Abstract
Background
There is increasing evidence that solid cancers contain cancer-initiating cells (CICs) that are capable of regenerating a tumor that has been surgically removed and/or treated with chemotherapy and/or radiation therapy. Currently, cell surface markers, like CD133 or CD44, are used to identify CICs in vitro; however, these markers cannot be used to identify and track CICs in vivo. The 26S proteasome is the main regulator of many processes within a proliferating cell, and its activity may be altered depending on the phenotype of a cell.
Methods
Human glioma and breast cancer cells were engineered to stably express ZsGreen fused to the carboxyl-terminal degron of ornithine decarboxylase, resulting in a fluorescent fusion protein that accumulates in cells in the absence of 26S proteasome activity; activities of individual proteases were monitored in a plate reader by detecting the cleavage of fluorogenic peptide substrates. Proteasome subunit expression in cells expressing the fusion protein was assessed by quantitative reverse transcription—polymerase chain reaction, and the stem cell phenotype of CICs was assessed by a sphere formation assay, by immunohistochemical staining for known stem cell markers in vitro, and by analyzing their tumorigenicity in vivo. CICs were tracked by in vivo fluorescence imaging after radiation treatment of tumor-bearing mice and targeted specifically via a thymidine kinase–degron fusion construct. All P values were derived from two-sided tests.
Results
Cancer cells grown as sphere cultures in conditions, which enrich for cancer stem cells (CSCs), had decreased proteasome activity relative to the respective monolayers (percent decrease in chymotryptic-like activity of sphere cultures relative to monolayers—U87MG: 26.64%, 95% confidence interval [CI] = 10.19 to 43.10, GL261, 52.91%, 95% CI = 28.38 to 77.43). The cancer cells with low proteasome activity can thus be monitored in vitro and in vivo by the accumulation of a fluorescent protein (ZsGreen) fused to a degron that targets it for 26S proteasome degradation. In vitro, ZsGreen-positive cells had increased sphere-forming capacity, expressed CSC markers, and lacked differentiation markers compared with ZsGreen-negative cells. In vivo, ZsGreen-positive cells were approximately 100-fold more tumorigenic than ZsGreen-negative cells when injected into nude mice (ZsGreen positive, 30 mice per group; ZsGreen negative, 31 mice per group), and the number of CICs in tumors increased after 72 hours post radiation treatment. CICs were selectively targeted via a proteasome-dependent suicide gene, and their elimination in vivo led to tumor regression.
Conclusion
Our results demonstrate that reduced 26S proteasome activity is a general feature of CICs that can easily be exploited to identify, track, and target them in vitro and in vivo.
Context and Caveats
Prior knowledge
It had become evident that in many solid cancers there are small subpopulations of cells with stem cell–like properties known as cancer initiating cells (CICs) (or cancer stem cells [CSCs]) that are relatively resistant to conventional cancer therapies. Methods to identify and track these cells in vivo were lacking.
Study design
Cancer cells were engineered to express a fluorescent protein that is a target of the 26S protesome, a multiprotein complex which appeared to have reduced proteolytic activity in CICs. The correlation of various CIC/CSC phenotypes in the engineered cells with fluorescence, and thus 26S proteosome activity, was assessed.
Contribution
This study found that reduced 26S proteosome activity was closely correlated with CIC phenotypes in glioma and breast cancer cells. Thus, engineering cells to express a substrate of the protease is a viable method to identify and track these cell populations in vivo.
Implications
The ability to identify and track CICs in animal models of cancer may allow better assessment of therapeutic approaches compared to conventional methods such as measuring tumor response.
Limitations
The CICs with low protease activity may themselves be a heterogenous population of cells that needs to be further defined.
From the Editors
Cancer cell propagation in vivo has been explained by the “stochastic model” (1,2), which claims that every cancer cell in a tumor can ultimately acquire a capacity for self-renewal and multilineage potency so that it can repopulate an entire tumor. The stochastic model of cancer has long been regarded as the only working model of cancer organization, largely because until recently it has not been possible to prospectively isolate the very small population of cancer stem cells (CSCs) from the bulk of an unselected tumor cell population for characterization. In addition, working with unselected cell populations is inexpensive and experimental responses of the bulk of these cells can be easily used as a rationale for pharmaceutical approaches against cancer. Unfortunately, and despite an enormous effort, these approaches based on the behavior of unselected cell populations, even those termed targeted cancer therapies, have not yet resulted in cancer cures.
An alternative view of cancer cell propagation that is now supported by an increasing body of experimental evidence is the hierarchical model (3). It assumes that most, if not all, solid cancers are characterized by a hierarchical organization in which there are small populations of cancer stem cells (CSCs) or cancer-initiating cells (CICs) that are capable of repopulating an entire tumor, but whose progeny lacks this ability (1). Although this model has long been postulated, it has only recently become possible to provide supporting experimental evidence because of technological advances, such as the availability of sophisticated fluorescence-activated cells sorting (FACS) instruments, which are capable of efficiently isolating viable populations of rare cells from the bulk of tumor cells based on their expression of several cell surface markers. An increasing number of cell surface markers have been used to distinguish CSCs and CICs from the bulk of the tumor cells and to demonstrate their self-renewal capacity, multilineage potency, lack of expression of differentiation markers, and increased tumorigenicity when injected into immune-deficient mice (4–10). CSCs/CICs identified in this way have been shown to be relatively resistant to conventional anticancer therapies, such as chemotherapy and/or radiation therapy (11–13), and there is now strong evidence to suggest that a successful cancer treatment strategy will have to be based on the elimination of this cell population via novel therapeutic approaches. Thus, although it is becoming increasingly important to identify, track, and target CSCs/CICs in vivo, available markers are not very suitable for the purpose. A reliable system that would allow identification and tracking of CSCs/CICs would be an invaluable tool to study the effects of established and novel therapies on CSCs/CICs, but one that requires identification of cellular factors specific to this population.
The 26S proteasome is a multicatalytic protease complex with at least three distinct kinds of proteolytic activities—chymotrypsin like, trypsin like, and caspase like. It accounts for almost 1% of total protein expressed in eukaryotic cells and is a key regulator of many cellular functions, including cell cycle control, DNA repair, cell death, and survival (14). Its inhibition causes apoptosis and sensitization of cells to chemotherapeutic agents (15) and ionizing radiation (16,17). Furthermore, conventional anticancer therapies, for example, ionizing radiation (18), chemotherapeutic drugs (19,20), and hyperthermia (21), inhibit the proteasome, suggesting that their anticancer activity may be mediated by effects on this protein complex.
A proteasome inhibitor, bortezomib, is in clinical use for patients suffering from multiple myeloma or mantle cell lymphoma (22). However, despite its excellent preclinical efficacy in animal models of other cancers, bortezomib failed to demonstrate antitumor activity as a single agent in patients with solid cancers in clinical trials, suggesting that CICs might not be affected by proteasome inhibition (23–27). The results of these clinical studies prompted us to investigate proteasome activity and subunit expression in CICs of solid cancers relative to that in monolayer cultures and to examine whether proteasome activity is a useful marker for CICs. To monitor proteasome activity in living cells, we generated cancer cell lines that stably expressed a fusion protein consisting of a fluorescent protein fused to a degron, a sequence that targets the fusion protein for destruction by the proteasome. We also generated cancer cell lines expressing the degron fused to the suicide gene thymidine kinase (TK) to specifically investigate the effect of eliminating cells with low proteasome activity on sphere-forming capacity and self-renewal in vitro and tumor growth in vivo.
Materials and Methods
Cell Culture
Human U87MG glioma cell line was a kind gift from Dr P. Michel (Department of Pathology, University of California, Los Angeles, Los Angeles, CA). Murine GL261 glioma and 67NR breast cancer cell lines were a kind gift from Dr Sandra DeMaria (Department of Pathology, New York University School of Medicine, New York, NY). Human U343 and MCF-7 breast cancer cell lines were purchased from American Type Culture Collection (Manassas, VA). All cells were cultured in log-growth phase in Dulbecco's Modified Eagle Medium (DMEM) (Invitrogen, Carlsbad, CA) (supplemented with 10% fetal bovine serum [Sigma, St Louis, MO] and penicillin and streptomycin cocktail [Sigma]) and were grown in a humidified incubator at 37°C at 5% CO2. To obtain CICs, MCF-7, 67NR, U87MG, U343, and GL261 cells were seeded into selection media (DMEM/F12, 0.4% bovine serum albumin (BSA) [Sigma], 10 mL per 500 mL B27 [Invitrogen], 5 μg/mL bovine insulin [Sigma], 4 μg/mL heparin [Sigma], 20 ng/mL fibroblast growth factor 2 [bFGF, Sigma], and 20 ng/mL epidermal growth factor [EGF, Sigma]) at a density of 1000 cells per mL. Under these conditions, only CICs and early progenitor cells survive and proliferate, whereas differentiated cells die (28).
Generation of Stable Cell Lines Expressing ZsGreen-cODC and TK-ZsGreen-cODC Fusion Proteins Using Retroviral Transduction
Proteasomal degradation of most proteins depends on their ubiquitination. However, a small number of proteins such as ornithine decarboxylase (ODC) contain amino acid sequences that are directly recognized by the proteasome, which leads to the immediate destruction of the proteins that contain them. Viral expression vectors in which the carboxyl terminus of the murine ornithine decarboxylase (cODC) degron was fused to reporter proteins were constructed as follows. ZsGreen-cODC: The degron from the carboxyl-terminal 37 amino acids of ODC fused to ZsGreen (ZsGreen-cODC) was digested with BglII and NotI from pZsProsensor-1 (BD Biosciences, San Jose, CA) and cloned into the BamHI and EcoRI sites of the retroviral vector pQCXIN (BD Biosciences) using the NotI-EcoRI DNA oligonucleotide adaptor (EZCLONE Systems, New Orleans, LA). TK-ZsGreen-cODC: ZsGreen-cODC was amplified from pZsProsensor-1 and cloned into the BamHI and EcoRI sites of pQCXIN vector (pQCXINBamH1-ZsGreen-ODC). The sequence of TK was amplified from pORF-HSVtk expression vector (InvivoGen, San Diego, CA) and cloned into pQCXINBamH1-ZsGreen-cODC using the NotI and BamHI sites (pQCXIN/TK-ZsGreen-ODC). pQCXIN/ZsGreen-cODC or pQCXIN/TK-ZsGreen-cODC was transfected into GP2-293 pantropic retroviral packaging cells (BD Biosciences). The retrovirus collected from the supernatant of the packaging cells was used to infect the different cell lines. Stable transfectants were selected with G418 (Invitrogen). The accumulation of ZsGreen-cODC protein was monitored by flow cytometry (FL-1 channel). To determine that the cells not accumulating the ZsGreen-cODC protein still contained the expression vector, the cells were incubated with 50 μM of the proteasome inhibitor MG-132 (Calbiochem, San Diego, CA) for 4 hours and the accumulation of the ZsGreen-cODC protein due to proteasome inhibition was analyzed by flow cytometry.
Primary Sphere Formation Assay
ZsGreen-cODC–expressing U87MG cells were grown in selection media as sphere cultures (primary spheres) and were sorted into ZsGreen-negative and -positive populations by FACS. Cells were defined as “ZsGreen positive” if the fluorescence in the FL-1 channel exceeded the fluorescence of nontransfected control cells by at least three orders of magnitude. The two cell populations were plated in selection media into 96-well plates, ranging from 1 to 100 cells per well. Growth factors, EGF and bFGF, were added every 3 days, and the cells were allowed to form spheres (secondary spheres) for 7–10 days. The number of spheres formed per well was then counted and expressed as a percentage of the initial number of cells plated. Four independent experiments were performed.
Proteasome Function Assays
Chymotryptic, tryptic, and caspase proteasome activities were measured as described previously (29) with a few minor modifications. MCF-7, 67NR, U87MG, U343, and GL261 cells were washed with phosphate buffered saline (PBS) and pelleted by centrifugation. Glass beads and homogenization buffer (25 mM Tris [pH 7.5], 100 mM NaCl, 5 mM ATP, 0.2% [vol/vol] Nonidet P-40 and 20% glycerol) were added to the cells, and the mixtures were vortexed for 1 minute. Beads and cell debris were removed by centrifugation at 4°C. Protein concentration in the resulting crude cellular extracts was determined by the Micro (bicinchoninic acid) protocol (Pierce, Rockford, IL) with BSA (Sigma) as standard. To measure 26S proteasome activity, 100 μg of protein from crude cellular extracts of each sample was diluted with buffer I (50 mM Tris [pH 7.4], 2 mM dithiothreitol, 5 mM MgCl2, 2 mM ATP) to a final volume of 1 mL (assayed in quadruplicate). The fluorogenic proteasome substrates Suc-LLVY-AMC (chymotryptic substrate; Biomol International, Plymouth Meeting, PA), Z-ARR-AMC (tryptic substrate; Calbiochem), and Z-LLE-AMC (caspase-like substrate; Biomol International) were dissolved in dimethyl sulfoxide DMSO and added to a final concentration of 80 μM in 1% DMSO. Proteolytic activities were continuously monitored by measuring the release of the fluorescent group, 7-amido-4-methylcoumarin (AMC), with the use of a fluorescence plate reader (Spectramax M5, Molecular Devices, Sunnyvale, CA) at 37°C, at excitation and emission wavelengths of 380 and 460 nm, respectively.
Luminescence ATP Detection Assay
Single-cell suspensions derived from U87MG-ZsGreen-cODC monolayer cultures (1000 cells) or 3-day-old primary sphere cultures were plated into 96-well plates of DMEM or DMEM/F12 selection medium (100 μL per well). The proteasome inhibitor PS341 (kind gift of Julian Adams, Millennium Pharmaceuticals, Cambridge, MA) was added at the indicated concentrations. After 5 days of incubation, 40 μL of ATP-lite substrate (Perkin-Elmer, Waltham, MA) was added to each well and luminescence was measured immediately using a fluorescence plate reader (Spectramax M5).
Quantitative Reverse Transcription–Polymerase Chain Reaction
Total RNA was isolated from U87MG monolayer cells and from sorted ZsGreen-cODC–positive and –negative cells using TRIZOL Reagent (Invitrogen). Complementary DNA (cDNA) synthesis was carried out using TaqMan Reverse Transcription Reagents (Applied Biosystems, Foster City, CA). Quantitative polymerase chain reaction (PCR) of proteasome subunit cDNAs was performed in an iQ5 Real-Time PCR Detection System (Bio-Rad, Hercules, CA) using the 2× iQ SYBR Green Supermix (Bio-Rad). Ct for each gene was determined after normalization to GAPDH, and ΔΔ Ct was calculated relative to the designated reference sample. Gene expression values were then set equal to 2–ΔΔCt as described by the iQ5 Optical System Software (Bio-Rad). All PCR primers were synthesized by Invitrogen and designed to amplify human GAPDH and the human proteasome subunits Lmp2, Y, Mecl1, Z, Lmp7, X, 11S PA28alpha, 11S PA28beta, 19S ATPase PSMC1, and 19S non-ATPase PSMD4 (all primer sequences are provided in the Supplementary Methods, available online).
Mice
Nude (nu/nu), 6- to 8-week-old female mice originally from The Jackson Laboratories (Bar Harbor, ME) were rederived, bred, and maintained in a Defined Flora environment in the Association for the Assessment and Accreditation of Laboratory Animal Care International–accredited animal facilities of the Department of Radiation Oncology, University of California, Los Angeles (Los Angeles, CA) in accordance with all local and national guidelines for the care of animals. We used 74 mice for the in vivo experiments: 61 mice were used for the tumorgenicity experiments, 10 mice were implanted with U87-TK-ZsGreen-ODC tumors, and three mice were used to analyze the effect of fractionated radiation on U87MG-ZsGreen-ODC tumors.
Tumor Xenotransplantation and Tumorigenicity
U87MG-ZsGreen–high or U87MG-ZsGreen–negative cells derived from 5- to 6-day-old spheres and sorted by FACS were injected subcutaneously into both thighs of nude mice (106, 105, 104, 103, or 102 cells per inoculum), and each tumor was considered as the unit of analysis. The number of tumors used for each group is summarized in Table 1. Tumor growth was monitored on a weekly basis, and the mice were killed by CO2 asphyxiation when the tumor size reached the protocol guidelines requiring euthanasia (1.3 cm in diameter).
Table 1.
Enhanced tumor formation by cells with low levels of 26S proteasome*
| Fraction (%) of injected mice that developed tumors | |||
| No. of cells injected | Injected with ZsGreen-high cells | Injected with ZsGreen-negative cells | Injected with ZsGreen-negative cells, and excluding ZsGreen-positive tumors† |
| 102 | 1/4 (25%) | 0/3 | 0/3 |
| 103 | 8/11 (73%) | 1/4 (25%) | 0/3 |
| 104 | 10/11 (91%) | 5/9 (56%) | 2/6 (33%) |
| 105 | 4/4 (100%) | 7/10 (70%) | 4/7 (57%) |
| 106 | — | 4/5 (80%) | 2/3 (67%) |
The ZsGreen protein was fused to the murine ornithine decarboxylase degron and its accumulation was thus an indicator of low proteasome activity.
When the tumors that arose from injections with ZsGreen-negative cells resulted in macroscopically green tumors, they were excluded from the total number of tumors formed from the ZsGreen-negative population.
Fractionated Radiation
Subcutaneous tumors (average diameter of 1 cm) generated from implanting 1 × 106 cells derived from unselected U87MG-ZsGreen-cODC monolayers into the thighs of nude mice were irradiated with 3 Gy for 5 consecutive days, using a cobalt-60 source (dose rate 0.6 Gy/min). The thighs of anesthetized mice bearing the tumor were placed in a 5 × 5-cm radiation field of the cobalt-60 source, whereas the rest of the body was shielded. The mice were anesthetized at different time points after irradiation and imaged for the macroscopic presence of ZsGreen-positive cells in the tumors using the Maestro In-Vivo Imaging System (Cambridge Research & Instrumentation, Woburn, MA).
Immunocytochemistry
Spheres from U87MG-ZsGreen-cODC cells that were 5–6 days old were transferred onto glass slides by cytocentrifugation (Cytospin; Shandon Elliot, London, UK) and fixed with 4% formaldehyde. The cells were incubated in permeabilization buffer (10% saponin, 0.5% BSA, in PBS) for 10 minutes, followed by incubation with the following primary antibodies for 30 minutes at room temperature: rabbit anti-human nestin (1:500) (Abcam, Cambridge, MA), rabbit anti-human glial fibrillary acidic protein (GFAP) (1:1000) (Abcam), mouse anti-human TUJ-1 (1:1000) (Abcam), mouse anti-human Sox2 (10 μg/mL) (R&D Systems, Minneapolis, MN), or mouse anti-human Musashi-1 (10 μg/mL) (R&D Systems). For staining with the mouse anti-human 19S regulator non-ATPase subunit Rpn2 (1:200) (Biomol International), the slides were first incubated with 5% goat serum in PBS and Triton to block nonspecific antibody binding, and then with the primary antibody overnight at 4°C. The secondary antibodies tetramethyl rhodamine isothiocynate (TRITC)-conjugated goat anti-rabbit IgG [1:200], and TRITC-conjugated goat anti-mouse Fab [1:80], both from Sigma) were diluted in PBS and 1% BSA/0.5% saponin and incubated with the cells for 1 hour. Hoechst 33342 (5 μg/mL; Invitrogen) solution was added for nuclear staining. The slides were visualized with an Olympus IX71 inverted fluorescent microscope.
Immunohistochemistry
Tumors were removed from sacrificed mice and fixed in buffered formalin. The tissue was then embedded in paraffin and cut into 4-μm sections. The tissue sections were deparaffinized in xylene (2 × 5 minutes) and rehydrated in a graded series of ethanol solutions (100%, 90%, 75%, 50%, and 25%) for 5 minutes each, followed by a final rinse in PBS. Antigen retrieval was performed by incubating the sections with CAREZYME I-Trypsin solution (BioCare Medical, Concord, CA). The sections were incubated with 10% goat serum in PBS to block nonspecific binding. The primary antibodies, rabbit anti-human Ki67 (1:100) (Abcam), and mouse anti-human CD31 (10 μg/mL) (Abcam) were diluted in 1% BSA and PBS and incubated with the sections overnight at 4°C. TRITC-conjugated secondary antibodies (TRITC-conjugated goat anti-rabbit IgG [1:80], and TRITC-conjugated goat anti-mouse Fab [1:400], both from Sigma) diluted in 1% BSA and PBS were added for 1 hour. Hoechst 33342 was added for 10 minutes at room temperature. The sections were visualized using an Olympus IX71 inverted fluorescent microscope.
Statistical Analysis
All results are expressed as mean values with 95% confidence intervals. For the proteasome activity assays, normal distributions of the data were confirmed using a Kolmogorov–Smirnov test. All statistical comparisons used a two-sided paired Student's t-test. The test was applied to normalized data to compensate for the variance of measurements between biologically independent replicates of the same experiments. Statistical significance was defined as P ≤ .05.
Results
Proteasome Activities of CICs in Breast Cancer and Glioma
We recently reported that sublethal doses of ionizing radiation increased the number of CICs in breast cancer (13) and sought to prevent this increase by blocking cell cycle progression using proteasome inhibitors. The approach was ineffective because CICs derived from mammospheres (data not shown) or glioma neurospheres were very resistant to proteasome inhibition (Supplementary Figure 1, A and B, available online). To monitor 26S proteasome activity in living cells, we stably transduced murine and human glioma and breast cancer cells lines with a fusion of a fluorescent protein, ZsGreen, and the degron from the cODC using retroviral vectors. The cODC is recognized by the 26S proteasome in an ubiquitin-independent manner (30), thus leading to immediate degradation of the ZsGreen fluorescent protein. Untreated monolayer cultures of U87MG exhibited low background fluorescence in more than 94% of the cells, but very few cells (<4%) displayed high levels of ZsGreen expression (Figure 1, A). Treatment of cultures with proteasome inhibitors, such as MG-132, caused 100% of cells to express ZsGreen (data not shown). Interestingly, primary spheres that are derived from U87MG cells under serum-free conditions and which are highly enriched for CICs (31,32) were greatly enriched for ZsGreen-positive cells (Figure 1, B), indicating that CICs may have low 26S proteasome activity.
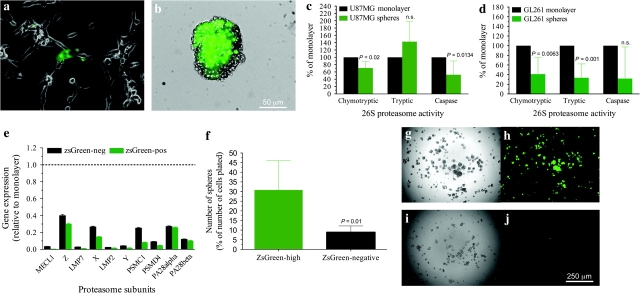
Figure 1.
Characterization of cancer-initiating cells based on their proteasome activity. A, B) Frequency of cells in U87MG monolayer cultures (A) and U87MG-derived primary spheres (B) with accumulation of ZsGreen-cODC, and thus low proteasome activity. C, D) Proteasome activities of the 26S proteasome in U87MG (C) and GL261 (D) monolayer and sphere cultures. Means ± 95% confidence intervals (CIs) derived from three to four independent experiments (four replicates per experiment). The Kolmogorov–Smirnov test was used to confirm the normal distribution of the data, and the Student's paired, two-tailed t-test were performed. E) Reverse transcription—polymerase chain reaction analysis of expression of mRNAs encoding proteasome components in ZsGreen-positive and ZsGreen-negative cells derived from spheres relative to expression in unselected monolayers (dotted line), three independent experiments. F) Number of spheres formed from the ZsGreen-high population vs the ZsGreen-negative population after sorting with flow cytometry into 96-well plates. Means ± 95% CIs from four independent experiments are shown. G, H) Secondary spheres derived from fluorescence-activated cells–sorted ZsGreen-high (G and H) and ZsGreen-negative cells (I, J). An image of bright field (left) and ZsGreen fluorescence (right) is shown for both populations.
To validate this observation, we performed fluorogenic proteasome function assays using monolayer and primary sphere cultures from U87MG human glioma and GL261 murine glioma cells (29). Chymotrypsin-like activity, the predominant activity of the 26S proteasome, as well as caspase-like activity were reduced in the sphere cultures, whereas the trypsin-like activity of U87MG cells was increased (percent decrease in proteasome activities of sphere cultures relative to monolayer cultures, mean of difference: chymotryptic-like activity, 26.64%, 95% CI = 10.19 to 43.10, P = .02, n = 3; trypsin-like activity, −39.46%, 95% CI = −88.18 to 9.266, P = .08, n = 4; caspase-like activity, 38.74%, 95% CI = 15.29 to 62.20, P = .01, n = 4; GL261: chymotryptic-like activity, 52.91%, 95% CI = 28.38 to 77.43, P = .006, n = 4; trypsin-like activity, 65.23%, 95% CI = 49.03 to 81.43, P = .001, n = 4; caspase-like activity, 52.88%, 95% CI = −18.22 to 124, P = .09, n = 3; Figure 1, C and D). Using the same fluorogenic peptide assays, reduced proteasome activity was further confirmed in U343 human glioma cells (proteasome activities of sphere cultures relative to the monolayer, mean of difference: chymotryptic-like activity, 37.74%, P = .01; trypsin-like activity, 26.25%, P = .03; caspase-like activity, 35.34%, P = .04; Supplementary Figure 1, C, available online), as well as in human and murine breast cancer cells, MCF-7 and 67NR (proteasome activities of sphere cultures relative to the monolayer, mean of difference—MCF-7: chymotryptic-like activity, 56.62%, P = .1; trypsin-like activity, 56.51%, P = .02; caspase-like activity, 36.92%, P = .041; 67NR: chymotryptic-like activity, 46.4%, P = .004; trypsin-like activity, 15.87%, P = .008; caspase-like activity, 55.45%, P = .055; Supplementary Figure 1, D and E, available online), indicating that reduced proteasome activity in CICs is a feature found across tumor entities of different species. Furthermore, quantitative reverse transcription—polymerase chain reaction revealed that several proteasome subunit mRNAs were decreased more than 100-fold in sphere cultures of CICs compared with cells in monolayer cultures (Figure 1, E). Confocal microscopy imaging for the Rpn2 subunit of the regulatory 19S cap of the 26S proteasome revealed a marked decrease in its expression in cells positive for ZsGreen (low proteasome activity), whereas cells lacking ZsGreen (high proteasome activity) expressed substantial levels of Rpn2 (Supplementary Figure 2, A–D, available online). Interestingly, a low 20S proteasome function has previously been reported for embryonic stem cells (33).
CICs, Proteasome Activity, and the CSC Phenotype
ZsGreen-high cells had a statistically significantly higher secondary sphere-forming capacity compared with the ZsGreen-negative population (average number of spheres formed, expressed as a percentage of the number of cells plated: ZsGreen-high cells versus ZsGreen-negative cells = 31% vs 9%; difference = 22%, 95% CI = 9.5% to 34.5%, P = .01, n = 4; Figure 1, F). The ZsGreen-high cells redistributed into ZsGreen-positive and -negative cells. By contrast, ZsGreen-negative cells formed only a few small spheres, which did not contain any ZsGreen-positive cells when analyzed by fluorescent microscopy (Figure 1, G–J).
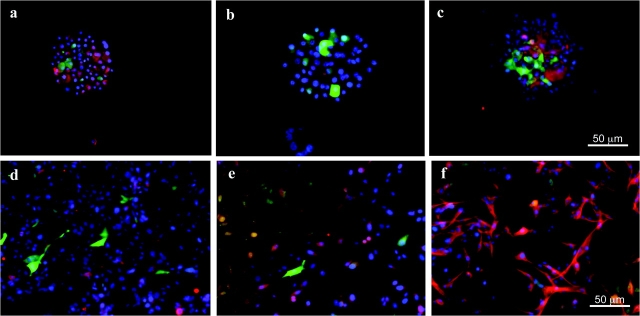
Further immunohistochemical characterization of the ZsGreen-positive cells derived from U87MG-ZsGreen-cODC spheres revealed that they were positive for the stem cell marker nestin (Figure 2, A, and Supplementary Figure 2, E, available online), which is a substrate of the 26S proteasome (34), and negative for the differentiation markers GFAP and neuron-specific class III β-tubulin (Figure 2, B and C). When U87MG-ZsGreen-cODC spheres were allowed to attach to the surface of the culture dishes and were exposed to serum-containing standard growth media (differentiating conditions), the number of ZsGreen-positive cells declined, and the attached cells became positive for GFAP and neuron-specific class III β-tubulin, indicating that they had undergone differentiation (Figure 2, D–F). Similarly, spheres derived from the U343 human glioma cells stably expressing the ZsGreen-cODC reporter also expressed stem cell markers, such as Musashi-1 and Sox2 (Supplementary Figure 2, F and G, available online), in the ZsGreen-positive cells, whereas the ZsGreen-negative cells expressed the differentiation markers GFAP and TUJ-1 (Supplementary Figure 2, H and I, available online).
Figure 2.
Cancer stem cell phenotype of cancer cells with low proteasome activity. A–C) Cytospins of U87MG-ZsGreen-cODC–derived neurospheres. ZsGreen-positive cells are positive for the stem cell marker nestin (red, A) but negative for the differentiation markers glial fibrillary acide protein (red, B) and TUJ-1 staining of neuron-specific class III β-tubulin (red, C). D–F) U87MG-ZsGreen-cODC–derived neurospheres differentiated in the presence of fetal bavine serum. Cells lost expression of ZsGreen and nestin (red, D) but expressed the differentiation markers GFAP (red, E) and exhibited TUJ-1 staining of neuron-specific class III β-tubulin (red, F). Counterstaining was performed with 4’,6-diamidino-2-phenylindole (blue).
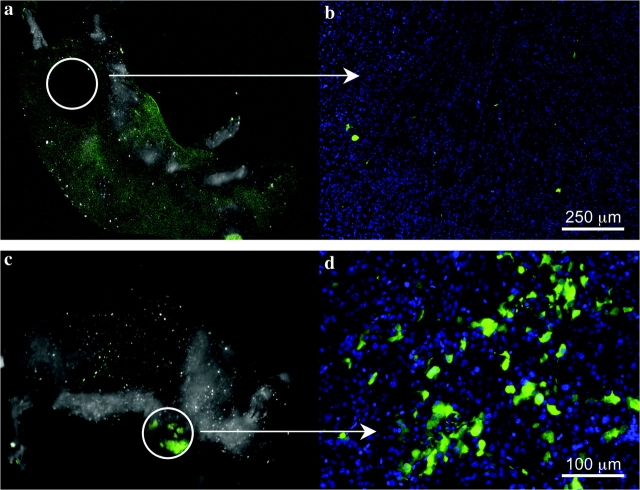
An important test for validating the identity of a CIC population is the demonstration of increased tumorigenicity in vivo. Therefore, the tumorigenicity of U87MG-ZsGreen-cODC cells was tested by first sorting them into ZsGreen-high and -negative populations using FACS. Different cell numbers from each population were injected subcutaneously into nude mice in numbers ranging from 102 to 106 cells per injection. Analysis of the sorted populations immediately after sorting revealed that 99.75% of ZsGreen-negative population were in fact negative for the fluorescent protein; thus, we estimated that one in 400 ZsGreen-negative cells injected was ZsGreen positive. Given that as few as 100 ZsGreen-positive cells could form a tumor (Table 1), we expected that some tumors would form in mice injected with at least 103 ZsGreen-negative cells and that those tumors would contain ZsGreen-positive cells. This was confirmed by in vivo fluorescent imaging, which demonstrated that some of the tumors arising from inoculation of sorted ZsGreen-negative cells contained ZsGreen-positive cells (Figure 3, C and D). Also, as mentioned above (Figure 1, G–J), ZsGreen-negative cells never repopulated into ZsGreen-positive cells in vitro; therefore, we concluded that formation of a ZsGreen-positive tumor in vivo from the injection of ZsGreen-negative cells was due to the less than 100% purity of this population after sorting. Mice were inspected for tumor formation on a daily basis, and the presence of ZsGreen-positive cells was monitored by in vivo imaging for all macroscopic tumors. When we excluded tumors deriving from ZsGreen-negative cells contaminated with ZsGreen-positive cells (visible in macroscopic imaging, Figure 3, C and D) from the total number of tumors formed, we found that the ZsGreen-positive cells exhibited approximately 100-fold increased tumorigenicity compared with the ZsGreen-negative cells, suggesting that glioma cells with reduced proteasome activity are highly enriched for CICs (Table 1).
Figure 3.
Tumorigenicity of cancer cells with low proteasome activity. A, B) ZsGreen-negative tumors derived from the ZsGreen-negative fraction of sorted sphere cells, viewed macroscopically or at high magnification. C, D) ZsGreen-positive tumors due to the contaminating presence of ZsGreen-positive cells in the negative fraction.
To examine if the ZsGreen-cODC reporter could be used to identify CICs in tumors, we stained sections of tumors derived from monolayer cultures of U87MG-ZsGreen-cODC cells with antibodies against the endothelial marker CD31 to identify vasculature. ZsGreen-positive cells were mainly found in perivascular regions and were absent in necrotic or avascular areas of the tumor (Supplementary Figure 3, A–C, available online). This observation was consistent with a previous report by Calabrese et al. (35), showing brain tumor stem cells to reside in a perivascular niche. In this context, it is important to note that the percentage of ZsGreen-positive cells in U87MG-ZsGreen-cODC tumors greatly exceeded the low percentage of ZsGreen-positive cells seen in vitro, which is consistent with earlier reports showing higher numbers of CICs in vivo than might be expected from in vitro observations (31).
Tracking and Targeting of CICs In Vivo Based on Proteasome Activity
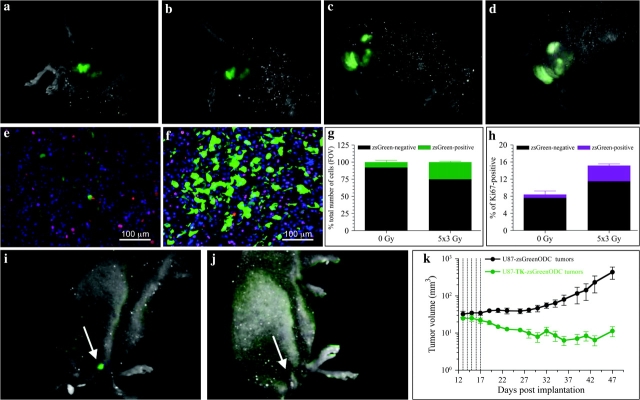
These findings motivated us to test if this reporter could be used to track CICs by in vivo imaging. We recently showed that daily local fractionated irradiation (5 × 3 Gy) causes an increase in CICs numbers in vitro (13) and hypothesized that this increase was the mechanism for accelerated repopulation (36). Therefore, we mimicked this schedule in vivo. Imaging of U87MG-ZsGreen-cODC tumor-bearing mice revealed a steady increase in the fluorescent signal, which was consistent with an increase in the ZsGreen-positive cell population (Figure 4, A–D). The fact that the increase in ZsGreen fluorescence was seen 72 hours after the last fraction of radiotherapy reflects changes only in ZsGreen-positive cell numbers and not general proteasome inhibition by the radiation (18). Furthermore, the location of the positive cells near blood vessels indicated that hypoxia-induced proteasome inhibition was not a factor in the increase in ZsGreen-positive cells. These experiments demonstrate that CICs in a tumor can be imaged macroscopically in vivo and that their response to radiation therapy can be followed temporally.
Figure 4.
The effect of fractionated radiation on cancer-initiating cells in vivo. A–D) U87MG-ZsGreen-cODC–expressing tumors subjected to fractionated radiation and imaged before treatment (A), after a 3-Gy radiation exposure (B), after 5- × 3-Gy radiation exposures (C), or 72 hours after the last fraction (D). E, F) High-magnification views of untreated tumors (E), in which the proliferating, Ki67-positive population of cells displays high proteasome activity with only a few low proteasome activity (ZsGreen-positive) cells, and tumors treated with daily fractions of 3 Gy (F), in which the number of ZsGreen-positive cells increased substantially (G) as did the percentage of cells that were positive for Ki67 (H). Counterstaining with 4’,6-diamidino-2-phenylindole (blue). Mean values and 95% confidence intervals (CIs) are shown for two independent experiments. I–K) Mice with tumors derived from U87MG cells expressing a fusion protein of thymidine kinase, ZsGreen and the carboxyl terminus of the murine ornithine decarboxylase degron, treated with ganciclovir (5 intraperitoneal injections of 50 mg/kg starting on day 12 after implantation [I] and 18 days after initiation of treatment [J]). K) Growth of the tumors in the mice treated with ganciclovir. Tumor volume (mm3) was assessed with calipers and are shown as means ± 95% CIs (n = 5 mice per group).
The localization of ZsGreen-positive cells adjacent to blood vessels, which is traditionally thought of as a proliferative zone (37), prompted us to test if these cells were indeed proliferating, using Ki67 staining as a marker. Only a minority of ZsGreen-positive cells was also Ki67 positive (Figure 4, E). However, after sublethal fractionated irradiation (5 × 3 Gy) of tumor-bearing mice, the number of ZsGreen-positive cells in tumors increased (Figure 4, F and G) and the number of double-positive (ZsGreen-positive and Ki67-positive) cells tripled (n = 2) (Figure 4, H), suggesting that accelerated tumor repopulation may derive from the ZsGreen-positive cell compartment.
To explore if constitutive reduction of proteasome activity in CICs could be exploited therapeutically, we expressed a fusion protein consisting of TK, ZsGreen, and the cODC degron in U87MG cells. These cells were tested for sphere-forming capacity (self-renewal) in vitro and tumor formation in vivo in the presence or absence of the TK substrate ganciclovir, which would be expected to eliminate ZsGreen-positive cells accumulating the ZsGreen-TK-cODC fusion protein. Treatment with ganciclovir selectively killed ZsGreen-positive cells, thus abrogating the sphere-forming capacity in vitro (Supplementary Figure 4, A–E, available online), and caused tumor regression in vivo (mean tumor volumes on day 48 post tumor implantation: ZsGreen-ODC tumors = 612.2 mm3, n = 5, TK-ZsGreen-ODC tumors = 9.495 mm3, n = 6; difference = 602.7 mm3, 95% CI = 164.3 to 1041 mm3, P = .0125, Student's t-test) (Figure 4, I–K).
Discussion
Here, we provide evidence that constitutively reduced 26S proteasome activity is a general feature of CICs in glioma and breast cancer cells. Consistent with this finding, Rpn2, which is thought to feed substrates into the 20S core particle (38), was nearly absent in CICs. Like normal stem cells, CICs are usually considered quiescent (39,40) with low protein turnover and reduced metabolism and thus they may not need an ATP-dependent protein degradation machinery (41).
Using a reporter construct for 26S proteasome activity, we were able to identify CICs in cell populations in vitro and in vivo. In vivo, these cells localized around blood vessels (Supplementary Figure 3, available online) consistent with a recent report by Calabrese et al. (35) showing that brain tumor stem cells reside in a perivascular niche.
Reduced proteasome activity coincided with the expression of stem cell markers and a lack of differentiation markers (Supplementary Figure 2, available online). Cells with low proteasome activity also showed increased self-renewal capacity and could form tumors in immunologically incompetent mice from as few as 100 cells (Figure 1 and Table 1). Taken together, our results indicate that the population of cells identified by reduced proteasome activity was identical or overlapped with CICs and that reduced 26S proteasome activity is a property of CICs that can cross species barriers.
Our study has several limitations. First, the fusion protein–negative cell populations in our study were not 100% pure, and purging of the remaining contaminating ZsGreen-positive cells via specific targeting with ganciclovir would have caused considerable toxicity. However, given this impurity, our study likely underestimates the difference in tumorgenicity between ZsGreen-positive and ZsGreen-negative cells. Second, we cannot rule out the possibility that ZsGreen-negative cells can eventually become ZsGreen-positive cells. Long-term experiments that follow single cells will be necessary to address this question. Finally, like all other investigators to our knowledge, we have required approximately 100 ZsGreen-positive cells to form tumors in vivo. Therefore, it is possible that ZsGreen-positive cells need to be further purified to obtain a pure CSC population.
Although the cell population with reduced proteasome activity was noncycling in vivo, it responded to fractionated doses of ionizing radiation by rapidly undergoing proliferation. This observation suggests that regulation of proteasome function in CICs can be turned on temporarily to allow progression through the cell cycle (a gain of proteasome function would not necessarily be detected by ZsGreen-cODC which has a half-life of >3.5 hours in cells). This switch in function may be highly dependent on developmental pathways as suggested by the fact that Notch signaling–dependent increases in stem cell numbers occur after sublethal doses of fractionated radiation in breast cancer (13) and may be an indicator of accelerated repopulation observed clinically in radiation therapy (an increased tumor growth rate during gaps in radiation treatment has been described in the context of clinical radiation therapy) (36). Because accelerated repopulation of tumors is a major cause of treatment failure, tracking of CICs in vivo may facilitate the search for novel therapeutic approaches that improve radiation therapy outcome. One possible application of being able to track CICs is the use of fusion of fluorescent proteins, suicide genes, and the cODC degron in gene therapy. Similar to our experiments with cells expressing the TK-ZsGreen-ODC fusion protein, the suicide gene can be substituted for the gene of interest, whereas the fluorescent component of the reporter will allow for the tracking of the cells accumulating the suicide protein, as well as their subsequent elimination. Using a TK-ZsGreen-cODC vector, we demonstrated that CICs could be targeted specifically which led to tumor control (Figure 4).
Our observations also offer a simple explanation for the high expression levels of known stem cell markers like BMI-1 and nestin in CICs that are both substrates of the proteasome (34,42). Furthermore, because the proteasome is also responsible for the generation of peptides presented to the immune system on Major histocompatibility complex -I molecules, our data suggest that CSCs may be immunologically silent or express antigens that may not be targeted by many current immunotherapy approaches. Finally, this system allows for screening of novel compounds that might modulate 26S proteasome function specifically in CICs, which could lead to novel targeted therapies against this therapeutically important cancer cell subpopulation.
Funding
E. Vlashi was supported by a training grant from the National Institute of Biomedical Imaging and BioEngineering (2 T32 EB002101-31). This work was supported by a grant from the Department of Defense (PC060599), by a grant of the California Breast Cancer Research Program (BC060077), and by a developmental grant from the In Vivo Cellular and Molecular Imaging Centre at University of California, Los Angeles (PI: H. Herschman, NIH P50CA08630608) to F. Pajonk.
Supplementary Material
Footnotes
The authors are grateful for the excellent technical support provided by the staff of the Defined Flora animal facility of the Department of Radiation Oncology at the University of California, Los Angeles. The authors take full responsibility for the study design; collection, analysis, and interpretation of the data; the decision to submit the manuscript for publication; and the writing of the manuscript.
References
- 1.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Lagasse E. Cancer stem cells with genetic instability: the best vehicle with the best engine for cancer. Gene Ther. 2008;15(2):136–142. doi: 10.1038/sj.gt.3303068. [DOI] [PubMed] [Google Scholar]
- 3.Park CY, Tseng D, Weissman IL. Cancer stem cell-directed therapies: recent data from the laboratory and clinic. Mol Ther. 2008 doi: 10.1038/mt.2008.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 6.Fang D, Nguyen TK, Leishear K, et al. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65(20):9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- 7.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65(23):10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 8.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445(7123):111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 9.Prince ME, Sivanandan R, Kaczorowski A, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA. 2007;104(3):973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67(3):1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 11.Eramo A, Ricci-Vitiani L, Zeuner A, et al. Chemotherapy resistance of glioblastoma stem cells. Cell Death Differ. 2006;13(7):1238–1241. doi: 10.1038/sj.cdd.4401872. [DOI] [PubMed] [Google Scholar]
- 12.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 13.Phillips TM, McBride WH, Pajonk F. The response of CD24(−/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98(24):1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 14.Pajonk F, McBride WH. The proteasome in cancer biology and treatment. Radiat Res. 2001;156(5):447–459. doi: 10.1667/0033-7587(2001)156[0447:tpicba]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 15.Cusack JC, Jr, Liu R, Houston M, et al. Enhanced chemosensitivity to CPT-11 with proteasome inhibitor PS-341: implications for systemic nuclear factor-kappaB inhibition. Cancer Res. 2001;61(9):3535–3540. [PubMed] [Google Scholar]
- 16.Pajonk F, Pajonk K, McBride WH. Apoptosis and radiosensitization of Hodgkin cells by proteasome inhibition. Int J Radiat Oncol Biol Phys. 2000;47(4):1025–1032. doi: 10.1016/s0360-3016(00)00516-2. [DOI] [PubMed] [Google Scholar]
- 17.Pajonk F, van Ophoven A, Weissenberger C, McBride WH. The proteasome inhibitor MG-132 sensitizes PC-3 prostate cancer cells to ionizing radiation by a DNA-PK-independent mechanism. BMC Cancer. 2005;5(1):76. doi: 10.1186/1471-2407-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pajonk F, McBride WH. Ionizing radiation affects 26s proteasome function and associated molecular responses, even at low doses. Radiother Oncol. 2001;59(2):203–212. doi: 10.1016/s0167-8140(01)00311-5. [DOI] [PubMed] [Google Scholar]
- 19.Fekete MR, McBride WH, Pajonk F. Anthracyclines, proteasome activity and multi-drug-resistance. BMC Cancer. 2005;5(1):114. doi: 10.1186/1471-2407-5-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piccinini M, Tazartes O, Mezzatesta C, et al. Proteasomes are a target of the anti-tumor drug vinblastine. Biochem J. 2001;356(pt 3):835–841. doi: 10.1042/0264-6021:3560835. Online Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pajonk F, van Ophoven A, McBride WH. Hyperthermia-induced proteasome inhibition and loss of androgen receptor expression in human prostate cancer cells. Cancer Res. 2005;65(11):4836–4843. doi: 10.1158/0008-5472.CAN-03-2749. [DOI] [PubMed] [Google Scholar]
- 22.Kane RC, Bross PF, Farrell AT, Pazdur R. Velcade: U.S. FDA approval for the treatment of multiple myeloma progressing on prior therapy. Oncologist. 2003;8(6):508–513. doi: 10.1634/theoncologist.8-6-508. [DOI] [PubMed] [Google Scholar]
- 23.Blaney SM, Bernstein M, Neville K, et al. Phase I study of the proteasome inhibitor bortezomib in pediatric patients with refractory solid tumors: a Children's Oncology Group study (ADVL0015) J Clin Oncol. 2004;22(23):4752–4757. doi: 10.1200/JCO.2004.12.185. [DOI] [PubMed] [Google Scholar]
- 24.Davis NB, Taber DA, Ansari RH, et al. Phase II trial of PS-341 in patients with renal cell cancer: a University of Chicago phase II consortium study. J Clin Oncol. 2004;22(1):115–119. doi: 10.1200/JCO.2004.07.165. [DOI] [PubMed] [Google Scholar]
- 25.Gomez-Abuin G, Winquist E, Stadler WM, et al. A phase II study of PS-341 (Bortezomib) in advanced or metastatic urothelial cancer. A trial of the Princess Margaret Hospital and University of Chicago phase II consortia. Invest New Drugs. 2007;25(2):181–185. doi: 10.1007/s10637-006-9009-4. [DOI] [PubMed] [Google Scholar]
- 26.Kondagunta GV, Drucker B, Schwartz L, et al. Phase II trial of bortezomib for patients with advanced renal cell carcinoma. J Clin Oncol. 2004;22(18):3720–3725. doi: 10.1200/JCO.2004.10.155. [DOI] [PubMed] [Google Scholar]
- 27.Shah MH, Young D, Kindler HL, et al. Phase II study of the proteasome inhibitor bortezomib (PS-341) in patients with metastatic neuroendocrine tumors. Clin Cancer Res. 2004;10(18 pt 1):6111–6118. doi: 10.1158/1078-0432.CCR-04-0422. [DOI] [PubMed] [Google Scholar]
- 28.Reynolds BA, Tetzlaff W, Weiss S. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J Neurosci. 1992;12(11):4565–4574. doi: 10.1523/JNEUROSCI.12-11-04565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glas R, Bogyo M, McMaster JS, Gaczynska M, Ploegh HL. A proteolytic system that compensates for loss of proteasome function. Nature. 1998;392(6676):618–622. doi: 10.1038/33443. [DOI] [PubMed] [Google Scholar]
- 30.Matsuzawa S, Cuddy M, Fukushima T, Reed JC. Method for targeting protein destruction by using a ubiquitin-independent, proteasome-mediated degradation pathway. Proc Natl Acad Sci USA. 2005;102(42):14982–14987. doi: 10.1073/pnas.0507512102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemmati HD, Nakano I, Lazareff JA, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100(25):15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ponti D, Costa A, Zaffaroni N, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65(13):5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 33.Hernebring M, Brolen G, Aguilaniu H, Semb H, Nystrom T. Elimination of damaged proteins during differentiation of embryonic stem cells. Proc Natl Acad Sci USA. 2006;103(20):7700–7705. doi: 10.1073/pnas.0510944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mellodew K, Suhr R, Uwanogho DA, et al. Nestin expression is lost in a neural stem cell line through a mechanism involving the proteasome and Notch signalling. Brain Res Dev Brain Res. 2004;151(1–2):13–23. doi: 10.1016/j.devbrainres.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 35.Calabrese C, Poppleton H, Kocak M, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11(1):69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 36.Withers HR, Maciejewski B, Taylor JM, Hliniak A. Accelerated repopulation in head and neck cancer. Front Radiat Ther Oncol. 1988;22:105–110. doi: 10.1159/000415101. [DOI] [PubMed] [Google Scholar]
- 37.Tannock IF. Population kinetics of carcinoma cells, capillary endothelial cells, and fibroblasts in a transplanted mouse mammary tumor. Cancer Res. 1970;30(10):2470–2476. [PubMed] [Google Scholar]
- 38.Rosenzweig R, Osmulski PA, Gaczynska M, Glickman MH. The central unit within the 19S regulatory particle of the proteasome. Nat Struct Mol Biol. 2008;15(6):573–580. doi: 10.1038/nsmb.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holtz MS, Forman SJ, Bhatia R. Nonproliferating CML CD34+ progenitors are resistant to apoptosis induced by a wide range of proapoptotic stimuli. Leukemia. 2005;19(6):1034–1041. doi: 10.1038/sj.leu.2403724. [DOI] [PubMed] [Google Scholar]
- 40.Holtz M, Forman SJ, Bhatia R. Growth factor stimulation reduces residual quiescent chronic myelogenous leukemia progenitors remaining after imatinib treatment. Cancer Res. 2007;67(3):1113–1120. doi: 10.1158/0008-5472.CAN-06-2014. [DOI] [PubMed] [Google Scholar]
- 41.Babbitt SE, Kiss A, Deffenbaugh AE, et al. ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle. Cell. 2005;121(4):553–565. doi: 10.1016/j.cell.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 42.Ben-Saadon R, Zaaroor D, Ziv T, Ciechanover A. The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol Cell. 2006;24(5):701–711. doi: 10.1016/j.molcel.2006.10.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.