Abstract
The successful use of small interfering RNAs (siRNAs) for therapeutic purposes requires safe and efficient delivery to specific cells and tissues. In this study, we demonstrate cell type–specific delivery of anti-human immunodeficiency virus (anti-HIV) siRNAs through fusion to an anti-gp120 aptamer. The envelope glycoprotein is expressed on the surface of HIV-1-infected cells, allowing binding and internalization of the aptamer–siRNA chimeric molecules. We demonstrate that the anti-gp120 aptamer–siRNA chimera is specifically taken up by cells expressing HIV-1 gp120, and that the appended siRNA is processed by Dicer; this releases an anti-tat/rev siRNA which, in turn, inhibits HIV replication. We show for the first time a dual functioning aptamer–siRNA chimera in which both the aptamer and the siRNA portions have potent anti-HIV activities. We also show that gp120 expressed on the surface of HIV-infected cells can be used for aptamer-mediated delivery of anti-HIV siRNAs.
INTRODUCTION
RNA interference (RNAi) is a process of sequence-specific post-transcriptional gene silencing triggered by small interfering RNAs (siRNAs). The silencing is sequence specific and one of the two strands of the siRNA guides the RNA-induced silencing complex (RISC) to the complementary target, resulting in cleavage and subsequent destruction of the target RNA.1 RNAi is rapidly becoming one of the methods of choice for gene function studies, and is also being exploited for therapeutic applications.2,3 The success of therapeutic applications of RNAi is critically dependent on efficient intracellular delivery of siRNAs.3 Currently, there are several methods of delivering siRNA in vivo. These can be classified as: (i) physical and mechanical methods (hydrodynamic tail vein injections in mice,4–6 electroporation,7–9 ultrasound,10 and the use of a gene gun11); (ii) local administration3 (intravenous injection,12 intraperitoneal injection, subcutaneous injection); and (iii) chemical methods (cationic lipids,13,14 polymers,15–20 and peptides21–24). However, the efficiency of delivery (desired dose), uncontrollable biodistribution, and delivery-related toxicitities need to be carefully analyzed. Recently, the cell type–specific delivery of siRNAs has been achieved using aptamer–siRNA chimeras.25 In this system, the aptamer portion of the chimera mediated binding to the prostate-specific membrane antigen (a cell-surface receptor), and the siRNAs linked to the aptamer were selectively delivered into prostate-specific membrane antigen–expressing cells, resulting in silencing of the target transcripts in both cell culture and in vivo after intratumoral delivery. In a similar study,26 a modular streptavidin bridge was used for connecting lamin A/C or glyceraldehyde-3-phosphate dehydrogenase siRNAs to the prostate-specific membrane antigen aptamer. Consequently, this system induced silencing of the targeted genes only in cells expressing the prostate-specific membrane antigen receptor.
In this study, we used the advantage offered by the gp120 glycoprotein27,28 binding properties of an anti-gp120 RNA aptamer to explore the potential of using this aptamer for delivery of anti-human immunodeficiency virus (anti-HIV) siRNAs into HIV-infected cells. On the basis of our own earlier studies,29,30 we tested the aptamer as a chimeric transcript with a Dicer substrate RNA duplex [25–30 nucleotides (nt)]. We utilized a 27-mer siRNA in which one strand is covalently attached to the aptamer and the second strand is base paired to the upper strand, and compared a 27-base pair double-stranded RNA with a 21-base pair double-stranded RNA fused to the aptamer in an analogous fashion. The binding of anti-gp120 aptamer to the R5 version of HIV-1 gp120 has been demonstrated earlier.31 is aptamer was shown to neutralize HIV-1 infectivity31–33 by directly binding to gp120 in virions. We wanted to determine whether the anti-gp120 aptamer could provide selective binding and subsequent internalization into HIV-infected cells that would be expressing gp120 on their surfaces. Although the aptamer, used by itself, provided some inhibitory function when tested in this setting, the siRNA chimeras provided more potent inhibition than the aptamer alone, thereby suggesting cooperation of action between the siRNA and aptamer portions in inhibiting the replication and spread of HIV. Our results demonstrate that the gp120 aptamer–siRNA chimeras are internalized in cells expressing gp120 either ectopically or from HIV infection, and, moreover, that the chimeric RNAs provide potent and lasting inhibition of HIV replication in T cells in culture. These results support the concept of using aptamer–siRNA conjugates for systemic treatment of HIV infection. This approach has the major advantage of not relying upon gene therapy, and the siRNAs can be changed or multiplexed to avert viral resistance.
Design of anti-gp120 aptamer–siRNA chimeras
In order to enhance the stability of the chimeric RNAs in cell culture and in vivo,4,34–37 the aptamer and sense-strand segment of the siRNAs contained nuclease-resistant 2′-fluoro uridine triphosphate and 2′-fluoro cytidine triphosphate, and were synthesized from corresponding double-stranded DNA templates by in vitro bacteriophage transcription (Figure 1). In order to prepare the siRNA-containing chimeras, in vitro–transcribed chimeric aptamer-sense strand polymers were annealed with equimolar concentrations of an unmodified antisense strand RNA. These 2′-fluoro-modified chimeras were stable in cell culture media for up to 48 hours whereas the unmodified control RNAs were quickly degraded within several minutes (data not shown).
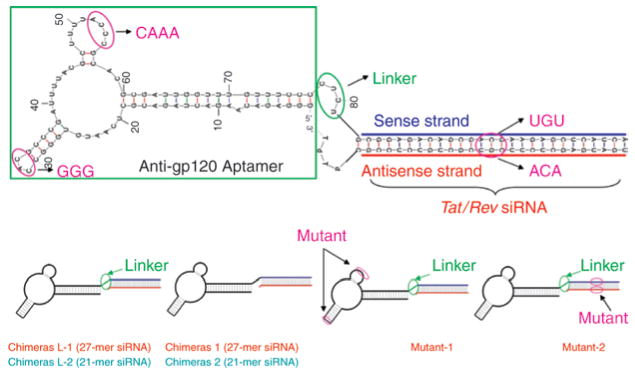
Figure 1. Predicted secondary structure of anti-gp120 aptamer–small interfering RNA (siRNA) chimeras.
The region of anti-gp120 aptamer responsible for binding to gp120 is outlined in green. The siRNA part of the chimera consists of 27 base pairs as an example here, targeting Site-I of human immunodeficiency type 1 tat/rev. Two mutated chimeras, M-1 (mutant aptamer) and M-2 (mutant siRNA), were constructed as experimental controls. Mutated regions are shown in magenta.
The gp120-binding aptamer that neutralizes R5 strains of HIV-1 has been earlier described and characterized.31 Given that the synthetic Dicer substrate duplexes of 25–30 nt have been shown to enhance RNAi potency and efficacy, we chose a 27-mer duplex RNA as the siRNA portion of our chimeric molecule. The 27-mer siRNA portion of the chimeras (Ch L-1 and Ch 1) targets the HIV-1 tat/rev common exon sequence. The designed chimeras, Ch L-2 and Ch 2, are identical to Ch L-1 and Ch 1 with the exception that the 27-mer duplex is replaced with a 21-base pair duplex. In the Ch L-1 and Ch L-2 designs, we inserted a 4-nt linker (CUCU) between the aptamer and siRNA portions to minimize steric interference of the aptamer portion with Dicer. Earlier studies using the anti-gp120 aptamer have identified the minimal region of the aptamer that is essential for binding gp120, and have shown that mutations within this region significantly lower the binding affinity. As controls for aptamer binding, we created the chimera designated as M-1. As a control for the siRNA-mediated silencing, we constructed an additional mutant in the siRNA portion, which is expected to abrogate RNAi-directed cleavage of the target, and this is designated as M-2.
Because of competition by the sense (passenger) strand with the antisense (guide) strand for RISC entry, the strand selectivity is an important factor in the evaluation of siRNAs. Therefore, we evaluated these chimeras RNAs using the siCHECK reporter system, which readily allows screening of the potencies of candidate short hairpin/siRNAs. The gene silencing of both the sense target (corresponding to the mRNA) and the antisense target were tested independently, and the selectivity ratios could be calculated as a measure of the relative incorporation of each strand into the RISC. The comparison (Supplementary Figure S1) demonstrated that the Ch L-1 succeeded in mediating ~86% knockdown of the sense target; however, the extent of knockdown of the antisense target was much less (~50%), which is indicative of good strand selection (R = 3.2). Ch 1 also indicated a similar extent of knockdown (~83%) of the sense target and strand selection (R = 2.9). In contrast, Ch L-2 and Ch 2 have poorer efficacy (<70%) and strand selectivity (R = 1.9 and 1.6, respectively). These data suggest that the aptamer–27-mer siRNA chimeras indeed enhance the RNAi efficacy and potency, in agreement with the results of earlier studies in our laboratory.29,30
Anti-gp120 aptamer–siRNA chimeras bind and are internalized by cells expressing HIV gp160
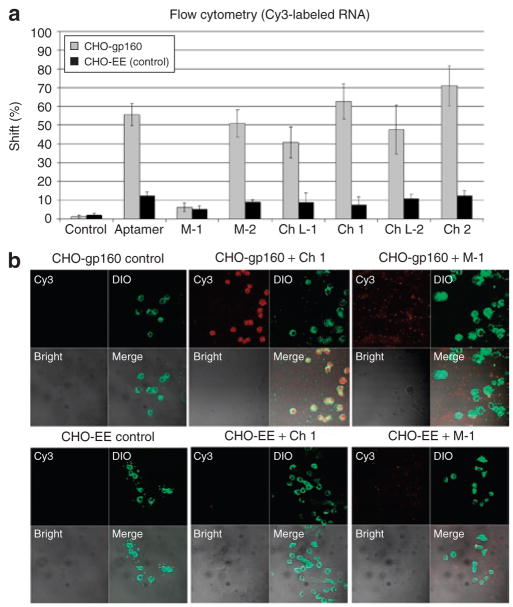
CHO-gp160 cells stably expressing the HIV envelope glycoprotein gp160 were used for testing uptake of the chimeric aptamer–siRNAs. These cells do not process gp160 into gp120 and gp41, because they lack the gag-encoded proteases required for envelope processing. As a control we used the parent CHO-EE cell line which does not express gp160. The anti-gp120 aptamer and the chimeras were labeled with Cy3 so that they could be followed to investigate their binding and potential internalization in gp160-expressing cells. Flow cytometric analyses (Figure 2a) revealed that the aptamer and chimeras did bind specifically to the CHO-gp160 cells but did not bind to the control CHO-EE cells. As anticipated, the M-1 dramatically reduced binding to the CHO-gp160-expressing cells.
Figure 2. Cell type–specific binding and internalization of the aptamer–small interfering RNA (siRNA) chimeras.
(a) Binding affinity assay. Cy3-labeled RNAs were tested for binding to CHO-gp160 cells and CHO-EE control cells. Cell surface binding of Cy3-labeled aptamer–siRNA chimeras was assessed using flow cytometry. (b) Binding and uptake of Ch 1 to CHO-gp160 cells. CHO-gp160 cells and CHO-EE control cells were grown on chamber slides and incubated with 20 nmol/l of Ch 1 in culture medium for 2 hours. The cells were washed in phosphate-buffered saline three times, fixed and stained with C18(3) 3,3′-diocta-decyloxacarbocyanine perchlorate (DIO) (a plasma membrane dye), washed again, and then analyzed using confocal microscopy.
In order to determine whether the bound aptamer and chimeras were internalized in the gp160-expressing cells, we carried out z-axis confocal microscopy and three-dimensional image reconstruction on the CHO-gp160 cells incubated with Cy3-labeled transcripts. Both the anti-gp120 aptamer (data not shown) and Ch 1 (Figure 2b) were selectively internalized within the CHO-gp160 cells but not within the CHO-EE control cells. The M-1 was also not internalized. Three-dimensional image reconstruction (Supplementary Figure S2) shows localization of the Cy3-labeled Ch 1 in a single cell. For visualizing the plasma membranes, the cells were stained with the carbocyanine dye C18(3) 3,3′-dioctadecyloxacarbocyanine perchlorate.
Anti-gp120 aptamer–siRNA chimeras are processed by Dicer
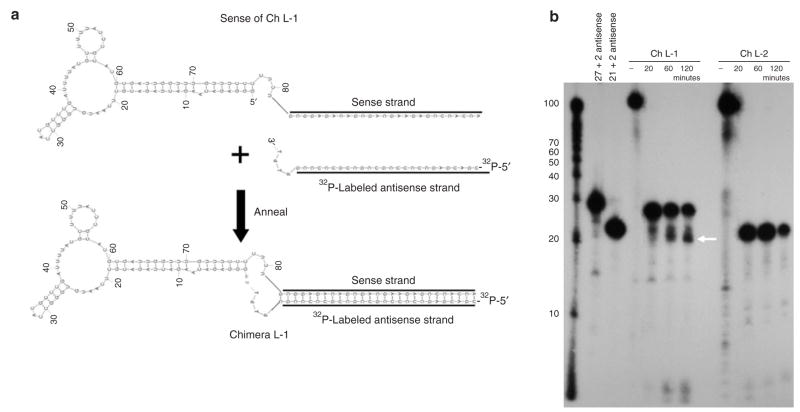
We next investigated whether the siRNAs could be processed from the chimeras using Dicer in whole cell extracts that contain good Dicer cleavage activity. The first set of experiments used a 5′-end 32P-labeled antisense strand to follow Dicer processing (Figure 3a). The size of the 32P-labeled cleavage product(s) indicates the direction from which Dicer enters the siRNA and achieves the cleavage. When Ch L-1 was incubated with the cytoplasmic lysate, we observed that the 27-nt antisense strand was processed into a 21–23-nt siRNA (Figure 3b). This result suggests that Dicer preferentially enters from the 5′-end of the antisense strand and cleaves 21–23 nt downstream, leaving the 5′-end of the antisense strand intact. In contrast, the 21-base siRNAs were not processed further in these extracts.
Figure 3. Analysis of chimera processing. 21–23-nucleotide (nt) RNA fragments are produced following incubation of chimera RNAs in HCT116 cell extracts.
(a) Chimera sense strands were annealed with equal molar equivalents of 5′-end 32P-labeled antisense oligos. (b) The cleavage products or denatured strands were visualized after denaturing polyacrylamide-gel electrophoresis. Note that the major Dicer product (marked by a white arrow) of the 27-mer aptamers is processed from the 5′-end of the antisense strand, as shown by the 21-base product harboring the 5′-32P label.
Anti-gp120 aptamer–siRNA chimeras specifically silence target gene expression
In order to evaluate whether these anti-gp120 aptamer–siRNA chimeras have the effect of triggering RNAi, we first transfected CHO-gp160 and CHO-EE cells with HIV pNL4-3 Luc. The HIV pNL4-3 has the firefly luciferase under the control of the HIV long-terminal repeat and is Tat responsive. The anti-tat/rev siRNA efficacy is monitored by inhibition of luciferase expression. Subsequent to the transfections, the cells were treated with the chimeras in the absence or presence of the transfection reagent Lipofectamine 2000.
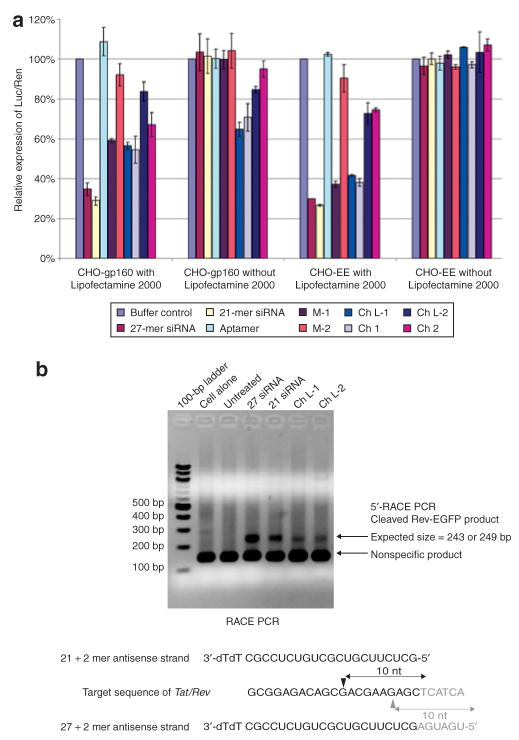
Luciferase expression was potently inhibited when Ch L-1 and Ch 1 were lipofected into both types of cells (Figure 4a). However, in the absence of lipofection, gene silencing from Ch L-1 and Ch 1 was specific to CHO-gp160-expressing cells, and no inhibition of luciferase was observed in CHO-EE cells. Interestingly, Ch L-1 and Ch 1, which are linked to the 27-mer duplex RNA, showed somewhat greater efficacy than Ch L-2 and Ch 2 did, consistent with our earlier observations that Dicer substrates enhance RNAi.29,30
Figure 4. Cell type–specific gene silencing mediated by aptamer–small interfering RNA (siRNA) chimeras via the RNA interference pathway.
(a) Aptamer–siRNA chimeras mediate silencing of pNL4-3 luciferase. CHO-gp160 cells or CHO-EE cells transfected with pNL4-3 luc were incubated with 200 nmol/l of the experimental RNAs in the presence or absence of the transfection reagent Lipofectamine 2000. In the absence of the transfection reagent, inhibition of pNL4-3 luc expression was observed only for CHO-gp160 cells. These results are consistent with the aptamer-mediated binding to gp160 and internalization of the chimera, followed by processing into siRNAs. The data were normalized to Renilla luciferase expression, and represent the average of three replicate assays. (b) 5′-Rapid amplification of cDNA ends (5′-RACE) PCR analysis. The cleaved product of mRNA from CHO-gp160 cells, previously transfected with saline (untreated), Tat-Rev site I 27-mer siRNA, 21-mer siRNA, Ch L-1, or Ch L-2 RNAs was ligated to an RNA adaptor and reverse transcribed using a gene-specific primer. An agarose gel electorphoresis of the 5′-RACE-PCR amplification products, using a primer specific to the RNA adaptor and a reverse primer (GSP-Rev-2) to Rev-EGFP, indicated specific siRNA-mediated cleavage products of Rev-EGFP mRNA. EGFP, enhanced green fluorescent protein.
In order to confirm that the siRNAs released from the chimeras were actually triggering RNAi, we transfected CHO-gp160 cells with a Rev-EGFP fusion construct harboring the siRNA targets. The transfected cells were then further transfected with Ch L-1, Ch L-2, 27-mer siRNA or 21-mer siRNA in the presence of Lipofectamine 2000. Thirty-six hours after transfection, total RNA was isolated and subjected to a modified 5′-rapid amplification of cDNA ends technique to identify the specific cleavage products in the Rev portion of the fusion transcript. We assumed that the Ago2-mediated cleavage was between bases 10 and 11 relative to the 5′-end of each siRNA. Our Dicer analyses of the 27-mer revealed that it is cleaved 21–23 nt downstream from the 5′-end of the antisense strand (antisense relative to the tat/rev target), whereas the 21-mer is not processed further (Figure 3b). We expected that the RNAi-mediated cleavage site in the target would be shifted by 6 bases between the 27-mer- and the 21-mer-derived siRNAs. Fragments of the predicted lengths were obtained from cells treated with the siRNAs or with chimeras (Figure 4b). Direct sequencing of the excised bands verified the expected PCR product, thereby demonstrating that cleavage did indeed occur at the predicted position for the siRNA duplex between positions 10 and 11 from the 5′-end of the siRNA anti-sense strand (Supplementary Figure S3). These data provide a formal demonstration that the chimeras produce siRNAs that are incorporated into RISC. As expected, no 5′-rapid amplification of cDNA ends PCR products were generated from the RNA isolated from cells that were not treated with the chimeras or with siRNAs.
Anti-gp120 aptamer–siRNA chimeras inhibit HIV gp120-mediated cell fusion and HIV-1 infection CEM T-cells
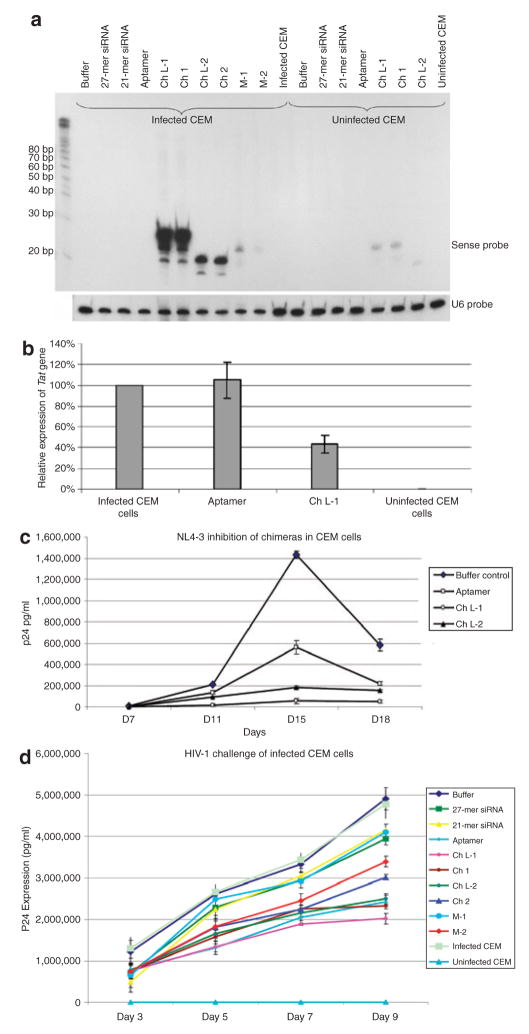
If aptamer–siRNA chimeras are to be used in treating HIV infection it is essential that the aptamer allows internalization of the chimeras in HIV-infected cells. We first demonstrated by northern blot analyses that chimera-delivered siRNAs were detectable in HIV-infected CEM cells that had been treated with the chimeras. The northern blot analyses data shown in Figure 5a demonstrate that the siRNAs from the chimeras are internalized in HIV-infected CEM cells. The 27-mer was processed to 21–23 base siRNAs in these cells but not in the gp120-negative uninfected CEM cells, thereby suggesting that the chimeras specifically delivered siRNA into the infected CEM cells through the anti-gp120 aptamer. As expected, the 21-mer or 27-mer duplex siRNAs, in the absence of the aptamers, were not detectable in the CEM cells because of lack of internalization (Figure 5a). Because nonspecific bindings existed to a small extent on the cell surfaces, tiny 21-mer or 27-mer RNA from the chimeras (Ch L-1, Ch 1, and Ch L-2) were also hybridized in uninfected CEM.
Figure 5. Dual inhibition on human immunodeficiency virus type 1 (HIV-1) infection mediated by small interfering RNA (siRNA) chimeras.
(a) Northern blots of infected CEM cells. Infected CEM cells were directly treated with siRNA and chimeras. The 27-chimera RNA is partially processed to a 21-mer siRNA after uptake into the CEM cells. Total RNAs were hybridized with a 21-mer 32P-labeled oligonucleotide probe. U6 RNA was used as an internal loading control. (b) Aptamer-mediated inhibition of expression of tat/rev in infected CEM cells. Cells were incubated with either the wild-type aptamer or Ch L-1 for 7 days before RNA extraction. Gene expressions for Tat/rev and glyceraldehyde-3-phosphate dehydrogenase were assayed using quantitative real-time PCR. The data shown represent the average of three replicate assays. (c) Chimera RNAs inhibit HIV infection. HIV-1 NL4-3 was incubated with the various RNAs at 37 °C for 1 hour. Subsequently, the treated virions were used for infecting CEM cells. The culture supernatant was collected at different time points (7, 11, 15, and 18 days) for p24 antigen analyses. The data shown represent the average of duplicate assays. (d) The siRNAs delivered by the chimera RNAs inhibit HIV-1 replication in previously infected CEM cells. Approximately 1.5 × 104 infected CEM cells and 3.5 × 104 uninfected CEM cells were incubated at 37 °C with the various RNAs at a final concentration of 400 nmol/l. The culture supernatant was collected at different time points (3, 5, 7, and 9 days) for p24 antigen analyses. The data shown represent the average of triplicate measurements of p24.
In order to further confirm siRNA function after internalization to infected CEM cells, quantitative real-time-PCR (qRT-PCR) was performed to evaluate the tat/rev gene expression. Either the aptamer or the chimeras were added directly to media containing infected CEM cells. After 7 days, treated cells were harvested, the total RNA was extracted, and the extent of tat/rev gene inhibition was determined using qRT-PCR expression assays. We found that the treatment of infected CEM cells with the chimeras was able to induce silencing of the tat/rev gene, while the use of the aptamer alone did not affect tat/rev gene expression (Figure 5b). These results provide further confirmation that the chimera-delivered siRNA triggers RNAi.
In HIV-1 infection, gp120 expressed at the cell surface will induce syncytia formation between infected and uninfected cells because of interactions between gp120 and CD4.27,28 We therefore sought to determine whether the aptamer and chimeras would have an impact on syncitia formation in cell culture. In this assay, the HIV-1-infected CEM cells were incubated with siRNAs or with chimera RNAs. Subsequently, the uninfected MT2 cells expressing CD4 were added to the infected CEM cells treated with RNA. After 48 hours of co-incubation at 37 °C, syncytia formation was analyzed microscopically. The treatment of the HIV-infected cultures with the aptamer and chimeras resulted in a clear reduction in syncytia formation (data not shown). We also investigated whether the aptamer and chimeras prevent HIV replication in an acute infection assay. We did this by monitoring HIV-1 gag p17 using an immunofluorescence assay (Supplementary Figure S4). These assays revealed a marked reduction in p17 expression in the HIV-infected cells treated with the anti-gp120 aptamer, and an even more pronounced reduction in those treated with the Ch L1 chimera.
In order to obtain further confirmation, anti-HIV activity of the chimeras in inhibition of HIV-1 replication, we carried out more assays. In the first assay, HIV-1 was first mixed with the chimeras or with the aptamer, and subsequently the viruses were used for infecting CEM cells. The infectivities of the aptamer- or chimera-treated virus were significantly reduced and viral replication was suppressed for up to 2 weeks (Figure 5c). Ch L-1 was the most effective inhibitory agent. In the second experiment, the aptamer or chimeras were incubated with HIV-infected CEM cells. At different time points after the treatment with the aptamer and chimeras, aliquots of the media were assayed for viral p24 antigen levels. The results of these analyses (Figure 5d) showed that all of the aptamer-containing RNAs inhibited p24 production, but the strongest inhibition was observed with Ch L-1 treatment, again consistent with our results from the other assays. These data, together with the observed inhibition of cell fusion and p17 expression, demonstrate that the anti-gp120 aptamer–siRNA chimera system can strongly inhibit HIV-1 replication and infection. Moreover, the suppression is attributable to the combined affect of the aptamer-binding gp120 and RNAi.
Anti-gp120 aptamer–siRNA chimeras do not trigger an interferon response
It has been reported earlier that siRNAs delivered by liposomes or polyplex reagents can nonspecifically activate inflammatory cytokine production (tumor necrosis factor-α, interleukin-6, and interleukin-12) as well as interferon (IFN)-responsive genes, and this, in turn, can trigger cellular toxicity.38–40 We therefore assessed the induction of type I IFN–regulated gene expression by our anti-gp120 aptamer–siRNA chimeras using qRT-PCR expression assays. As a positive control, we incubated the target cells with poly(IC). We find that the treatment of HEK293 cells with the chimeras did not significantly induce expression of the IFN-β and p56 genes (Supplementary Figure S5a). Given that CEM cells are difficult to transfect with control molecules such as poly(IC), we used IFN-α as a positive control to confirm upregulation of p56 and OAS1 gene expression. As we observed in the HEK293 transfection assays, treatment of CEM cells with the chimeras did not induce type I IFN responses (Supplementary Figure S5b). Similar results were obtained when using HIV-infected CEM cells treated with the chimeras, thereby suggesting that the gp120-mediated internalization of the chimeras does not trigger toxic IFN responses.
DISCUSSION
Aptamers are nucleic acid species that have been engineered through repeated rounds of in vitro selection to bind to various molecular targets such as small organic molecules, proteins, nucleic acids, and even cells.41–43 Because aptamers are capable of binding with high specificity to their ligands at low nanomolar to picomolar dissociation constants, they can be used as molecular drugs for both basic research and clinical purposes.44–48
The success of RNAi-based clinical applications is dependent on the efficiency of siRNA delivery to target cells. In this report, we have capitalized upon the exquisite specificity of a gp120 aptamer to deliver anti-HIV siRNAs into HIV infected cells, with the net result that replication and spread of HIV is strongly inhibited by the combined action of the aptamer and siRNA targeting the tat/rev common exon of HIV-1.
We utilized the HIV-1 envelop protein gp120 as a model receptor for targeted intracellular delivery of anti-HIV siRNAs. Cell type–specific binding and uptake of chimeric aptamer–siRNA conjugates were achieved through the interaction of the aptamer portion with gp120 on the cell surface of infected cells. In order to ensure the stability of our RNA chimeric molecules in sera, we utilized the RNA stabilizing 2 ′-fluoro backbone modifications of pyrimidines on the aptamer and siRNA sense strand. The antisense strand was not chemically modified but was, in fact, stabilized by virtue of its base pairing to the modified sense strand.
Notably, the cell type–specific gene silencing revealed that the siRNAs were successfully delivered into cells and entered into the RNAi pathway through the interaction of the anti-gp120 aptamer with gp120 expressed on the cell surface. Interestingly, the chimeras containing a 27-mer duplex RNA showed better efficacy in gene silencing than the corresponding 21-mer duplex containing chimeras. The 27-mer duplex by itself was also more potent than the 21-mer duplex when these RNAs were delivered by lipofection. We attribute this increased potency to Dicer processing of the 27-mer, wherein the processed 21–23-mer siRNAs are more readily handed off to RISC. It is of interest that we never observed complete processing of the 27-mer into 21–23 mers in our northern blot analyses of cells treated with the chimeras. This may, in part, be a consequence of the high intracellular concentrations achieved by aptamer delivery, but may also reflect the possibility that the design of our blunt-ended duplexes is suboptimal for Dicer processing. We observed that Dicer cleavage was initiated by entering the duplex from the blunt end rather than from the two base 3′-overhang. On the basis of this finding, we are testing other structures of the siRNA portion of the aptamers with a view to achieving more complete Dicing.
An interesting observation was that analyses of the target cleavage products by a 5′-rapid amplification of cDNA ends technique further demonstrated that neither the 27-mer nor the 21-mer siRNAs underwent processing to trigger duplexes with two base 3 ′-overhangs on both ends of the siRNAs. In fact for both the 21-mer- and 27-mer-derived siRNAs, the target mRNA was cleaved between positions 10 and 11 relative to the blunt 5 ′-end of the siRNA antisense strand. These results suggest that unprocessed 27-mer as well as Dicer-processed 27-mer antisense strands may be incorporated into RISC. Given the results from the cell extract Dicing reaction, which revealed that the 27-mer is not processed at the 5 ′-end of the antisense but only at the 3′-end, it is not possible to determine whether all, some, or none of the activated RISC was derived from intact 27-mer antisense being incorporated directly into RISC.
Aptamers that bind to viral or cellular proteins with high affinity and specificity are useful for therapeutic applications. In this study, both aptamer and chimeras were shown, through a variety of assays, to suppress the replication and production of HIV-1 dramatically. These results demonstrate important attributes of the anti-gp120 aptamer as being both an inhibitor of HIV through direct binding to virions or intracellular gp120 and a cell type–specific delivery vector for therapeutic siRNAs.
Because the anti-gp120 aptamer is responsible for the targeted delivery of siRNAs, gp120 expression is necessary for cell type–specific transport. This is, in essence, a safety feature which could be capitalized upon to deliver siRNAs that target HIV, or even cellular messages that are essential for viral replication. Given that only HIV-infected cells would be affected by the inhibitory action of the siRNA, this approach greatly minimizes potential off-target effects by the siRNAs.
The dual inhibitory potential of the aptamer–siRNA fusion is an important point of discussion. Both the aptamer and the chimeras produced strong inhibition of syncitial cell formation as well as of expression of HIV-1 gag p17, and replication and spread of HIV in HIV-1-infected CEM T lymphocytes. The anti-gp120 aptamer neutralizes HIV-1 infectivity by blocking the interaction of gp120 and CD4, and the siRNA silences tat/rev expression. In this manner, the anti-gp120/HIV chimeras serve a double-function and therefore provide greater efficacy than either the aptamer or the siRNA applied alone. Finally, we show that the aptamer-mediated delivery of siRNAs through binding to gp120 and subsequent internalization does not trigger type I IFN gene responses in various cell lines.
In summary, this strategy provides a new paradigm for delivery of anti-HIV siRNAs by allowing selective delivery to HIV-infected cells and providing dual-function inhibition of HIV replication and spread. Moreover, the aptamer and siRNAs can be readily changed to accommodate genetic changes in the virus, making this an attractive approach for systemic anti-HIV therapy.
MATERIALS AND METHODS
Generation of aptamer and chimera RNAs by in vitro transcription
Double-stranded DNA templates were directly generated by PCR, and the resulting PCR products were recovered using a QIAquick Gel purification Kit (Qiagen, Valencia, CA). Chimera sense strands were transcribed from its PCR-generated DNA templates using the DuraScription Kit (Epicentre, Madison, WI) in accordance with the manufacturer’s instruction. In the transcription reaction mixture, the canonical cytidine triphosphate and uridine triphosphate were replaced with 2 ′-fluoro-cytidine triphosphate and 2 ′-fluoro-uridine triphosphate to produce RNA that is resistant to RNase A degradation. The reactions were incubated at 37 °C for 6 hours, and subsequently purified using Bio-Spin 30 Columns (Bio-Rad, Hercules, CA) after phenol extraction and ethanol precipitation. RNA was treated by calf intestinal phosphatase to remove the initiating 5 ′-triphosphate. For preparation of the chimeras, the chimera harboring only the sense strand RNA was combined with the appropriate antisense RNA in HEPES buffer saline (10 mmol/l 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid pH 7.4, 150 mmol/l NaCl, 1 mmol/l CaCl2, 1 mmol/l MgCl2, 2.7 mmol/l KCl), heated to 95 °C for 3 minutes and then cooled to 37 °C slowly. Incubation continued at 37 °C for 10 minutes. Fluorescent aptamer and chimeras were generated using the Silencer siRNA labeling kit (Ambion, Austin, TX) in accordance with the manufacturer’s instructions.
Cell culture
HEK 293 cells and CEM cells were purchased from American Type Culture Collection and cultured in Dulbecco’s modified Eagle’s medium and Roswell Park Memorial Institute medium-1640 supplemented with 10% fetal bovine serum respectively, in accordance with their respective data sheets. CHO-WT and CHO-EE cells were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. These cells were grown in Gibco’s modified Eagle’s medium-S. The cells were cultured in a humidified 5% CO2 incubator at 37 °C.
Cell-surface binding of aptamer–siRNA chimeras (flow cytometry analysis)
CHO-gp160 or CHO-EE cells were washed with phosphate-buffered saline (PBS), trypsinized, and detached from the plates. The cells were washed two times with 500 μl of binding buffer [10 mmol/l 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid pH 7.4, 150 mmol/l NaCl, 1 mmol/l CaCl2, 1 mmol/l MgCl2, 2.7 mmol/l KCl, 0.01% bovine serum albumin]. The cell pellets were resuspended in binding buffer and incubated at 37 °C for 30 minutes. The cells were then pelleted and resuspended in 50 μl of binding buffer (prewarmed to 37 °C) containing either 400 nmol/l Cy3-labeled aptamer or chimera RNAs. After incubation at 37 °C for 40 minutes, the cells were washed three times with 500 μl of binding buffer prewarmed to 37 °C, again resuspended in 350 μl of binding buffer prewarmed to 37 °C, and analyzed using flow cytometry.
Cellular binding and uptake studies (confocal microscopy analysis)
The CHO-gp160 and CHO-EE cells lines were grown in 8-well chamber slide with seeding at 1 × 105 in Gibco’s modified Eagle’s medium-S to allow 50–70% confluence in 24 hours. On the day of the experiments, the cells were washed with 250 μl of prewarmed PBS and incubated with 250 μl of prewarmed complete growth medium for 30 minutes at 37 °C. Cy3-labeled RNAs at 20 nmol/l of final concentration were added into the medium and incubated at 37 °C for 1.5 hours. Subsequently, the cells were washed three times with 250 μl of prewarmed PBS and then fixed with 4% formaldehyde for 10 minutes. The cells were stained by treatment with 100 μl of Vybrant cell-labeling solution (C18(3) 3,3′-dioctadecyloxacarbocyanine perchlorate membrane dye; Molecular Probes, Invitrogen, Carlsbad, CA) in accordance with the manufacturer’s instructions. The images were collected using a Zeiss LSM 510 upright 2-photon confocal microscopy system under water immersion at ×40 magnifications. The images were combined and decon-voluted to reconstruct a three-dimensional image.
Analysis of chimera processing
Sense RNAs were annealed with equal moles of 5′-end-labeled antisense strands in HEPES buffer saline in order to form chimeric double-stranded RNA. The chimeras or double-stranded RNAs were incubated in HCT116 cell lysates at 10 nmol/l of final concentration, in the absence of target mRNA, for varying times (20 minutes, 60 minutes, and 120 minutes). The reactions were stopped by phenol/ chloroform extraction and the RNAs were collected for electrophoresis in a denaturing 20% polyacrylamide gel. The gels were subsequently dried and exposed to X-ray film.
Dual luciferase assays
(Day 1) CHO-gp160 and CHO-EE cells were transfected with pNL4-3.Luc.R-.E- (NIH AIDS Research and Reagent Program, Germantown, MD) and pRSV-Renilla, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) in accordance with the manufacturer’s instructions. pNL4-3.Luc.R-.E- is an Env-Vpr-noninfectious clone containing the firefly luciferase (F-Luc) gene inserted into the nef gene. (Day 2) Cells which transiently expressed pNL4-3.Luc were seeded in 24-well plates at 50–70% confluency. (Day 3) For siRNA, cells were transfected with 200 nmol/l RNA using Lipofectamine 2000. For aptamer-mediated siRNA delivery (also on day 3) cells were incubated in 400 μl refresh complete growth medium for 30 minutes at 37 °C. The chimera RNAs were added directly to the medium (400 μl) at a final concentration of 200 nmol/l chimeras. (Day 4) The cells were harvested for analysis. The expression of the pNL4-3.Luc and normalizing control Renilla luciferase were detected using the Dual-luciferase reporter assay system (Promega, Madison, WI), in accordance with the manufacturer’s instructions. All samples were transfected in triplicate, and the experiment was performed a minimum of three times.
5′-Rapid amplification of cDNA ends PCR assay
Total RNA (5 μg) from CHO-gp160 cells treated with different siRNAs and chimeras was ligated to a GeneRacer adaptor (Invitrogen, Carlsbad, CA) without prior treatment. Ligated RNA was reverse transcribed using a gene-specific primer 1 (GSP-Rev 1: 5′-TCACCCTCTCCACTGACAGAGAACTT-3′). In order to detect cleavage products, PCR was preformed using primers complementary to the RNA adaptor (5′-cDNA primer: 5′-GGACACTGACATG GACTGAAGGAGTA-3′) and gene-specififc primer 2 (GSP-Rev 2: 5′-TA ACCTCTCAAGCGGTGGTAGCTGAA-3′). The amplification products were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining. The identity of the specific PCR products was confirmed by sequencing of the excised bands.
Northern blot analysis
CEM cells were infected with HIV NL4-3 for 10 days. Before adding the various RNAs, the infected CEM cells were gently washed three times to clear out free virus. Approximately 5 × 104 cells were incubated with refolded RNAs at 400 nmol/l final concentrations in 96-well plates at 37 °C. The total RNAs were harvested on the seventh day after application for analysis using STAT-60 (TEL-TEST “B”, Friendswood, TX) in accordance with the manufacturer’s instructions. Two micrograms of total RNAs were electrophoresed in a 15% polyacryl-amide-8 mol/l urea gel and then transferred to a Hybond N+ membrane (Amersham pharmacia Biotech, Piscataway, NJ). Prehybridization and hybridization were carried out using PerfectHyb Plus hybridization buffer (Sigma, St. Louis, MO) at 37 °C with 3 pmol of a 27-mer DNA oligonucleotide probe end-labeled with T4 polynucleotide kinase and γ-[32P]-ATP. Before autoradiography, the filters were washed three times at 37 °C for 15 minutes. We also probed for human U6 small nuclear RNA as an internal RNA loading standard.
qRT-PCR analysis
CEM cells were infected with HIV NL4-3 for 10 days. Before being analyzed, the infected CEM cells were gently washed three times to eliminate free virus. The infected CEM cells were treated directly with the aptamer or Ch L-1 at 400 nmol/l of final concentration. After 7 days, total RNAs were isolated using STAT-60 (TEL-TEST “B”, Friendswood, TX). The expression of the tat/rev coding RNAs was analyzed by qRT-PCR using 2× iQ SYBR green mastermix (Bio-Rad, Hercules, CA) and specific primer sets at a final concentration of 400 nmol/l. The primers used were: tat/rev forward primer: 5′-GGCGTTACTCGACAGAGGAG-3′; tat/rev reverse primer: 5′-TGCTTTGATAGAGAAGCTTGATG-3′; glyceral-dehyde-3-phosphate dehydrogenase forward primer: 5′-CATTGACCT CAACTACATG-3′; glyceraldehyde-3-phosphate dehydrogenase reverse primer: 5′-TCTCCATGGTGG TGAAGAC-3′.
RNA-Stat60 was used for extracting total RNA in accordance with the manufacturer’s instructions (TEL-TEST “B”, Friendswood, TX). Residual DNA was digested using the DNA-free kit as per the manufacturer’s instructions (Ambion, Austin, TX). cDNA was produced using 2 μg of total RNA moloney murine leukemia virus reverse transcriptase and random primers in a 15 μl reaction in accordance with the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Glyceraldehyde-3-phosphate dehydrogenase expression was used for normalization of the qPCR data.
HIV-1 challenges and p24 antigen assay
Method 1: NL4-3 virus was incubated with refolded RNAs at 37 °C for 1 hour. Subsequently, the viruses were gently washed with PBS and used for infecting CEM cells. The culture supernatants were collected at different time points after infection (7, 11, 15, and 18 days) for p24 antigen analyses. Method 2: CEM cells were infected with HIV NL4-3 for 10 days. Before the RNA treatments, the infected-CEM cells were gently washed with PBS three times to eliminate free virus. Approximately 1.5 × 104 infected CEM cells and approximately 3.5 × 104 uninfected CEM cells were incubated with refolded RNAs at 400 nmol/l of final concentration in 96-well plates at 37 °C. The culture supernatants were collected at different time points (3, 5, 7, and 9 days). The p24 antigen analyses were performed using a Coulter HIV-1 p24 antigen assay (Beckman Coulter, Fullerton, CA) in accordance with the manufacturer’s instructions.
Supplementary Material
Figure S1. Gene silencing activity and strand selectivity of chimera RNAs and siRNA.
Figure S2. Images were combined and deconvoluted to reconstruct a three-dimensional image.
Figure S3. RACE PCR sequence.
Figure S4. Immunofluorescence assay of HIV-1 p17.
Figure S5. IFN assays. IFN-β, the interferon responsive P56 (CDKL2) and OAS1 mRNAs were measured by quantitative RT-PCR.
Acknowledgments
We thank Daniela Castanotto, Guihua Sun, Harris Soifer, Hoshang Unwalla, and Masayuki Sano for helpful discussions. This work was supported by grants from the National Institutes of Health awarded to J.J.R. The CHO-EE and CHO-gp160 cell line49,50 and the pNL4-3 luc vector were obtained from the NIH AIDS Reagent and Repository.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 3.Behlke MA. Progress towards in vivo use of siRNAs. Mol Ther. 2006;13:644–670. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Layzer JM, McCaffrey AP, Tanner AK, Huang Z, Kay MA, Sullenger BA. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morrissey DV, Blanchard K, Shaw L, Jensen K, Lockridge JA, Dickinson B, et al. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology. 2005;41:1349–1356. doi: 10.1002/hep.20702. [DOI] [PubMed] [Google Scholar]
- 6.Lewis DL, Wolff JA. Delivery of siRNA and siRNA expression constructs to adult mammals by hydrodynamic intravascular injection. Methods Enzymol. 2005;392:336–350. doi: 10.1016/S0076-6879(04)92020-4. [DOI] [PubMed] [Google Scholar]
- 7.Kishida T, Asada H, Gojo S, Ohashi S, Shin-Ya M, Yasutomi K, et al. Sequence-specific gene silencing in murine muscle induced by electroporation-mediated transfer of short interfering RNA. J Gene Med. 2004;6:105–110. doi: 10.1002/jgm.456. [DOI] [PubMed] [Google Scholar]
- 8.Akaneya Y, Jiang B, Tsumoto T. RNAi-induced gene silencing by local electroporation in targeting brain region. J Neurophysiol. 2005;93:594–602. doi: 10.1152/jn.00161.2004. [DOI] [PubMed] [Google Scholar]
- 9.Inoue A, Takahashi KA, Mazda O, Terauchi R, Arai Y, Kishida T, et al. Electro-transfer of small interfering RNA ameliorated arthritis in rats. Biochem Biophys Res Commun. 2005;336:903–908. doi: 10.1016/j.bbrc.2005.08.198. [DOI] [PubMed] [Google Scholar]
- 10.Tsunoda S, Mazda O, Oda Y, Iida Y, Akabame S, Kishida T, et al. Sonoporation using microbubble BR14 promotes pDNA/siRNA transduction to murine heart. Biochem Biophys Res Commun. 2005;336:118–127. doi: 10.1016/j.bbrc.2005.08.052. [DOI] [PubMed] [Google Scholar]
- 11.Kim TW, Lee JH, He L, Boyd DA, Hardwick JM, Hung CF, et al. Modification of professional antigen-presenting cells with small interfering RNA in vivo to enhance cancer vaccine potency. Cancer Res. 2005;65:309–316. [PubMed] [Google Scholar]
- 12.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 13.Hassani Z, Lemkine GF, Erbacher P, Palmier K, Alfama G, Giovannangeli C, et al. Lipid-mediated siRNA delivery down-regulates exogenous gene expression in the mouse brain at picomolar levels. J Gene Med. 2005;7:198–207. doi: 10.1002/jgm.659. [DOI] [PubMed] [Google Scholar]
- 14.Spagnou S, Miller AD, Keller M. Lipidic carriers of siRNA: differences in the formulation, cellular uptake, and delivery with plasmid DNA. Biochemistry. 2004;43:13348–13356. doi: 10.1021/bi048950a. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Lee RJ, Cauchon G, Gorenstein DG, Low PS. Delivery of antisense oligodeoxyribonucleotides against the human epidermal growth factor receptor into cultured KB cells with liposomes conjugated to folate via polyethylene glycol. Proc Natl Acad Sci USA. 1995;92:3318–3322. doi: 10.1073/pnas.92.8.3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner E, Zenke M, Cotten M, Beug H, Birnstiel ML. Transferrin-polycation conjugates as carriers for DNA uptake into cells. Proc Natl Acad Sci USA. 1990;87:3410–3414. doi: 10.1073/pnas.87.9.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiffelers RM, Ansari A, Xu J, Zhou Q, Tang Q, Storm G, et al. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 2004;32:e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pun SH, Bellocq NC, Liu A, Jensen G, Machemer T, Quijano E, et al. Cyclodextrin-modified polyethylenimine polymers for gene delivery. Bioconjug Chem. 2004;15:831–840. doi: 10.1021/bc049891g. [DOI] [PubMed] [Google Scholar]
- 19.Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer Res. 2005;65:8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- 20.Weissleder R, Kelly K, Sun EY, Shtatland T, Josephson L. Cell-specific targeting of nanoparticles by multivalent attachment of small molecules. Nat Biotechnol. 2005;23:1418–1423. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- 21.Sorgi FL, Bhattacharya S, Huang L. Protamine sulfate enhances lipid-mediated gene transfer. Gene Ther. 1997;4:961–968. doi: 10.1038/sj.gt.3300484. [DOI] [PubMed] [Google Scholar]
- 22.Simeoni F, Morris MC, Heitz F, Divita G. Insight into the mechanism of the peptide-based gene delivery system MPG: implications for delivery of siRNA into mammalian cells. Nucleic Acids Res. 2003;31:2717–2724. doi: 10.1093/nar/gkg385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muratovska A, Eccles MR. Conjugate for efficient delivery of short interfering RNA (siRNA) into mammalian cells. FEBS Lett. 2004;558:63–68. doi: 10.1016/S0014-5793(03)01505-9. [DOI] [PubMed] [Google Scholar]
- 24.Simeoni F, Morris MC, Heitz F, Divita G. Peptide-based strategy for siRNA delivery into mammalian cells. Methods Mol Biol. 2005;309:251–260. doi: 10.1385/1-59259-935-4:251. [DOI] [PubMed] [Google Scholar]
- 25.McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 26.Chu TC, Twu KY, Ellington AD, Levy M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006;34:e73. doi: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, et al. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure. 2000;8:1329–1339. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- 29.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 30.Rose SD, Kim DH, Amarzguioui M, Heidel JD, Collingwood MA, Davis ME, et al. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005;33:4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khati M, Schüman M, Ibrahim J, Sattentau Q, Gordon S, James W. Neutralization of infectivity of diverse R5 clinical isolates of human immunodeficiency virus type 1 by gp120-binding 2′F-RNA aptamers. J Virol. 2003;77:12692–12698. doi: 10.1128/JVI.77.23.12692-12698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dey AK, Khati M, Tang M, Wyatt R, Lea SM, James W. An aptamer that neutralizes R5 strains of human immunodeficiency virus type 1 blocks gp120-CCR5 interaction. J Virol. 2005;79:13806–13810. doi: 10.1128/JVI.79.21.13806-13810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dey AK, Griffiths C, Lea SM, James W. Structural characterization of an anti-gp120 RNA aptamer that neutralizes R5 strains of HIV-1. RNA. 2005;11:873–884. doi: 10.1261/rna.7205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Czauderna F, Fechtner M, Dames S, Aygün H, Klippel A, Pronk GJ, et al. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Braasch DA, Jensen S, Liu Y, Kaur K, Arar K, White MA, et al. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 36.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 37.Allerson CR, Sioufi N, Jarres R, Prakash TP, Naik N, Berdeja A, et al. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J Med Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 38.Kim DH, Longo M, Han Y, Lundberg P, Cantin E, Rossi JJ. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- 39.Schlee M, Hornung V, Hartmann G. siRNA and isRNA: two edges of one sword. Mol Ther. 2006;14:463–470. doi: 10.1016/j.ymthe.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Robbins MA, Li M, Leung I, Li H, Boyer DV, Song Y, et al. Stable expression of shRNAs in human CD34+ progenitor cells can avoid induction of interferon responses to siRNAs in vitro. Nat Biotechnol. 2006;24:566–571. doi: 10.1038/nbt1206. [DOI] [PubMed] [Google Scholar]
- 41.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 42.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 43.Fitzwater T, Polisky B. A SELEX primer. Methods Enzymol. 1996;267:275–301. doi: 10.1016/s0076-6879(96)67019-0. [DOI] [PubMed] [Google Scholar]
- 44.Tuerk C, MacDougal S, Gold L. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci USA. 1992;89:6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hicke BJ, Stephens AW. Escort aptamers: a delivery service for diagnosis and therapy. J Clin Invest. 2000;106:923–928. doi: 10.1172/JCI11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pestourie C, Tavitian B, Duconge F. Aptamers against extracellular targets for in vivo applications. Biochimie. 2005;87:921–930. doi: 10.1016/j.biochi.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 47.Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: an emerging class of therapeutics. Annu Rev Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 48.Proske D, Blank M, Buhmann R, Resch A. Aptamers—basic research, drug development, and clinical applications. Appl Microbiol Biotechnol. 2005;69:367–374. doi: 10.1007/s00253-005-0193-5. [DOI] [PubMed] [Google Scholar]
- 49.Weiss CD, White JM. Characterization of stable Chinese hamster ovary cells expressing wild-type, secreted, and glycosylphosphatidylinositol-anchored human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1993;67:7060–7066. doi: 10.1128/jvi.67.12.7060-7066.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vodicka MA, Goh WC, Wu LI, Rogel ME, Bartz SR, Schweickart VL, et al. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology. 1997;233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Gene silencing activity and strand selectivity of chimera RNAs and siRNA.
Figure S2. Images were combined and deconvoluted to reconstruct a three-dimensional image.
Figure S3. RACE PCR sequence.
Figure S4. Immunofluorescence assay of HIV-1 p17.
Figure S5. IFN assays. IFN-β, the interferon responsive P56 (CDKL2) and OAS1 mRNAs were measured by quantitative RT-PCR.