oxygen (for reviews see Refs. 12 and 13). Thus, hemes must be tightly bound to proteins during their synthesis, transport, function, and degradation.
The most common heme found in proteins is the b-type, an iron protoporphyrin IX noncovalently bound to the protein. It is thought that all heme derivates use b-type hemes as template.14,15 A common modification is attaching a b-type heme covalently to the protein forming thereby a c-type heme.16 The covalent bonds made between the heme vinyls at the position 8 and 13 of the porphyrin macrocycle and two specific cysteine residues form a classical Cys-Xaa-Xaa-Cys-His (CXXCH) c-type heme-binding motif. Here the histidine acts as one of the heme ligands and XX are two arbitrary amino acids other than Cys.17 Other modifications of b-type hemes lead to a-, o-, and d-type hemes,14 which are found, for example, in the terminal oxidases of bacteria and eukaryotes.
Proteins can provide axial ligands to the heme via groups that contain oxygen, nitrogen, or sulfur atoms such as amino acid side chains or peptide chain termini. Each group has a different heme affinity,18,19 which will modify the resultant stability of the folded cofactor bound protein.20 In addition, each ligand will have a different affinity for the oxidized and reduced heme, which will shift the resultant in situ heme electrochemistry.20
Several surveys of heme-binding proteins taken from the Protein Data Bank21 have examined ligand orientation,22 binding motifs,23–25 and electrochemistry.26 Some of those survey results can be accessed through Internet databases.26–28 However, none of these use a nonredundant dataset, which is needed to obtain statistically relevant comparisons. This is especially true for heme proteins where some protein families are highly over-represented in the structural databank.29
One open question in understanding heme-binding motifs is which factors determine the histidine heme ligand orientation and are therefore responsible for ligand orientation dependent physical chemical properties of the heme.30,31 Possibilities include heme ruffling,32,33 the ligand-heme orbital interactions which distinguish between relative parallel and perpendicular orientations of two heme ligands,31,34 the heme propionate ionization state,22,35 and hydrogen bonds from the protein to the heme ligating histidine.36,37 These factors have been examined using different techniques, such as site directed mutagenesis,37 EPR, NMR, and Mössbauer spectroscopy,38–40 classical40 and quantum mechanic calculations,41–43 and the synthesis of model heme complexes.40,44 However, until now there has been no clear ranking of which factors contribute most to the histidine ligand orientation which could provide guidelines in bioinspired protein design.
The survey reported here uses a nonredundant dataset of b- and c-type heme-binding proteins. The b- and c-type hemes are found to have different propensities for different ligation motifs and for the orientations of the histidine heme ligands relative to the heme plane. Electrostatic and molecular mechanics calculations are used to explain why certain histidine ligand orientations are never found. Additionally, the use of a nonredundant dataset allows the estimation of the importance of various properties that may influence heme ligation using a knowledge-based approach.45–47
METHODS
Protein data sets
A redundant set of 1503 heme proteins was created in March 2007 using the RCSB protein database search facility with a resolution cutoff of 2.5 Å.48 These proteins were grouped into protein families using the Dunbrack PISCES web server.49 Default PISCES parameters were used: 25% identity cutoff, R-factor better than 0.3, and the resolution better than 2.5 Å. PISCES compares not only sequence but also protein folds and topology. This allows recognition of families with low sequence identity, which are common in heme-binding proteins.
PISCES classified the 1503 heme proteins into 153 protein families, each with one protein chain as the representative head of the family (see Ref. 50 for PISCES output files). The 153 heads of the families were investigated to see whether they contribute significantly to heme-binding. A heme was associated with the head of the family and was therefore part of the nonredundant heme-binding protein set if the head of the family protein chain provided at least 30% of all residues within a 10 Å sphere around the heme iron (see Ref. 50 for log files). This resulted in 89 protein families containing 135 nonredundant heme-binding sites. Of all nonredundant heme-binding sites, 5% have hemes with ligands provided by two separate protein side chains. As this survey focuses on b- and c-type heme binding sites, two protein families providing binding sites to two a-type and one o-type heme-binding sites have been excluded (1V54, 1XME). Therefore the nonredundant heme-binding protein set contains 87 protein families providing a total of 132 heme-binding sites (see Table S1). We will hereafter refer to b- and c-type heme-binding sites as heme-bindings sites in general.
The ligands to the heme were identified by having a nitrogen, oxygen, or sulfur atom within 2.5 Å of the heme iron. Protein database structure files were examined by p3d, a python framework to examine pdb structures developed by C.F. in the laboratory.
Geometric properties of heme-binding motifs
The histidine heme ligand orientation is defined by the alpha angle (see Fig. 1). It is described by the angle between the heme basis vector Fe-CHA and the histidine vector Cγ-Nδ, when the histidine is ligating the heme via its Nε atom. If the heme is ligated via the Nδ atom, the Cε and Nε atoms define the histidine vector (e.g., Cyt c554). Ligand orientations towards the heme nitrogen NA and ND are +45° and −45°, respectively. Alpha is unambiguously defined as the angle between the projected histidine vector onto the heme plane and the Fe-CHA basis vectors. A dihedral angle, for example, Nδ-Nε-FE-CHA, as used in previous studies22 results in ambiguous values of the ligand orientation, because a histidine orientated towards NA would result in +45° for a ligand on one side of the heme plane whereas the same orientation would result in −45° for a ligand on the other side of the heme plane. In contrast, using the projection of the ligand onto the heme plane gives the same angle for both of these histidine orientations.
Figure 1.
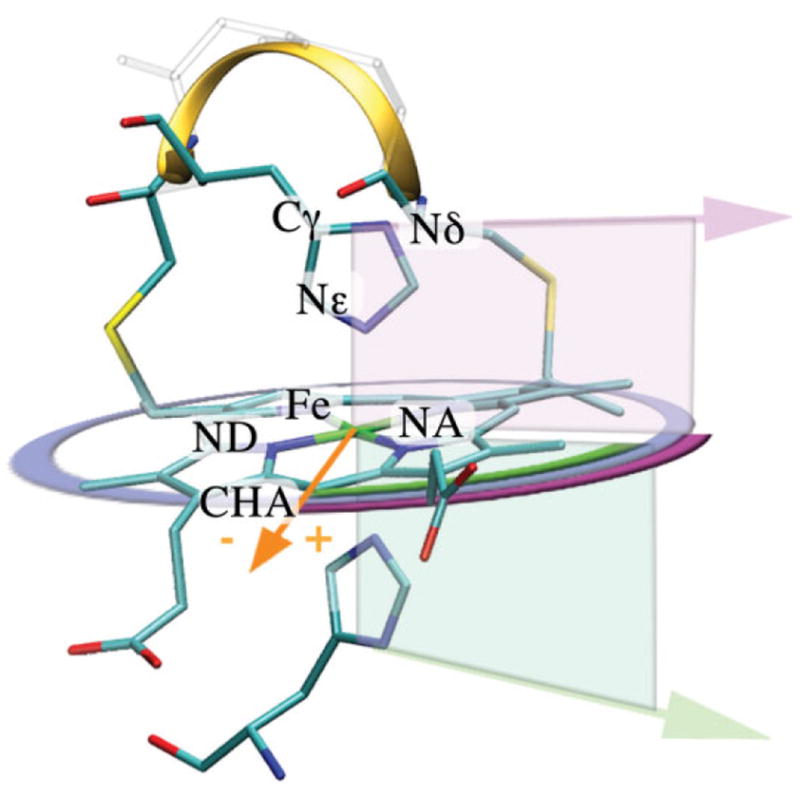
Heme structure with definitions used in this paper. Structure of a typical c-type heme-binding motif is shown (1Z1N). The heme is ligated by two histidines and covalently attached to the protein via two thioether bonds made between two cysteine side chains and two heme vinyls. Ligation and attachment forms the classical c-type heme-binding motif Cys-Xaa-Xaa-Cys-His (CXXCH) illustrated as a ribbon. The orange vector (Fe-CHA) represents the heme basis vector (0°). The orientation of the histidine ligand (purple and green arrows), alpha is defined after the histidine vector (Cγ-Nδ) is projected onto the heme plane. The resulting projected ligand vectors unambiguously describe their orientation (purple and green arcs). The right hand rule (NA-NB-NC) defines the top (purple) and the bottom (green) side of the heme plane. Alpha of +45° and −45° is pointing towards the heme nitrogen NA and ND, respectively.
As hemes are commonly distorted, a plane was fitted through the heme nitrogens to mathematically describe the heme. This was achieved by averaging the four possible planes defined by the right hand rule, by three nitrogens in alphabetical order (i.e., NA-NB-NC, NB-NC-ND, NC-ND-NA, and ND-NA-NB) and by using standard linear algebra to describe the plane (e.g., plane normal = (NA-NC) × (NA-NB)). The resultant plane normal defines the top and bottom of the heme plane.
Molecular mechanics calculations
Histidine ligand orientation dependent potential energy of a c-type heme with its binding motif (CXXCH) was determined using the molecular mechanics program CHARMM (Brooks, MacKerell). Not all 61 c-type heme-binding sites and their corresponding binding motif residues (CXXCH) in the nonredundant data set could be investigated with the standardized setup used here. Three binding sites have no His ligand (see Table I) and additionally, two binding sites (PDB id: 1J0P, heme id: 1001, 1004) have been excluded, as their binding motif are CXXXXCH. The coordinates of the remaining 56 c-type heme-binding motifs were extracted and the protein sequence was converted to CGGCH.
Table I.
Ligand Type Distribution in b- and c-type Hemes
|
b-Type heme |
c-Type heme |
Sum |
|||||
|---|---|---|---|---|---|---|---|
| Occurrence | [%] | Occurrence | [%] | Occurrence | [%] | ||
| Redundant | His-His | 149 | 6.5 | 603 | 61.2 | 752 | 23.0 |
| His-Met | 10 | 0.4 | 247 | 25.1 | 257 | 7.9 | |
| His-Xaa | 14 | 0.6 | 31 | 3.1 | 45 | 1.4 | |
| His-Ex | 1514 | 66.4 | 68 | 6.9 | 1582 | 48.4 | |
| Xaa-Xaa | 4 | 0.2 | 0 | 0.0 | 4 | 0.1 | |
| Xaa-Ex | 529 | 23.2 | 26 | 2.6 | 555 | 17.0 | |
| Other | 61 | 2.7 | 10 | 1.0 | 71 | 2.2 | |
| Total | 2281 | 985 | 3266 | ||||
| Nonredundant | His-His | 17 | 23.9 | 42 | 68.9 | 59 | 44.7 |
| His-Met | 1 | 1.4 | 10 | 16.4 | 11 | 8.3 | |
| His-Xaa | 2a | 2.8 | 2a,b | 3.3 | 4 | 3.0 | |
| His-Ex | 31 | 43.7 | 4 | 6.6 | 35 | 26.5 | |
| Xaa-Xaa | 1c | 1.4 | 0 | 0.0 | 1 | 0.8 | |
| Xaa-Ex | 17 | 23.9 | 2d | 3.3 | 19 | 14.4 | |
| Other | 2 | 2.8 | 1 | 1.6 | 3 | 2.3 | |
| Total | 71 | 61 | 132 | ||||
Distribution of ligands to b- and c-type hemes in the redundant and nonredundant heme protein data sets used in this survey. Xaa, amino acids other than His and Met. These include Asn, Asp, Cys, Tyr, Glu, and Lys in the redundant data set. Nonredundant data set:
His-Tyr,
Lys-Met,
His-Cys,
Lys-Ex. Ex: denotes hemes without a 6th ligand or an exchangeable, nonamino acid ligand (e.g., O2).
Molecular mechanics calculations were performed using CHARMM (Brooks) with topology and parameter sets 22 (MacKerell). The additional CHARMM topology, patches, and parameters files that were created to be used here can be found in the supplementary material. The calculations were carried out in the absence of solvent to focus on the geometric constraints on ligand rotation. To avoid unwanted hydrogen bond interactions between the protein backbone and the N- or C-terminus, the N-terminus was methylated and the C-terminus removed and replaced by hydrogen (see Fig. S1). A harmonic force of 48 kcal/(mol Å) was applied to the N-terminus and to the propionates. The propionates were patched to be protonated to minimize the electrostatic interactions with the histidine or heme. The histidine imidazole ring was rotated by one degree around the axis defined by the iron and the ligating Nε atom. The hydrogens on the histidine side chain were then rebuilt and the structure relaxed keeping the imidazole ring and the heme center atoms fixed. The fixed heme center atoms are the Fe, the nitrogens, the two carbons adjacent to each nitrogen and the four bridging methine carbons. The total potential energy was calculated at each angle. To avoid conformational local minima, rotation first increased alpha to 180°, then decreased it to −180° and finally increased it to the original orientation. This covers the range of the ligand orientation twice, reaching each angle from two directions. Before the results of the 56 c-type heme-binding motifs were averaged; each energy profile was normalized by subtracting its smallest energy. As a result, naturally occurring differences in the overall potential energy due to more or less optimized structures are diminished. The trajectory and a movie of a typical series of calculations can be found on the web.50
Calculations of the electrostatic interactions between the heme and its ligand
CHARMM22 calculations and rotations of imidazole ligands on a generic flat b-type heme were performed as described earlier, but with a rotation step size of 10° with both propionic acids protonated and with one or both ionized. For each rotation step a new protonated, but unrelaxed pdb file was created, which was used as input for the continuum electrostatic calculations, using MCCE Version 2.4beta8 (Multiple Conformer Continuum Electrostatics).51 Within the MCCE package DELPHI Version 452 is used to perform the continuum electrostatic calculations. Charge sets for the oxidized heme and imidazole ligand (see Suppl. Mat.) were calculated using Jaguar Version 7.0.53 Van der Waals (VDW) interactions were calculated with the AMBER force field.54 The pairwise electrostatic and VDW interactions between the imidazole and the heme were reported as a function of ligand angles relative to the heme plane.
Knowledge-based potential function
From classical thermodynamics, the frequency of an observation (e.g., ligand orientation, alpha) is governed by the relative energies of the different orientations. This can be expressed by a Boltzmann like equation45–47:
whereas fobserved, is the frequency of a given ligand orientation (here in 10° bins) and funbiased, the frequency that would be expected, if all states/bins have the same relative energy (i.e., observations/number of bins). The scaling factor 0.6 is to express the energies in kcal/mol.
All figures were plotted using Profit 6.1.4.55 Visualizations of pdb files were done using VMD 1.8.6.56
RESULTS
This survey of heme proteins analyses the type of heme ligation and the orientation of histidine ligands relative to the heme plane. The aim was to determine and to quantify the factors that influence ligand orientation in naturally occurring heme-binding motifs within the observed context. These results may then aid in bioinspired protein design.
A group of 87 nonredundant heme-binding proteins containing 132 hemes (Table S1) was derived from the 1503 heme containing proteins in the protein database in March 2007 using PISCES.49 The 87 nonredundant protein structures are each the representative or the head of a given protein structural family. Those families have 1–561 members (Table S1). Because the number of structures in each family varies so widely, a nonredundant protein data set is obligatory to draw significant statistical conclusions. Furthermore this nonredundant data set can then be used to quantify the relative importance of different factors on ligand orientation, for example, electrostatic interactions. The redundant data set can then be used to estimate variability of found patterns.
This survey focuses on b- and c-type heme-binding motifs. Other heme type binding motifs, that is, a-, o-and d-type hemes were found during the survey. However due to their small number, they were not investigated further, as they cannot provide statistically significant information.
Type of heme ligation
Table I shows the ligand type distribution in the redundant and nonredundant heme protein sets for b- and c-type heme-binding sites. In the nonredundant data set, the most common ligand is histidine, with 82.6% of the 132 hemes having at least one histidine ligand. Still, b- and c-type hemes have different ligand preferences. Nearly all c-type hemes (95.1%) and most b-type hemes (71.8%) have at least one His ligand. The higher prevalence of histidine ligands to c-type hemes is no surprise since one His is needed to form the classical c-type CXXCH binding motif. The exceptions are two proteins that use a Lys and an exchangeable molecule as heme ligands. Each shows a different way to achieve Lys ligation. The tetrathionate reductase from Shewanella oneidensis (1SP3),57 although having a CXXCH sequence, employs a distant Lys as the heme ligand while the nitrite reductase of Wolinella succinogenes (1FS7) employs an altered version of the classical c-binding motif that incorporates already the needed Lys, CXXCK.58
The remaining 28% of b-type heme-binding motifs in the nonredundant data set without a His ligand have Lys-Met (1.4%), Cys-Ex (17.0%), Met-Ex (1.4%), or Tyr-Ex (5.6%), whereas Ex indicates an empty binding site or exchangeable ligand. The unusual Lys-Met binding motif is found in the ethylbenzene dehydrogenase from Aromatoleum aromaticum (2IVF).59
The bis-His binding motif is found in 68.9% of all c-type but only in 23.9% of all b-type hemes in the non-redundant heme protein data set. His-Met ligation is found in 16.4% of the c-type but only in 1.4% of the b-type hemes. Exchangeable small molecules act as heme ligands in 70.4% of all b-type hemes, while only 11.5% of all c-type hemes do not have two ligands provided by the protein.
The ligand types are conserved within the protein families in 96% of all cases. This excludes new ligation motifs within a family due to introduced mutations, redox mediated swapping, or dissociation of the ligand. The redundant set of heme proteins shows additional b-type heme ligation motifs ligands, such as Cys-Glu (1SMI), Cys-His (2IJ3), and Cys-Lys (2IJ4). Additional c-type heme ligation motifs are Asn-His (1DW0), His-Lys (2J7A), and Met-Ex (2FKZ). Most of these additional ligation motifs are the result of site directed mutagenesis. Some heme ligation motifs, such as the recently reported Asp-Met60 are listed under others because the ligand heme distance is longer than 2.5 Å, the threshold used in this survey.
Orientation of the histidine ligand to the heme
This survey focuses on His heme ligands, which are found in 83% of all heme-binding proteins in the non-redundant dataset. Figure 1 illustrates the definitions of a typical histidine ligated heme-binding motif, here a bis-His c-type heme (1Z1N), used in this paper. The ligand orientation, alpha, is unambiguously described via projection onto the heme plane. The ligands were divided into those: (i) ligating b-type hemes (51 hemes), (ii) ligating c-type hemes where the histidine ligand is not part of the CXXCH motif (42 hemes), and (iii) ligating c-type hemes where the histidine ligand is part of the CXXCH binding motif (58 hemes).
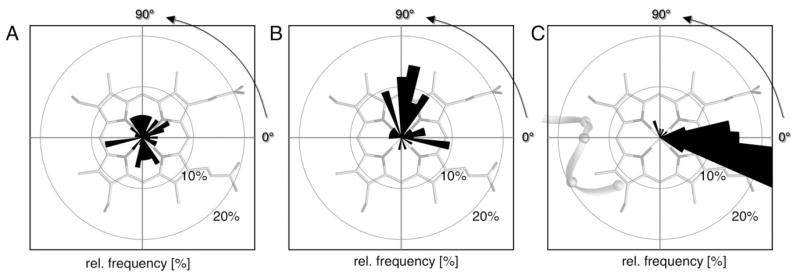
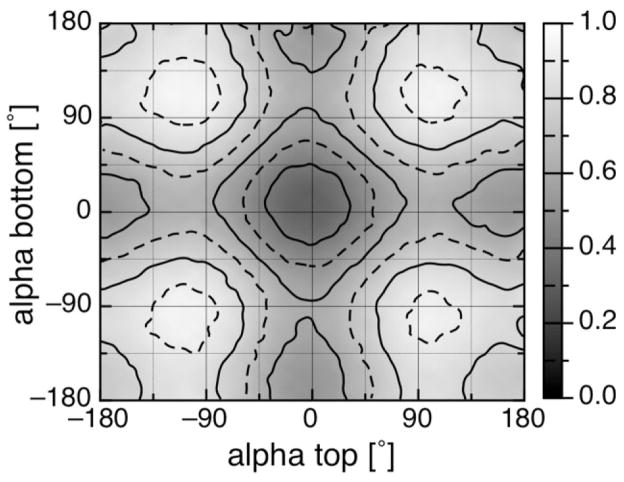
The distributions of the alpha angles are shown in Figure 2. The observed orientations were grouped in 10° bins. His ligands in b-type hemes [Fig. 2(A)] are found in nearly all orientations and have a smaller preference for a particular value of alpha. No 10° bin contains more than 7.4% of all b-type heme histidine ligands, having this maximum at −170 ± 5°. Similar, His ligands of c-type hemes that are not in the CXXCH motif are distributed over the entire range of angles [Fig. 2(B)]. Alphas near −10 ± 5° (9.5%), +80 ± 15° (33.3%), and 110 ± 5° (9.5%) are most likely. In contrast, c-type heme His ligands that are part of the CXXCH motif show a remarkable, asymmetric alpha distribution [Fig. 2(C)]. There are no histidines with alpha less than −35°. Approximately 86% are found around 0 ± 25° with 51.7% at −15 ± 10° (sum of −10° and −20° bin). The orientation +90 ± 25° contains ≈7% of all His ligands in the CXXCH motif (Table S2). Independent of heme type, those orientations that eclipse heme nitrogens (±45°, ±135°) are less frequently observed. Using the knowledge-based approach on all histidine ligand orientation except the c-type in CXXCH, the penalty for orientations that eclipse the heme nitrogens can be estimated using Eq. (1) to be ~1 kcal/mol (data not shown). The redundant data set shows similar possible alphas, however certain orientations are highly over represented (Fig. S2).
Figure 2.
Distribution of histidine ligand orientation for different heme types. The relative frequency of different histidine ligand orientations (alpha) taken from the nonredundant heme protein data set overlaid over the heme plane. Angles are grouped in 10° bins. (A) b-type heme histidine ligands show a relatively even distribution with a maximal relative frequency of ≈9% at −170° (n = 68). (B) Ligands of c-type hemes that are not part of the CXXCH motif are distributed over the whole range with 17% near +80° (n = 42). (C) Ligands of c-type hemes ligands in the CXXCH motif show only two preferred orientations with ≈90% between +20° and −30 and ≈10% between +40° and +90° (n = 58).
Asymmetric distribution of c-type heme histidine ligand orientation
Molecular mechanics calculations were used to investigate the underlying causes of the absence of His (in CXXCH) ligand orientation smaller than −35° in c-type heme-binding sites [Fig. 2(C)]. The hemes and their corresponding CXXCH motif were taken from all 56 c-type hemes in the nonredundant data set*. The two amino acids between the Cys, XX were changed to Gly, providing the least constraints on the backbone.
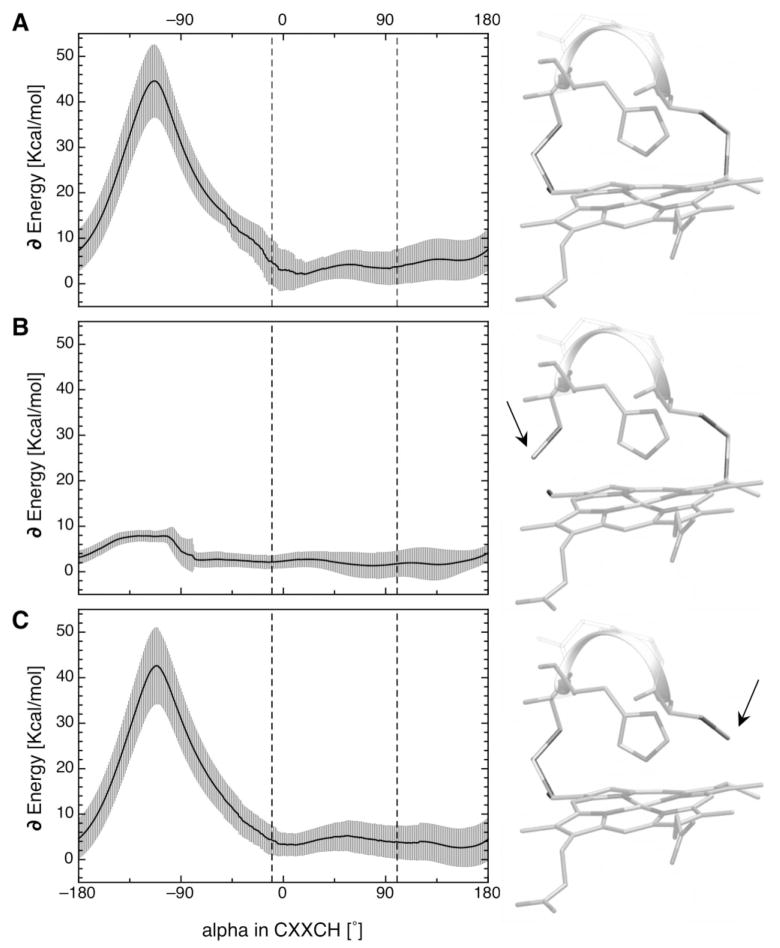
CHARMM22 was used to calculate the alpha angle dependent potential energies of each system [Fig. 3(A)]. This includes the energies from distortions of bond lengths and angles and from the van der Waals (VDW) interactions. The energy profiles for each heme were normalized by subtracting their lowest energy. The 56 energy profiles were then averaged. The standard deviation of the potential energy at each alpha angle is shown as grey error bars. They range from 2.5 to 8.0 kcal/mol depending on the alpha angle. This is due to the differences between the 56 structures that are kept fixed during the series of calculations, such as the heme-ligand distance, ligand tilt, or deformation of the heme center. The most observed ligand orientations [Fig. 2(C)], indicated with dashed lines in Figure 3, are close to the shallow minimum in the energy profile. The system energy is unfavorable for alphas between −45° and +180° with the maximum, near −120° of over 40 kcal/mol. At this angle the histidine is very close to the Cys side chain connected to the heme ring forcing significant distortions of the heme plane (see Fig. S1). The length of all connecting bonds starting from the covalent link between the Cys and the heme to the histidine-heme ligation are simply too short to accommodate an alpha of less than −45°.
Figure 3.
Orientation dependent potential energy profile for c-type heme histidine ligand in the CXXCH motif. Averaged alpha angle dependent potential energy profiles for all 56 standard c-type hemes with their CXXCH motif in the nonredundant data set calculated using CHARMM22. The grey swath shows the standard deviation. Structures used with (A) both cysteines attached to the heme plane, (B) cysteine adjacent to the histidine (position i–1) detached, and (C) cysteine at position i–4 detached from the heme. Dashed lines mark the most observed ligand orientations for the histidines in the CXXCH motif [Fig. 2(C)].
Calculations without the covalent attachment between the heme and the Cys that is adjacent to the histidine ligand in the CXXCH motif resulted in no significant increase in the alpha dependent potential energy surface [Fig. 3(B)]. With these structural constraints loosened, the rotation of the histidine ligand causes little change in the potential energy. The small increase of about 6.5 kcal/mol giving rise to the shoulder around −125° is due to a rearrangement of the backbone. A series of calculations performed with a setup in which the other Cys in the CXXCH motif is detached from the heme [Fig. 3(C)] results in same large potential barrier, as if both Cys are attached [Fig. 3(A)]. The images and the arrows next to the potential energy graphs illustrate the setup and detachment of the cysteines.
Thus the absence of c-type histidine ligands in the CXXCH motif with alpha less than −45° in the proteins found in the database [Fig. 2(C)] can be explained by the calculated potential energy profile [Fig. 3(A)].
Relative ligand orientations in bis-His heme-binding motifs
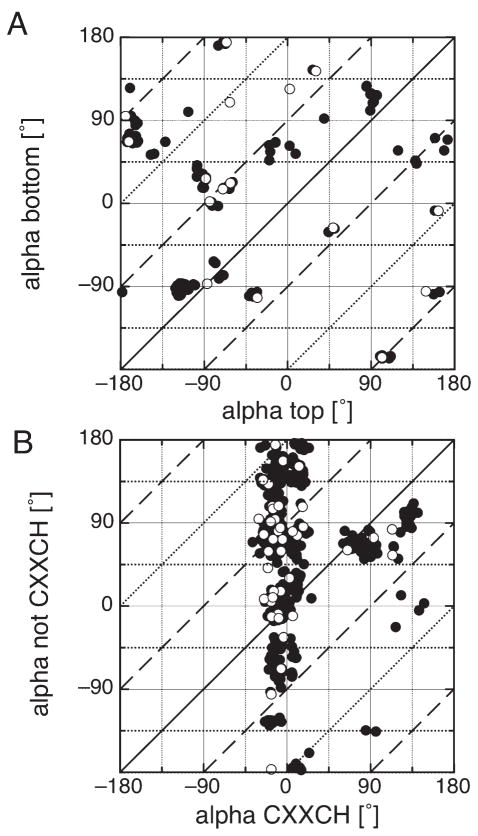
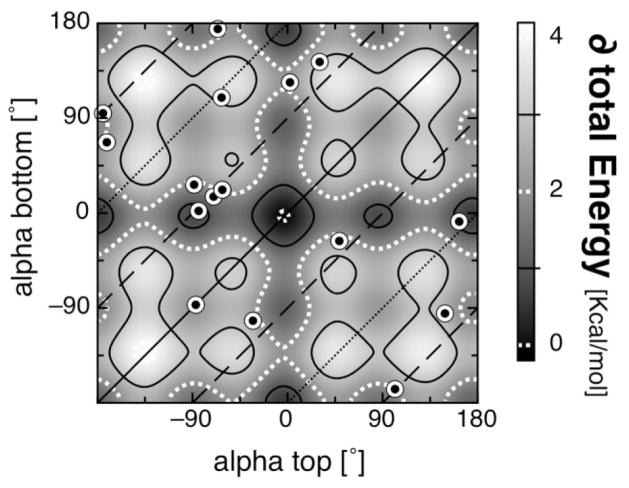
Figure 4 shows the relative ligand orientation in the bis-His b-type [Fig. 4(A)] and c-type hemes [Fig. 4(B)] as a correlation plot of the two heme ligating histidines. Shown are the redundant (closed circles) and the non-redundant (open circles) data sets. Alpha for the ligand on the top of the heme plane is plotted against alpha for the ligand on the bottom heme plane. In c-type hemes, the ligand in the CXXCH motif was found to be always on the top of the heme plane. The lines represent relative ligand orientations that are perpendicular (dashed), parallel (solid), or anti-parallel (dotted) to each other. The redundant protein data set was used to illustrate the variability of ligand orientations.
Figure 4.
Ligand orientations of bis-his heme binding motifs. Correlation plot of the two alpha angles for bis-His heme systems are shown for (A) b-type heme binding motifs and (B) c-type heme binding motifs, nonredundant set (open circles) and redundant set (closed circles). The top and bottom histidine is defined by the right hand rule (see text). The histidine in the CXXCH motif is always on the top of the heme. The lines represent the relative orientation of the two His ligands: parallel (solid), perpendicular (dashed) or anti-parallel (dotted).
The b-type heme ligation motifs appear to have a preference for the histidines to be oriented perpendicular to each other. In the nonredundant protein set, the ratio between parallel and perpendicular ligand orientation is 4:13. Some of the clustered points such as around (−90°, −90°) or (−180°, +90°) represent families with numerous members with their ligands in a narrow range of orientations. The preference for avoiding positions that occlude the heme nitrogens at ±35° and ±145° is clearly visible.
The bis-His c-type orientations show the same features as their nonredundant counterpart [Fig. 2(B,C)]. The His ligands that are not in the CXXCH motif show weak preferences for particular alphas in contrast to the His ligands in the CXXCH motif. There are, however, some correlations between the orientations of the two c-type heme ligating histidines. The motifs in which the His ligand in CXXCH has an alpha near +90° tend to be paired with a His ligand (not in CXXCH) that also has an alpha near +90°. There, the two ligands are relatively parallel to each other and perpendicular to the propionates (alpha = 0°). This explains the higher preference for alpha around +90° in the nonredundant set of histidine orientation not in CXXCH [Fig. 2(B)]. On the other hand, ligands in the CXXCH motif with alpha around −10° seem not to correlate with any particular alpha for the ligand on the other side of the heme. The ratio parallel to perpendicular ligand orientation is ~1:1.
The degree of conservation of the alpha angles within a given family in the redundant database for bis-His heme binding motifs was investigated (Fig. S3). Several family specific features are found that might be correlated to their function. The myoglobin family (1A6M) shows broad variation of alpha on the top of the heme plane with a conserved orientation of the histidine ligand on the bottom of the heme plane around +61 ± 12°. Other protein families, such as sufite oxidase (1MJ4), bc1 complex (1PPJ), neuroglobin (1Q1F), nitrate reductase (1Y5I), quinol-fumerate reductase (2BS2), succinate dehydrogenase (2H88) show a broader variability in alpha within their families, however their relative alpha angle between the ligating His seems to be conserved. c-Type binding protein families, such as nitrite reductase (1FS7) or cytochrome c (1H21) show the same patterns found in Figure 2, that is to say alpha on the top is very rigid and confined, whereas the alpha on the bottom of the heme shows broad variation.
Electrostatic interaction between the heme and its ligands
It has been proposed that His heme ligands have a strong preference for alpha around 0° due to the strong charge-dipole electrostatic interactions between the heme propionic acid and the His ligand.22 This argument was based on earlier surveys of heme-binding motifs and calculations that assumed both the propionic acids were ionized.
However, the survey presented here on a nonredundant data set shows, apart from His ligands in the CXXCH motif only a weak preference for His heme ligands to point between the two propionic acids with an alpha of 0° [Fig. 2(A,B)]. Moreover, the pH dependence of cytochrome Ems and electrostatic calculations61 suggest that these acids need not be fully deprotonated at physiological pH. Thus, MCCE calculations were performed to quantify the interaction energy and moreover its dependence on the ionization state of the propionates.
To simplify the calculations, the histidine ligands were reduced to their imidazole rings. The propionates were placed in one of the most occupied conformations found in this heme survey (see Fig. 1). The energy as a function of the alpha angles for the two imidazoles on a flat b-type heme was calculated using MCCE51 and CHARMM62 (MacKerell). The two methods show similar results (not shown). The main difference is that CHARMM can minimize the bond lengths and angles that are fixed in MCCE. Thus using CHARMM diminishes the overall nonelectrostatic system energy while retaining its dependence on alpha (not shown).
The total interaction energy between a heme and its ligand is composed of electrostatic, VDW, and quantum mechanical terms. The latter is, however, not considered in classical calculations. In an MCCE calculation where bond lengths and angles are fixed the VDW energy varies from −14.5 to −17 kcal/mol as the imidazole is rotated. The 2.5 kcal/mol increase in energy is due to the interaction between the imidazole and the heme nitrogens at alpha ±45° and ±135°. The change in VDW energy is however independent of the propionate ionization state. The electrostatic interaction between imidazole and the heme is calculated using DELPHI52 with a uniform dielectric constant of 6. This approximates the effective dielectric constant for the histidine to propionate interaction in a protein surrounded by water. This value may overestimate the interaction when the proteins are small63 and underestimate it when the heme and propionates are well buried.64
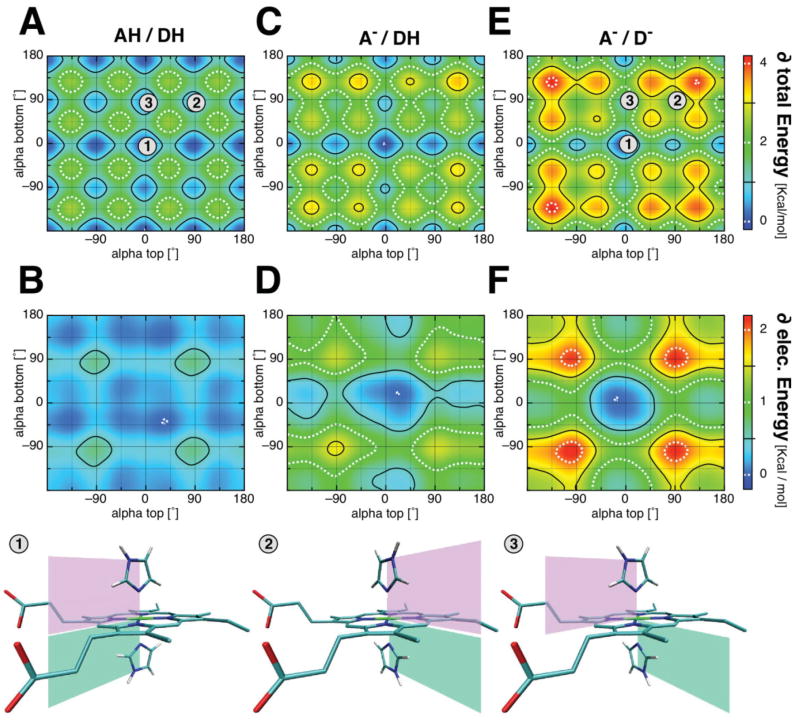
Figure 5 shows contour plots of the total interaction energy (top row) and its electrostatic contribution (middle row) as a function of ligand orientations, as a function of alphas. For the sake of clarity, the change in energy is plotted (see Table II for absolute values) with propionates protonated [Fig. 5(A,B)], one propionate deprotonated [Fig. 5(C,D)], and both propionates deprotonated [Fig. 5(E,F)]. Three different ligand orientations are marked in the total energy plots: (1) the two ligands are parallel to each other with alpha of 0°, that is, pointing between the propionates; (2) the ligands are parallel to each other and perpendicular to the propionates, that is both with an alpha of +90°; (3) the two ligands are perpendicular to each other at alpha 0° and +90°. The bottom row of Figure 5 illustrates these ligand configurations in the model complex.
Figure 5.
Dependence of the interaction energy on the histidine orientation and propionic acid ionization state for a bis-imidazole heme model complex. Interaction energies between imidazole ligands and the heme were calculated with MCCE at ε = 6. Shown are the total interaction energy (top row) and its electrostatic contribution (middle row); (A, B) when both propionates are protonated (AH, DH); (C, D) when the propionatic acid attached to ring A is deprotonated (A−, DH) and, (E, F) when both propionates are deprotonated (A−, D−). The change in the energy landscape with ligand orientation, alpha is illustrated (see Table II for absolute values). Bottom row shows three ligand conformations marked in the total energy plots with the numbers 1–3.
Table 2.
Calculated Interaction Energy of the Heme as a Function of Alpha, the Orientation of the Histidine Ligand
| [Kcal/mol] | AH/DH | A−/DH | A−/D− |
|---|---|---|---|
| Total interaction energy | |||
| Maximum | −15.6 | −20.6 | −17.2 |
| Minimum | −18.2 | −24.0 | −21.3 |
| Electrostatic interaction energy | |||
| Maximum | −1.2 | −6.2 | −2.7 |
| Minimum | −1.9 | −7.8 | −4.9 |
| Parallel (alpha 0°) – pos. 1 | −1.7 | −7.6 | −4.9 |
| Parallel (alpha 90°) – pos. 2 | −1.2 | −6.4 | −2.7 |
| Perpendicular (alpha 0° & 90°) – pos. 3 | −1.5 | −7.0 | −3.8 |
The total and the electrostatic interaction energies between the imidazole ligands and the heme. The plotted interactions in Figure 4 show only the ligand orientation energy difference for the sake of clarity. A is the propionate attached to the heme ring A and D to ring D. The propionatic acids are referred to as AH and DH, while the propionate is referred to as A− and D−.
Independent of the propionic acid ionization, the system shows a minimum of electrostatic and total interaction energy when both histidines have an alpha near 0°. However, when both propionates are protonated [Fig. 5(A,B)], the system shows a rather smooth energy landscape with minima at alpha 0°, ±90°, and ±180° and maxima at positions closest to the heme nitrogens. The maximal electrostatic contribution is only 0.7 kcal/mol, varying by only 0.46 kcal/mol between position 1 where both alphas are at 0° and position 2 where they are at +90°. The difference in total energy between position 1 and position 2 are in same order of only 0.56 kcal/mol.
In the case when one propionate is deprotonated (here the propionate on ring A), the system energy landscape shows a cross like shape with a local minimum at (0°, 0°) [Fig. 5(C)]. The energy landscape is slightly asymmetric favoring conformations where at least one ligand points towards the ionized acid, at alpha near 20°. The maximum change in total interaction energy is 3.4 kcal/mol with 1.6 kcal/mol from the alpha dependence of the electrostatic interaction [Fig. 5(D)]. The difference in electrostatic interaction when both ligands are pointing between the propionates (pos. 1) or perpendicular to the propionates (pos. 2) is 1.2 kcal/mol. The difference between having the ligands parallel (pos. 1) and perpendicular (pos. 3) to each other is of the order of 0.6 kcal/mol in this classical calculation.
When both propionates are ionized, the energy landscape exhibits a deeper, more symmetric shape [Fig. 5(E,F)]. The maximum change in total energy is 4.1 kcal/mol. The maximum difference in electrostatic interaction energy is 2.2 kcal/mol. Both ligands now prefer to be pointing between the two propionatic acids, with a penalty of 2.1 kcal/mol if they have both an alpha of 90° (pos. 1 vs. 2). Overall the pattern of interaction energies shown in Figure 5(E) agrees with previous DFT calculations of Knapp and coworkers35 obtained when both acids are deprotonated. However our values are somewhat smaller, most likely due to the fact that classical calculations are based on VDW and electrostatic interactions, whereas DFT calculations include quantum effects, such as orbital–orbital interactions. However, in terms of electrostatic interactions, the doubly deprotonated propionatic state is most likely an artificial state (see below).
Alpha dependent pKa of the heme propionatic acids
Interaction energy between the ligand and the heme can also be seen from a propionatic acid point of view. There a given alpha angle will induce a change in the pKa of the propionates. MCCE was used to calculate the alpha dependent shift in pKa. Figure 6 shows the change in pKa for both propionates as a function of both alphas. On average the pKa of each of the acids will be lowered by ≈0.5 pH unit by rotating both histidines from 90° to 0° (pos. 1 vs. pos. 2).
Figure 6.
Ligand orientation dependent effects on the pKas of the propionates for a bis-imidazole heme model complex. The pKa of the propionatic acids depends on the ligand orientation, alpha as a direct result of the interaction energy shown in Figure 4. The interaction energy as a function of ligand orientations (here bis-imidazole system) changes the propionatic acids pKa by as much as one pKa unit.
DISCUSSION
Heme cofactors play integral roles in proteins with very diverse functions1–12 and its large hydrophobic surface helps to stabilize many proteins in the folded state. As such, hemes have become a favored cofactor in bio-inspired and de novo designed proteins (for reviews see Refs. 65–67). Factors influencing heme ligand orientation and the effects of these have been a focus of debate.22,31–42,44 Surveys of native protein structures reveal that heme-binding patterns can help to evaluate factors via statistical knowledge-based approaches.45–47 The results can help to describe the constraints on the conformational space of designed heme-binding proteins.
In this work, heme ligation motifs were investigated using a nonredundant and representative protein data set of 87 proteins (Table S1). This survey highlights systematic differences between b- and c-type heme binding proteins. The focus is on His ligands and their orientation, as they are found to be the ligand to 83% of all hemes in the nonredundant dataset (Table I). A knowledge-based approach was used to estimate the importance of the different factors that can influence the orientation of the His ligands. Molecular mechanics calculations explain the asymmetric distribution of the observed His c-type heme ligands that are part of the CXXCH motif. The influences on the ligand orientation, such as electrostatic interactions, heme ruffling, hydrogen bond to the His ligands, and quantum effects are discussed on the basis of the observed ligand orientations.
Ligand preference in b- and c-type heme
In theory, a protein can provide ligands to the heme via peptide chain termini or amino acid side chains as long as oxygen, nitrogen, or sulfur atoms are involved. However, not all of the possible amino acids have been found to serve as ligands (see Table I). This might be due to several reasons. The simplest explanation is that certain heme ligation types have simply not yet been discovered; proteins with novel heme ligation motifs are still being found.60 Another reason is the difference in chemical properties of the possible ligands which influence the affinity and electrochemistry of the heme.19,20 For example, in the nonredundant database, b-type hemes rarely have His-Met ligation, while it is found in 16.4% of all c-type heme ligation. To understand this difference in preference it is worth looking into the properties of Met heme ligation. Met binds more weakly to hemes than His especially in the oxidized heme form.20 Because of this, His-Met ligation will raise the heme Em by ≈150 mV relative to a bis-His heme in the same environment.30 It is therefore no wonder that Nature has developed and kept the c-type binding motif to compensate for a weaker ligation, eliminate cofactor loss,26 ligand diffusion or competition, diminish protein unfolding68 and at the same time to benefit from the Em shift. Additionally, there have been other proposals for why Nature has kept the energy-expensive c-type heme-binding motifs (for a review see Ref. 69). One of which is based on the observed heme to amino acid ratio that is needed to form stable b- or c-type heme binding proteins. There the covalent attachment of c-type hemes provides the possibility to achieve higher heme packing with less protein structure, thus all multi heme proteins involve c-type hemes. An overall mixture of all factors might explain why c-type hemes are “still” in use.
In the nonredundant database, the ligation types of c-type hemes indicate that they are mainly associated with electron transfer reactions where there is no need for ligand exchange. These hemes have two amino acid ligands in 89% of all cases. In contrast, 68% of all b-type heme-binding proteins have only a single amino acid as a ligand, allowing them to function in substrate transport and substrate redox chemistry. Nature’s choice here seems counterintuitive, since the proteins that use noncovalently attached hemes do not exhibit bis-His ligation motifs, which would ensure tighter binding to the protein.70 Thus, in natural systems, the protein does not rely on the strong heme ligand interaction to bind the heme. This hypothesis is clearly supported by site directed mutagenesis assays in sperm whale myoglobin reported by Barrick.71 There, although the histidine heme ligand was changed to glycine, the apoprotein was still able to bind heme in its usual pocket, indicating that the largest component of the binding energy is provided by the hydrophobic protein interaction. In contrast, protein design studies do make use of the strength of bis-His ligation to help maintain a folded protein.70
Ligand orientation
The His ligand orientation is thought to be influenced by several factors22,31–42,44 and additionally the orientation has been proposed to influence the physiochemical properties of the heme, such as the redox potential.30,31 The distribution of histidine ligand orientation relative to the heme plane, denoted the alpha angle, was determined. There was no strong preference for particular values of alpha found for b-type heme-binding motifs. Histidine ligands in the c-type CXXCH heme-binding motif however have a strong preference to be oriented near −10° and less commonly near +90° [Fig. 2(C)]. These preferences were first described by Finzel et al. in 1985, when there were only seven X-ray structures of c-type heme proteins available.72 CHARMM molecular mechanics calculations provide the rationale for the observed distribution of alpha angles (see Fig. 3): the constraint imposed by the covalent attachment of the heme to the cysteine adjacent to the ligating histidine limits the ligand orientation to alphas between −45° and +135° [Fig. 3(A)]. The allowed region is further reduced by the smaller van der Waals (VDW) penalty that the heme nitrogens impose on the ligands, thus creating the two observed favorable regions. This restriction on the allowed alpha angles explains why ligands to c-type hemes are far more confined than those of the b-type hemes [Fig. 2(A)].
A previous survey of heme structures reported a strong preference for the His heme ligands for alphas near 0°.22 However, those authors used a redundant protein set and did not distinguish whether the His was in a CXXCH motif or not. It had been argued that a preference for alpha near 0° is due to the strong electrostatic interaction between the His dipole moment and the charge on the propionates.22,35 In general this preference for the histidine ligand to point between the propionates relies on both propionates being ionized [Fig. 5(E)], buried within a protein and not shielded by counter ions. In this case, having both ligands oriented with alpha around 0° [Fig. 5(F), pos 1] can be at most 2 kcal/mol more favorable than when they are both at +90° [Fig. 5(F), pos 2]. However, if the ionized propionates are exposed to the solvent and thereby screened by water63 or paired with counter ions the net effect of the propionates on alpha is diminished. The interaction shrinks to ≈0.5 kcal/mol when the acids are protonated [Fig. 5(B)]. Mao et al. reported in a study of the redox potentials of 13 heme protein that only 30% of all investigated hemes are calculated to have fully deprotonated propionates.61 More recent and extended calculations show only ~16% of all 142 investigated heme proteins have fully deprotonated propionatic acids in the oxidized and reduced state (Zheng and Gunner in preparation).
This survey, using a nonredundant data set, shows no preferred orientation of the histidine ligands for b- or c-type (not in CXXCH) heme-binding motifs. The only preferred alphas found were for c-type heme-binding motifs in the CXXCH motif. Their orientation could be shown to be strongly confined to alphas larger than −35° and smaller than 180° due to geometrical constrains (>40 kcal/mol). However, within this restriction the two observed and allowed orientations show a higher population for alpha around 0° (~90%) compared to +90° (~10%). This difference would be equal to a difference of −1.4 kcal/mol using the knowledge-based approach (eq. 1). This is comparable to the calculated difference in energy between these two ligand configurations due to electrostatic interaction if only one propionatic acid is deprotonated [pos.1 vs. pos.3, Fig. 5(C,D), Table II].
What is important? Observed ligand orientations versus known influencing factors
The observed absence of certain alpha angles in c-type heme histidine ligands within the CXXCH motif shown in Figures 2(C) and 4(B) can be explained by covalent constraints (see Fig. 3). These constraints are in the order of 40 kcal/mol and impose a forbidden ligand orientation region. In contrast, histidines in c- (not in CXXCH) and in b-type heme-binding sites show no preferred alpha angles. These heme-binding sites can be used to help to quantify other factors that may influence the histidine ligand orientation. Figure 7 compares the ligand orientations for all bis-his b-type hemes in the nonredundant dataset with the strongest interaction energy surface, including electrostatic and VDW interactions when both propionatic acids are deprotonated [Fig. 5(E)]. The lines representing relative perpendicular (dashed), parallel (solid), or anti-parallel (dotted) ligand orientations are shown. A striking feature is that none of the observed ligation motifs show alphas near (0°, 0°), which is the lowest energy configuration in the electrostatic/potential energy surface for a flat heme. Moreover, 50% of the heme-binding sites show ligand configurations that are in unfavorable regions with at least 2 kcal/mol over the calculated minimum (see Fig. 7). Nevertheless, each b-type heme avoids ligand orientations that eclipse the heme nitrogens. By analyzing the frequency distribution of alpha using Eq. (1), one can estimate the energy penalty for ligands to eclipse the heme nitrogens to be ~1 kcal/mol. Together, these findings clearly indicate that the ligand orientation is not dictated by the electrostatic and/or VDW interaction.
Figure 7.
Electrostatic interaction energy surface overlaid with observed ligand orientations in bis-His b-type heme-binding sites. The alpha angles for bis-His b-type hemes taken from the nonredundant protein set are plotted. The lines represent the relative orientation of the two ligands: parallel (solid), perpendicular (dashed) or anti-parallel (dotted). The underling contour map is the total interaction energy for a bis-imidazole heme model complex, both propionates ionized [see Fig. 5(E)].
It has been proposed that the parallel ligand orientation is lower in energy than the perpendicular orientation.31,34,40,42,43,73 A sensitive tool to interrogate ligand orientation in a heme system is EPR. A relatively perpendicular ligand orientation has been associated with the so called gmax74 or HALS75 feature in the EPR spectrum. When heme proteins that exhibit such a gmax feature are treated with detergent, they loose this feature in their EPR spectra.76,77 It has been argued that relaxation of the protein lets the ligands move into their lower energy, relatively parallel ligand conformation.78 However, the observed reorientation of the heme ligands in a looser protein could have a different origin, as many other factors stabilizing the ligand orientation are also loosened during treatment with detergent. Munro et al.40 reported the stabilization of a relative parallel ligand orientation of up to 3 kcal/mol compared to the perpendicular ligand arrangement, most likely due to Jahn-Teller effect and heme ring ruffling.31–34 Other quantum mechanics,41,42 electronic orbital calculations,43 and thermodynamic analysis of EPR/Mössbauer spectra73 estimated an energy difference of only ~1 kcal/mol between the two orientations.
A stabilization of 1–3 kcal/mol due to quantum effects in favoring the parallel ligand orientation would be observable as a 10- to 100-fold higher frequency of parallel over perpendicular ligand orientation, if these were the limiting factors. However, this is not the case in the observed heme-binding sites (see Fig. 7). Surprisingly, the opposite was found: in the 17 bis-His b-type heme-binding sites in the nonredundant data set the ratio of the observed parallel to perpendicular ligand orientations is 3:14 (see Fig. 7). Thus, other factors or the sum of other factors dictate the ligand orientation. For example, the first heme model complex reported that showed a perpendicular ligand orientation was achieved by enforcing heme ruffling, which put constraints on the ligands.44 Another possible factor is the hydrogen bond to the heme ligating nitrogen,36 which can contribute up to 5 kcal/mol per ligand to stabilize the observed ligand orientations.79 Factors that influence the histidine ligand orientation can be ranked into factors that have a strong, intermediate or weak influence on alpha. Strong influences on alpha include the CXXCH motif (>40 kcal/mol, Fig. 3) and other VDW terms such as heme ruffling.31–34 Intermediate influences on alpha are the hydrogen bonds to the heme ligand (~5 kcal/mol)79 and quantum effects due to orbital interactions and ligand field distortions (1–3 kcal/mol).40,73 The smallest influences on alpha are due to electrostatic interactions [~1 kcal/mol, Fig. 5(D)] and due to the VDW barriers imposed by the heme nitrogens [~1 kcal/mol, Fig. 2(A), Eq. (1)].
Implications for protein design
The factors listed here and the estimate in stabilization will aid bioinspired protein design. It is clear from the observed orientation (alphas) of the c-type heme histidine ligands that are part of the CXXCH motifs, that in designing such motifs one must take the forbidden region for alphas into account (see Fig. 3). The effects of forcing the heme ligand into an orientation with alphas less than −40° will impose heme deformation on a designed protein. This is an interesting feature since the deformation and change of the electronic structure might tune the redox potential.32
Apart from the special histidine ligand in the CXXCH motif, the other observed histidine heme ligands showed few constraints on their orientation. It is clear that electrostatic interactions have a maximal impact of 1 kcal/mol or even less in proteins. Nonetheless, electrostatic interactions should not have a significant impact on the stability of designed proteins. However, this interaction can lead to changes in the pKa of the propionates of up to 1 pKa unit (see Fig. 6). Thus, an alpha dependent pKa of the propionates might help to modulate the Em of designed heme-binding proteins by as much 60 mV. Reported ligand–ligand interactions and ligand heme interactions that favor a parallel over a perpendicular ligand orientation are not found and seem therefore not to dictate the observed ligand orientations in proteins. Additional factors, such as hydrogen bonds to the heme ligating histidine and heme distortion which were not treated here remain factors that could have a stronger impact on the ligand orientation and are therefore probably worthwhile taking into account for protein design.
Supplementary Material
Acknowledgments
The authors thank Drs. Brian Gibney, Ron Koder, Jessica Norman, Yifan Song for very fruitful discussions and suggestions.
Grant sponsor: NSF; Grant number: MCB-0517589; Grant sponsor: NIH; Grant number: 5G12 RR03060; Grant sponsor: Alexander von Humboldt foundation.
Abbreviations
- A and D
deprotonated propionates
- AH and DH
heme propionatic acids at ring A and ring D, respectively, (position 2,18)
- alpha
Histidine ligand orientation, 0° between the heme propionates
- bis-His heme
heme ligated by two histidines
- Em
redox potential versus SHE
- Ex
Exchangeable small molecule (e.g., O2)
- Heme
iron protoporphyrin IX
- VDW
van der Waals
- Xaa
any amino acid
Footnotes
Five c-type heme-binding motif were excluded, see M&M.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Chapman SK, Daff S, Munro AW. Heme: the most versatile redox centre in biology. Struct Bond. 1997;88:39–70. [Google Scholar]
- 2.Gray HB, Winkler JR. Electron transfer in proteins. Annu Rev Biochem. 1996;65:537–561. doi: 10.1146/annurev.bi.65.070196.002541. [DOI] [PubMed] [Google Scholar]
- 3.Perutz MF. Regulation of oxygen affinity of hemoglobin: influence of structure of the globin on the heme iron. Annu Rev Biochem. 1979;48:327–386. doi: 10.1146/annurev.bi.48.070179.001551. [DOI] [PubMed] [Google Scholar]
- 4.Kendrew JC, Bodo G, Dintzis HM, Parrish RG, Wyckoff H, Phillips DC. A three-dimensional model of the myoglobin molecule obtained by X-ray analysis. Nature. 1958;181:662–666. doi: 10.1038/181662a0. [DOI] [PubMed] [Google Scholar]
- 5.Perutz MF, Miurhead H, Cox JM, Goaman LC, Mathews FS, McGandy EL, Webb LE. Three-dimensional Fourier synthesis of horse oxyhaemoglobin at 2.8 Å resolution. I. X-ray analysis. Nature. 1968;219:29–32. doi: 10.1038/219029a0. [DOI] [PubMed] [Google Scholar]
- 6.Padmanaban G, Venkateswar V, Rangarajan PN. Haem as a multi-functional regulator. Trends Biochem Sci. 1989;14:492–496. doi: 10.1016/0968-0004(89)90182-5. [DOI] [PubMed] [Google Scholar]
- 7.Hon T, Dodd A, Dirmeier R, Gorman N, Sinclair PR, Zhang L, Poyton RO. A mechanism of oxygen sensing in yeast. Multiple oxygen-responsive steps in the heme biosynthetic pathway affect Hap1 activity. J Biol Chem. 2003;278:50771–50780. doi: 10.1074/jbc.M303677200. [DOI] [PubMed] [Google Scholar]
- 8.Mense SM, Zhang L. Heme: a versatile signaling molecule controlling the activities of diverse regulators ranging from transcription factors to MAP kinases. Cell Res. 2006;16:681–692. doi: 10.1038/sj.cr.7310086. [DOI] [PubMed] [Google Scholar]
- 9.Otyepka M, Skopalik J, Anzenbacherova E, Anzenbacher P. What common structural features and variations of mammalian P450s are known to date? Biochim Biophys Acta. 2007;1770:376–389. doi: 10.1016/j.bbagen.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Anzenbacher P, Anzenbacherova E. Cytochromes P450 and metabolism of xenobiotics. Cell Mol Life Sci. 2001;58:737–747. doi: 10.1007/PL00000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faller M, Matsunaga M, Yin S, Loo JA, Guo F. Heme is involved in microRNA processing. Nat Struct Mol Biol. 2007;14:23–29. doi: 10.1038/nsmb1182. [DOI] [PubMed] [Google Scholar]
- 12.Abraham NG, Kappas A. Heme oxygenase and the cardiovascular-renal system. Free Radic Biol Med. 2005;39:1–25. doi: 10.1016/j.freeradbiomed.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Vitek L, Schwertner HA. The heme catabolic pathway and its protective effects on oxidative stress-mediated diseases. Adv Clin Chem. 2007;43:1–57. doi: 10.1016/s0065-2423(06)43001-8. [DOI] [PubMed] [Google Scholar]
- 14.Hansson M, von Wachenfeldt C. Heme b (protoheme IX) is a precursor of heme a and heme d in Bacillus subtilis. FEMS Microbiol Lett. 1993;107:121–125. doi: 10.1016/0378-1097(93)90365-9. [DOI] [PubMed] [Google Scholar]
- 15.Gonzales DH, Neupert W. Biogenesis of mitochondrial c-type cytochromes. J bioenerg biomembr. 1990;22:753–768. doi: 10.1007/BF00786929. [DOI] [PubMed] [Google Scholar]
- 16.Scott RA, Mauk AG, editors. Cytochrome c soucebook. Mill Valley: University Science Books; 1996. [Google Scholar]
- 17.Allen JWA, Ferguson SJ. The Escherichia coli cytochrome c maturation (Ccm) apparatus can mature cytochromes with an extra cysteine within or adjacent to the CXXCH motif. In: Bedmar EJ, Delgado MJ, Moreno-Vivian C, editors. The 11th nitrogen cycle meeting; Granada, Spain: Estaci’ on Experimental del Zaid’in; 2005. pp. 91–93. [DOI] [PubMed] [Google Scholar]
- 18.Kadish KM. Electrochemistry of metalloporphyrins in nonaqueous media. Prog Inorg Chem. 1986;34:435–605. [Google Scholar]
- 19.Nesset MJM, Shokhirev NV, Enemark PD, Jacobson SE, Walker FA. Models of the cytochromes. Redox properties and thermodynamic stabilities of complexes of ‘hindered’ iron(III) and iron(II) tetraphenylporphyrinates with substituted pyridines and imidazoles. Inorg Chem. 1996;35:5188–5200. [Google Scholar]
- 20.Zhuang J, Amoroso JH, Kinloch R, Dawson JH, Baldwin MJ, Gibney BR. Evaluation of electron-withdrawing group effects on heme binding in designed proteins: implications for heme a in cytochrome c oxidase. Inorg Chem. 2006;45:4685–4694. doi: 10.1021/ic060072c. [DOI] [PubMed] [Google Scholar]
- 21.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaric SD, Popovic DM, Knapp EW. Factors determining the orientation of axially coordinated imidazoles in heme proteins. Biochemistry. 2001;40:7914–7928. doi: 10.1021/bi010428q. [DOI] [PubMed] [Google Scholar]
- 23.Huang SS, Koder RL, Lewis M, Wand AJ, Dutton PL. The HP-1 maquette: from an apoprotein structure to a structured hemoprotein designed to promote redox-coupled proton exchange. Proc Natl Acad Sci USA. 2004;101:5536–5541. doi: 10.1073/pnas.0306676101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reedy CJ, Gibney BR. Heme protein assemblies. Chem Rev. 2004;104:617–649. doi: 10.1021/cr0206115. [DOI] [PubMed] [Google Scholar]
- 25.Schneider S, Marles-Wright J, Sharp KH, Paoli M. Diversity and conservation of interactions for binding heme in b-type heme proteins. Nat Prod Rep. 2007;24:621–630. doi: 10.1039/b604186h. [DOI] [PubMed] [Google Scholar]
- 26.Reedy CJ, Elvekrog MM, Gibney BR. Development of a heme protein structure electrochemical function database. Nucleic Acids Res. 2008;36 (Database issue):D307–D313. doi: 10.1093/nar/gkm814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Degtyarenko KN, North AC, Findlay JB. PROMISE: a database of bioinorganic motifs. Nucleic Acids Res. 1999;27:233–236. doi: 10.1093/nar/27.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castagnetto JM, Hennessy SW, Roberts VA, Getzoff ED, Tainer JA, Pique ME. MDB: the metalloprotein database and browser at the scripps research institute. Nucleic Acids Res. 2002;30:379–382. doi: 10.1093/nar/30.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin ACR, Orengo CA, Hutchinson EG, Jones S, Karmirantzou M, Laskowski RA, Mitchell JBO, Taroni C, Thornton JM. Protein folds and function. Structure. 1998;6:875–884. doi: 10.1016/s0969-2126(98)00089-6. [DOI] [PubMed] [Google Scholar]
- 30.Wallace CJ, Clark-Lewis I. Functional role of heme ligation in cytochrome c. Effects of replacement of methionine 80 with natural and non-natural residues by semisynthesis. J Biol Chem. 1992;267:3852–3861. [PubMed] [Google Scholar]
- 31.Walker FA. Models of the bis-histidine-ligated electron-transferring cytochromes. Comparative geometric and electronic structure of low-spin ferro- and ferrihemes. Chem Rev. 2004;104:589–615. doi: 10.1021/cr020634j. [DOI] [PubMed] [Google Scholar]
- 32.Hobbs JD, Shelnutt JA. Conserved nonplanar heme distortions in cytochromes c. J prot chem. 1995;14:19–25. doi: 10.1007/BF01902840. [DOI] [PubMed] [Google Scholar]
- 33.Shelnutt JA, Song X-Z, Ma J-G, Jia S-L, Jentzen W, Medforth CJ. Nonplanar porphyrins and their significance in proteins. Chem Soc Rev. 1998;27:31–41. [Google Scholar]
- 34.Scheidt W, Lee Y. Recent advances in the stereochemistry of metal-lotetrapyrroles. In: Buchler JW, editor. Metal complexes with tetrapyrrole ligands I. Berlin/Heidelberg: Springer; 1987. pp. 1–70. [Google Scholar]
- 35.Galstyan AS, Zaric SD, Knapp EW. Computational studies on imidazole heme conformations. J Biol Inorg Chem. 2005;10:343–354. doi: 10.1007/s00775-005-0642-8. [DOI] [PubMed] [Google Scholar]
- 36.Valentine JS, Sheridan RP, Allen LC, Kahn PC. Coupling between oxidation state and hydrogen bond conformation in heme proteins. Proc Natl Acad Sci USA. 1979;76:1009–1013. doi: 10.1073/pnas.76.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Y, Chien EY, Sligar SG, La Mar GN. Influence of proximal side mutations on the molecular and electronic structure of cyanomet myoglobin: an 1H NMR study. Biochemistry. 1998;37:6979–6990. doi: 10.1021/bi9728295. [DOI] [PubMed] [Google Scholar]
- 38.Bertini I, Luchinat C, Parigi G, Walker FA. Heme methyl 1H chemical shifts as structural parameters in some low-spin ferriheme proteins. J Biol Inorg Chem. 1999;4:515–519. doi: 10.1007/s007750050337. [DOI] [PubMed] [Google Scholar]
- 39.Walker FA. The heme environment of mouse neuroglobin: histidine imidazole plane orientations obtained from solution NMR and EPR spectroscopy as compared with X-ray crystallography. J Biol Inorg Chem. 2006;11:391–397. doi: 10.1007/s00775-006-0095-8. [DOI] [PubMed] [Google Scholar]
- 40.Munro OQ, Serth-Guzzo JA, Turowska-Tyrk I, Mohanrao K, Shokhireva TK, Walker FA, Debrunner PG, Scheidt WR. Two crystalline forms of low-spin [Fe(TMP)(5-MeHIm)2]ClO4. Relative parallel and perpendicular axial ligand orientations. J Am Chem Soc. 1999;121:11144–11155. [Google Scholar]
- 41.Ghosh A, Gonzalez E, Vangberg T. Theoretical studies of low-spin six-coordinate iron(III) porphyrins relevant to cytochromes b: variable electronic configurations, ligand noninnocence, and macrocycle ruffling. J Phys Chem B. 1999;103:1363–1367. [Google Scholar]
- 42.Smith DM, Rosso KM, Dupuis M, Valiev M, Straatsma TP. Electronic coupling between heme electron-transfer centers and its decay with distance depends strongly on relative orientation. J Phys Chem B Condens Matter Mater Surf Interfaces Biophys. 2006;110:15582–15588. doi: 10.1021/jp057068r. [DOI] [PubMed] [Google Scholar]
- 43.Scheidt WR, Chipman DM. Preferred orientation of imidazole ligands in metalloporphyrins. J Am Chem Soc. 1986;108:1163–1167. [Google Scholar]
- 44.Scheidt WR, Kirner JF, Hoard JL, Reed CA. Unusual orientation of axial ligands in metalloporphyrins. molecular structure of low-spin bis(2-methylimidazole)(meso-tetraphenylporphinato)iron(III) perchlorate. J Am Chem Soc. 1987;109:1963–1968. [Google Scholar]
- 45.Grzybowski BA, Ishchenko AV, DeWitte RS, Whitesides GM, Shakhnovich EI. Development of a knowledge-based potential for crystals of small organic molecules: calculation of energy surfaces for C=0···H–N hydrogen bonds. J Phys Chem B. 2000;104:7293–7298. [Google Scholar]
- 46.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 47.Morozov AV, Kortemme T, Tsemekhman K, Baker D. Close agreement between the orientation dependence of hydrogen bonds observed in protein structures and quantum mechanical calculations. Proc Natl Acad Sci USA. 2004;101:6946–6951. doi: 10.1073/pnas.0307578101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berman HM, Henrick K, Nakamura H. Announcing the worldwide protein data bank. Nat Struct Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 49.Wang G, Dunbrack RL., Jr PISCES: recent improvements to a PDB sequence culling server. Nucleic Acids Res. 2005;33:W94–W98. doi: 10.1093/nar/gki402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fufezan C. 2007 http://www.fufezan.net/heme_survey.php.
- 51.Alexov EG, Gunner MR. Incorporating protein conformational flexibility into the calculation of pH-dependent protein properties. Biophys J. 1997;72:2075–2093. doi: 10.1016/S0006-3495(97)78851-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rocchia W, Alexov E, Honig B. Extending the applicability of the nonlinear Poisson-Boltzmann equation: multiple dielectric constants and multivalent ions. J Phys Chem B. 2001;105:6507–6514. [Google Scholar]
- 53.Schrodinger LLC, Portland, OR. http://www.schrodinger.com/rProductDescription.php?mID=6&sID=9&cID=0.
- 54.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 55.QuantumSoft. http://www.quansoft.com.
- 56.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;1433–38:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 57.Mowat CG, Rothery E, Miles CS, McIver L, Doherty MK, Drewette K, Taylor P, Walkinshaw MD, Chapman SK, Reid GA. Octaheme tetrathionate reductase is a respiratory enzyme with novel heme ligation. Nat Struct Mol Biol. 2004;11:1023–1024. doi: 10.1038/nsmb827. [DOI] [PubMed] [Google Scholar]
- 58.Einsle O, Stach P, Messerschmidt A, Simon J, Kroger A, Huber R, Kroneck PM. Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 A resolution, inhibitor binding, and heme-packing motifs. J Biol Chem. 2000;275:39608–39616. doi: 10.1074/jbc.M006188200. [DOI] [PubMed] [Google Scholar]
- 59.Kloer DP, Hagel C, Heider J, Schulz GE. Crystal structure of ethyl-benzene dehydrogenase from Aromatoleum aromaticum. Structure. 2006;14:1377–1388. doi: 10.1016/j.str.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 60.Rodrigues ML, Oliveira TF, Pereira IA, Archer M. X-ray structure of the membrane-bound cytochrome c quinol dehydrogenase NrfH reveals novel haem coordination. Embo J. 2006;25:5951–5960. doi: 10.1038/sj.emboj.7601439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mao J, Hauser K, Gunner MR. How cytochromes with different folds control heme redox potentials. Biochemistry. 2003;42:9829–9840. doi: 10.1021/bi027288k. [DOI] [PubMed] [Google Scholar]
- 62.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A Program for macromolecular energy, minimization, and dynamics calculations. J comput chem. 1983;4:187–217. [Google Scholar]
- 63.Gunner MR, Alexov E, Torres E, Lipovaca S. The importance of the protein in controlling the electrochemistry of heme metalloproteins: methods of calculation and analysis. J Biol Inorg Chem. 1997;2:126–134. [Google Scholar]
- 64.Song Y, Mao J, Gunner MR. Calculation of proton transfers in Bacteriorhodopsin bR and M intermediates. Biochemistry. 2003;42:9875–9888. doi: 10.1021/bi034482d. [DOI] [PubMed] [Google Scholar]
- 65.Discher BM, Koder RL, Moser CC, Dutton PL. Hydrophilic to amphiphilic design in redox protein maquettes. Curr Opin Chem Biol. 2003;7:741–748. doi: 10.1016/j.cbpa.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 66.Hecht MH, Das A, Go A, Bradley LH, Wei Y. De novo proteins from designed combinatorial libraries. Prot Sci. 2004;13:1711–1723. doi: 10.1110/ps.04690804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haehnel W. Chemical synthesis of TASP arrays and their application in protein design. Mol Divers. 2004;8:219–229. doi: 10.1023/b:modi.0000036252.33928.14. [DOI] [PubMed] [Google Scholar]
- 68.Tezcan FA, Winkler JR, Gray HB. Effects of ligation and folding on reduction potentials of heme proteins. J Am Chem Soc. 1998;120:13383–13388. [Google Scholar]
- 69.Barker PD, Ferguson SJ. Still a puzzle: why is haem covalently attached in c-type cytochromes? Structure. 1999;7:R281–R290. doi: 10.1016/s0969-2126(00)88334-3. [DOI] [PubMed] [Google Scholar]
- 70.Koder RL, Valentine KG, Cerda J, Noy D, Smith KM, Wand AJ, Dutton PL. Nativelike structure in designed four alpha-helix bundles driven by buried polar interactions. J Am Chem Soc. 2006;128:14450–14451. doi: 10.1021/ja064883r. [DOI] [PubMed] [Google Scholar]
- 71.Barrick D. Replacement of the proximal ligand of sperm whale myoglobin with free imidazole in the mutant His-93→Gly. Biochemistry. 1994;33:6546–6554. doi: 10.1021/bi00187a023. [DOI] [PubMed] [Google Scholar]
- 72.Finzel BC, Weber PC, Hardman KD, Salemme FR. Structure of ferricytochrome c’ from Rhodospirillum molischianum at 1.67 A resolution. J Mol Biol. 1985;186:627–643. doi: 10.1016/0022-2836(85)90135-4. [DOI] [PubMed] [Google Scholar]
- 73.Walker FA, Boi Hanh H, Scheidt WR, Osvath SR. Models of the cytochromes b. Effect of axial ligand plane orientation on the EPR and Moessbauer spectra of low-spin ferrihemes. J Am Chem Soc. 1986;108:5288–5297. [Google Scholar]
- 74.Walker FA, Reis D, Balke VL. Models of the cytochromes b. 5. EPR studies of low-spin iron(III) tetraphenylporphyrins. J Am Chem Soc. 1984;106:6888–6898. [Google Scholar]
- 75.Migita CT, Iwaizumi M. Low-temperature EPR studies of highly anisotropic low-spin (protoporphyrinato)iron(III) complexes. J Am Chem Soc. 1981;103:4378–4381. [Google Scholar]
- 76.T’Sai AL, Palmer G. Purification and characterization of highly purified cytochrome b from complex III of baker’s yeast. Biochim Biophys Acta. 1982;681:484–495. doi: 10.1016/0005-2728(82)90191-8. [DOI] [PubMed] [Google Scholar]
- 77.Salerno JC, McGill JW, Gerstle GC. The electron paramagnetic resonance spectra of partially purified cytochrome b6f complex from spinach. FEBS Lett. 1983;162:257–261. [Google Scholar]
- 78.Palmer G. The electron paramagnetic resonance of metalloproteins. Biochem Soc Trans. 1985;13:548–560. doi: 10.1042/bst0130548. [DOI] [PubMed] [Google Scholar]
- 79.Weiss MS, Brandl M, Suhnel J, Pal D, Hilgenfeld R. More hydrogen bonds for the (structural) biologist. Trends Biochem Sci. 2001;26:521–523. doi: 10.1016/s0968-0004(01)01935-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.