Abstract
Alternative splicing of the mu opioid receptor genes to create multiple mu receptor subtypes has been demonstrated in animals and humans. Previously, we identified a number of C-terminal variants in mice, rats and human, followed by several N-terminal variants associated with a new upstream exon in mice (exon 11). Behavioral studies in exon 11 knockout mice suggest an important role for the exon 11 variants in the analgesic actions of heroin and morphine-6β-glucuronide, but not morphine or methadone. We now have identified a homologous human exon 11 and three similar human exon 11-associated variants, suggesting conservation of exon 11 and its associated variants across species. hMOR-1i has an additional 93 amino acids at the tip of the N-terminus but is otherwise identical to hMOR-1. When expressed in Chinese hamster ovary cells, the additional 93 amino acids in hMOR-1i had little effect on opioid binding, but significantly altered agonist-induced G-protein activation. hMOR-1G1 and hMOR-1G2 predicted six transmembrane domain variants, similar to those seen in mice. The regional expression of these exon 11-associated variants, as determined by RT-PCR, varied markedly, implying region-specific alternative splicing. The presence of exon 11-associated variants in humans raises questions regarding their potential role in heroin and morphine-6β-glucuronide actions in people as they do in mice.
Keywords: G protein, mu opioid receptor (MOR), morphine, mu opioid receptor-1, opioid receptor, splicing
Pharmacological studies have classified three families of opioid receptors, mu, delta and kappa (Martin et al. 1976; Lord et al. 1977; Pasternak 2004). Of these three families, the mu opioid receptors are particularly important because they mediate the actions of morphine and most clinical analgesic agents, as well as heroin (Pasternak 1993). Clinically, patients often respond quite differently to various mu opioids in terms of analgesia, side effects and tolerance, leading to the need to individualize therapy (Foley 1985; Payne and Pasternak 1992; Portenoy 1996). Indeed, the existence of incomplete cross tolerance among mu opioids has led to the utilization of Opioid Rotation to regain analgesic activity in highly tolerant patients (Cherny et al. 2001). Similar observations have been made in animal models (Ling et al. 1984, 1985; Moulin et al. 1988; Chang et al. 1998; Pasternak 2001, 2004). These observations raised the possibility of subtypes of mu receptors, which were originally proposed based upon classical pharmacological and receptor binding studies (Wolozin and Pasternak 1981; Rossi et al. 1995a,b, 1997).
The cloning of the mu opioid receptor, MOR-1 (Chen et al. 1993; Thompson et al. 1993; Wang et al. 1993), provided the opportunity to explore the actions of mu opioid receptors at the molecular level. Only a single mu opioid receptor gene (OPRM1) has been isolated, but that does not eliminate the possibility of multiple mu receptors. Alternative splicing provides a mechanism for creating protein diversity. Genome-wide surveys reveal that approximately 70–75% of human genes generate alternatively spliced products (Johnson et al. 2003; Stamm et al. 2006; Vukusic et al. 2007), including a number of G protein-coupled receptors, such as dopamine D2 (Monsma et al. 1989), somatostatin 2 (Vanetti et al. 1998), prostaglandin EP3 (Namba et al. 1993; Sugimoto and Narumiya 2007) and serotonin receptor subtypes including 5-hydroxytryptamine (5-HT)2A (Guest et al. 2000), 5-HT2C (Canton et al. 1996), 5-HT3 (Hope et al. 1993), 5-HT4 (Blondel et al. 1998; Bender et al. 2000), 5-HT6 (Olsen et al. 1999) and 5-HT7 (Jasper et al. 1997). The strong evidence from pharmacological and receptor binding assays coupled with the existence of a single gene led us to hypothesize that multiple mu opioid receptor subtypes originally proposed might result through alternative pre-mRNA splicing of the OPRM1 gene. This possibility was supported by our earlier anti-sense mapping studies (Rossi et al. 1995a,b) and by an exon 1 knockout (KO) mouse (Schuller et al. 1999). In this mouse, disruption of exon 1 of the OPRM1 gene completely abolished morphine analgesic response, but not that of morphine-6β-glucuronide (M6G) and heroin, raising the possibility of that the residual M6G and heroin analgesia seen in the MOR-1 KO animals might be because of alternatively spliced transcripts that might normally lack exon 1.
In addition to the early isolation of two splice variants, hMOR-1A and rMOR-1B (Bare et al. 1994; Zimprich et al. 1995), over 28 splice variants from the mouse OPRM1 gene under the control of two diverse promoters have been isolated (Pan et al. 1999, 2000, 2001, 2005b; Pan 2002, 2005; Doyle et al. 2007a,b), with similar splicing patterns in rats (Pasternak et al. 2004), and humans (Pan et al. 2003, 2005a). The vast majority of these variants yield receptors differing from each other only at the tip of the C-terminus, a result of alternative splicing between exon 3 and more than 10 downstream exons, yielding at least 16 carboxyl terminal variants. Expression of most of these C-terminal variants is under the control of the exon 1 promoter. Although these full length variants share a common binding pocket within each species, they displayed significant differences in mu agonist-induced G protein activation, suggesting an important role of the C-terminus in these actions (Bolan et al. 2004; Pasternak et al. 2004; Pan et al. 2005a,b). Furthermore, these variants displayed different regional distributions within the brain, within cells and associate differently with another neurotransmitter, calcitonin gene-related peptide (CGRP) (Abbadie et al. 2000a,b,c, 2001, 2004; Schnell and Wessendorf 2004; Zhang et al. 2006).
There is also 5′-splicing. Several years ago we uncovered exon 11 and its associated splice variants in the mouse OPRM1 gene (Pan et al. 2001; Pan 2002; Abbadie et al. 2004). Exon 11 is located at 30 kb upstream of exon 1. Alternative splicing from exon 11 to its downstream exons produces nine exon 11-associated splice variants. Of these, three transcripts generate the same protein as MOR-1 while other transcripts predict a protein with six transmembrane (TM) domains, and or a protein with a single TM. The exon 11 promoter is located upstream of exon 11 has been directs the expression of nine exon 11-associated variants (Pan 2002). The significance of the exon 11 variants is supported by recent studies with an exon 11 KO mouse. In contrast to the exon 1 KO model that lost morphine analgesia and retained heroin and M6G analgesia, disruption of exon 11 greatly diminished heroin and M6G analgesia with little effect on morphine analgesia, suggesting a major role for the exon 11 variants in the actions of heroin and M6G, but not morphine (YX Pan, J Xu, M Xu, GR Rossi, J Matulonis and GW Pasternak, submitted). These observations raised the question of whether or not an analogous exon and splicing pattern existed in other species, particularly humans. We have recently isolated a mouse exon 11 homolog in the rat OPRM1 gene and seven rat exon 11-associated splice variants (J Xu, M Xu, GW Pasternak and YX Pan in preparation). We now report the identification and characterization human homolog of the mouse exon 11 and three human exon 11-associated splice variants.
Materials and methods
Genomic database searching
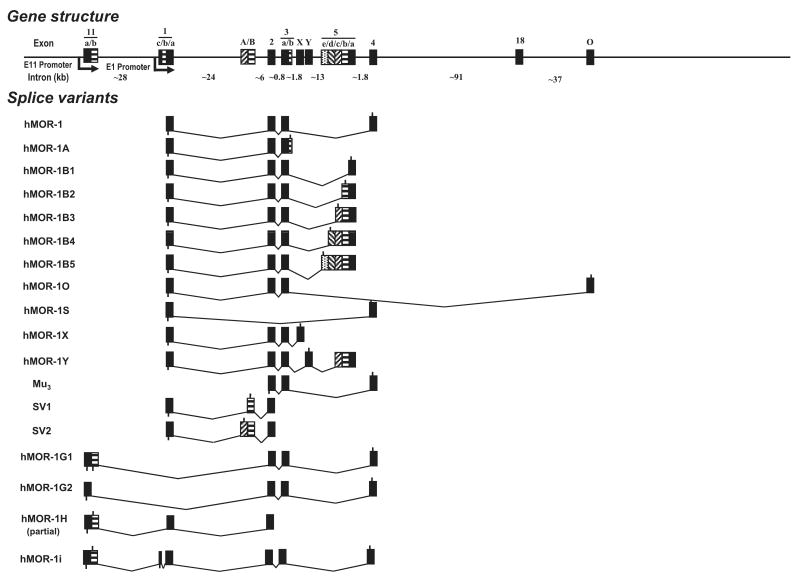
A human exon 11 homologous to the mouse exon 11 in the human OPRM1 gene was identified in the Ensembl human genome database by aligning the mouse exon 11 sequence. There is 63% identity at the nucleotide level or 33% identity at the predicted amino acid level between the human exon 11 and the mouse exon 11 sequences. The human exon 11 was located approximately 28 kb upstream of exon 1 in the human OPRM1 gene, a distance very similar to that between the mouse exons 11 and 1 (∼30 kb) (Fig. 1).
Fig. 1.
Schematic of the gene structure and alternative splicing of the human OPRM1 gene. Exons and introns are showed by boxes and horizontal lines, respectively. Translational start and termination sites are indicated by downward and upward lines on exon boxes, respectively. The exons are numbered by the order of their isolation and/or similarity to exons identified in the mouse: hMOR-1A (Bare et al. 1994), Mu3 (Cadet et al. 2003), SV1 and SV2 (Choi et al. 2006), hMOR-1B1, hMOR-1B2, hMOR-1B4, hMOR-1B5, and hMOR-1Y (Pan et al. 2005a), hMOR-1o and hMOR-1X (Pan et al. 2003), hMOR-1S (Du et al. 1997; Pan 2005).
RT-PCR cloning
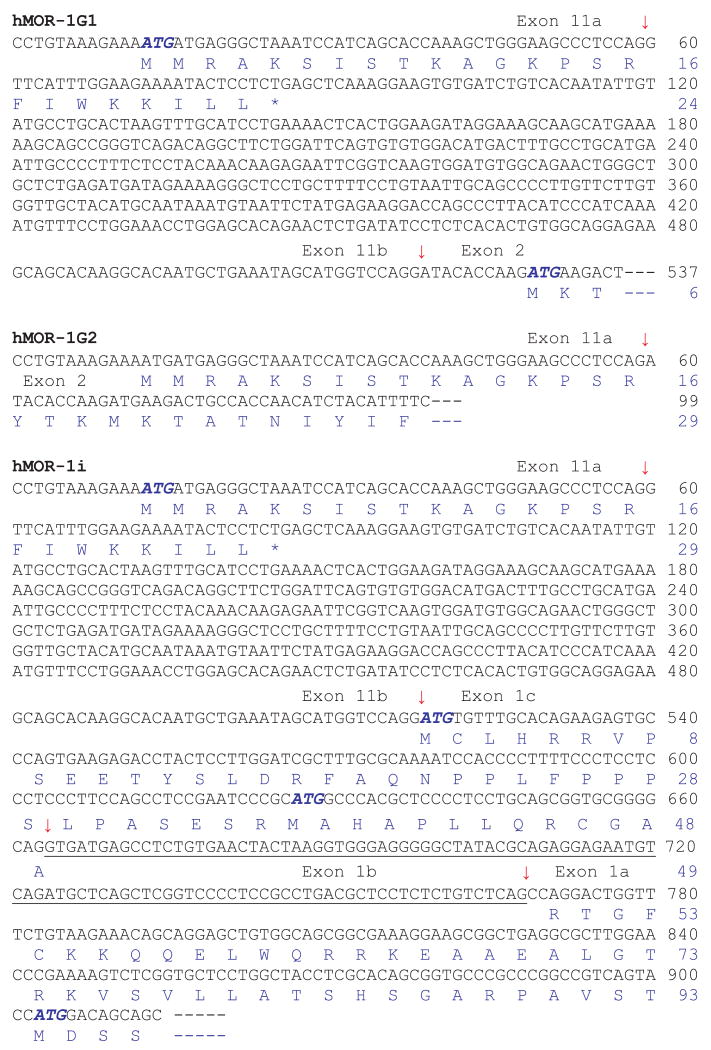
Total human brain RNAs (ClonTech, Palo Alto, CA, USA and Ambion, Austin, TX, USA) were reverse transcribed with random hexmers and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA). The first-strand cDNA was then used as templates in nested PCRs with a sense primer designed from the human exon 11 (E11-SE1, 5′-CCTGTAAAGAAAATGATGAGGG CTAAATCC-3′) and two anti-sense primers from exon 2 (E2-AN1: 5′-CATGTTCCCATTAGGTAATTCACACTCTGG-3′ and E2-AN2: 5′-GGGCAGGGTACTGGTGGCTAAGGC-3′). Three PCR fragments (A, B and C) containing the partial cDNA sequences were isolated and sequenced. Sequence analysis of the cDNA fragment A (1106 bp) contained a 600 bp exon 11, a 424 bp exon 1 and the 82 bp exon 2, a splicing patter similar to that of mouse MOR-1H. The upstream splice site of the 424 bp exon 1 was similar to that of mouse exon 1a. Therefore, the 424 bp exon 1 was designated as exon 1a. The fragment B sequence (1251 bp) had the same sequence as the fragment A except that there was a 145 bp insertion between exons 11 and 1a that was located 105 bp upstream of exon 1a and was designated as exon 1c. The 105 bp sequence was assigned as exon 1b. The splicing pattern of the fragment B was analogous to that of mouse mMOR-1I. Fragment C (141 bp) contained a 59 bp exon 11 followed by a 82 bp exon 2, a splicing pattern similar to those of mouse mMOR-1G, mMOR-1M and mMOR-1N. The 59 bp was identical to the first 59 bp in the fragments A and B, but the remaining 541 bp exon 11 sequence was spliced out by using a consensus splice site downstream of the 59 bp. We assigned the 541 bp as exon 11b and its upstream sequence as exon 11a. We then isolated three full length of cDNA fragments using nested PCRs with three sense primers from exon 11 (E11-SE1: see above, E11-SE2: 5′-GAAAATGATGAGGGCTAAATCCATCAGCACCAAAG CTGGGAAGCCCTCCAGATACAC-3′, and E11-SE3: 5′-GAAAT AGCATGGTCCAGGATACACCAAGATG-3) and two anti-sense primers from exon 4 (E4-AN1: GAGCCTCCTACACATTCTTGA AGCAAC-3′ and E4-AN2: 5′-GCTTGGTGAAGGTCGGAATGGC ATG-3′). The cDNA fragments were subcloned into pcDNA3.1/V5/His-TOPO vector (pcDNA3.1-TOPO) (Invitrogen) and sequenced with appropriate primers in both directions. We named the clones as hMOR-1G1, hMOR-1G2 and hMOR-1i based upon their similarities to the mouse homologs in terms of exon composition (Fig. 1). Like mMOR-1G, hMOR-1G1 contained exons 11a/11b, 2, 3 and 4. hMOR-1G2 had the same exon composition except that exon 11b was skipped. hMOR-1i contained exons 11a/11b, 1c, 1a, 2, 3 and 4, a pattern reassembling that of mMOR-1I.
In vitro transcription coupled translation
As pcDNA3.1-TOPO vector had an upstream T7 promoter, hMOR-1G1/pcDNA3.1-TOPO, hMOR-1G2/pcDNA3.1-TOPO and hMOR-1i/pcDNA3.1-TOPO plasmids, as well as hMOR-1/pcDNA3.1(-), were transcribed and translated in vitro with a TnT T7 coupled reticulocyte lysate system (Promega, Madison, WI, USA) following the manufacturer's protocol. Briefly, the plasmids were incubated with T7 RNA polymerase and reticulocyte lysate in the presence of 0.04 mCi of [35S]methionine (> 1000 Ci/mmol; PerkinElmer, Waltham, MA, USA) at 26°C for 90 min. The translated products were separated on a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the gel was treated with Amplify (GE Life, Piscataway, NJ, USA), dried and exposed to Kodak BioMax MR film (Kodak, New York, NY, USA).
Regional expression of the human MOR-1 variant mRNAs
Several human brain regions and spinal cord were obtained from The National Institute of Child Health and Human development (NICHD) Brain and Tissue Bank, University of Maryland or from the Forensic Medicine Department at the Karolinska Institutet, Stockholm, Sweden, under guidelines approved by the ethics committee and the Swedish Board of Health and Social Welfare. Total RNA was isolated from these frozen tissues using the guanidinium thiocyanate phenol-chloroform extract method (Chomczynski and Sacchi 1987) in conjunctions with RNAlater-ICE reagent (Ambion), reverse-transcribed with Superscript II reverse transcriptase and random hexamers. The first-strand cDNAs were used in PCR to amplify a 122 bp hMOR-1G1 fragment with E11-SE3 sense primer and E2-AN1 anti-sense primer, a 117 bp hMOR-1G2 fragment with E11-SE4 sense primer (5′-GGAAGCCCTCCAGATACACCAAG-3′) and E2-AN1 anti-sense primer, a 430 bp hMOR-1i fragment with E11-SE5 sense primer (5′-GAAACCTGGAGCACAGAACTCTGATATC-3′) and E1a-AN1 anti-sense primer (5′-GCCAGGAGCACCGAGACTTTTCG-3′), and a 443 bp exons 1 and 2 fragment representing hMOR-1 with E1a-SE1 sense primer (5′-CGGTGCTCCTGGCTACCTCGCAC-3′) and E2-AN1 anti-sense primer. RNA loading was estimated by a parallel PCR to amplify a 0.45 kb glyceraldehyde-3-phosphate dehydrogenase fragment with a pair of Amplimers (ClonTech). Platinum Taq DNA polymerase (Invitrogen) was used in PCR with different parameters. Following an initial 2 min at 94°C, 45 cycles were carried out for hMOR-1G1 with each cycle consisting of a 20 s denaturing step at 94°C, a 15 s annealing step at 68°C and a 15 s at 72°C extension, for hMOR-1G2 with each cycle consisting of a 20 s denaturing step at 94°C, a 20 s annealing step at 63°C and a 15 s at 72°C extension and for hMOR-1i with each cycle consisting of a 20 s denaturing step at 94°C, a 20 s annealing step at 63°C and a 26 s at 72°C extension, and 35 cycles for hMOR-1 with each cycle consisting of a 20 s denaturing step at 94°C, a 20 s annealing step at 65°C and a 40 s at 72°C extension and 25 cycles for glyceraldehyde-3-phosphate dehydrogenase with each cycle consisting of a 20 s denaturing step at 94°C, a 20 s annealing step at 63°C and a 1 min at 72°C extension. The PCR products were analyzed in 1.5% agarose gel, stained with ethiduim bromide and photographed with FluorChem 8000 Image system (Alpha Innotech, San Leandro, CA, USA).
Expression of hMOR-1i in Chinese hamster ovary cells
The hMOR-1i/pcDNA3.1-TOPO and hMOR-1/pcDNA3.1(-) plasmids were used to transfect Chinese hamster ovary (CHO) cells by LipofectAMINE reagent (Invitrogen). Stable transformants were obtained 10–14 days after selection with G418 and screened with [3H] [D-Ala2,MePhe4,Gly(ol)5]enkephalin binding assay.
Receptor binding assays
Membranes were prepared from stable transfectants as described previously (Pan et al. 1995, 1996). Saturation and competition binding assays were performed with [3H]DAMGO at 25°C for 60 min in 50 mM potassium phosphate buffer, pH 7.4, containing 5 mM magnesium sulfate. Specific binding was defined as the difference between total binding and non-specific binding, determined in the presence of 10 μM levallorphan. KD and Ki values were calculated by non-linear regression analysis (GraphPad Prism, La Jolla, CA, USA). Protein concentrations were determined as previously described (Lowry) using bovine serum albumin as the standard.
[35S]GTPγS binding assay
Membranes prepared from stable transfectants were incubated in the presence and absence of indicated opioids for 60 min at 30°C in the assay buffer (50 mM Tris-HCl, pH 7.7, 3 mM MgCl2, 0.2 mM EGTA, 10 mM NaCl) containing 0.05 nM [35S]GTPγS (PerkimElmer) and 60 μM GDP, as previously reported (Bolan et al. 2004; Pasternak et al. 2004). Basal binding was determined in the presence of GDP and absence of drug. The reaction was terminated by rapid filtration under vacuum through glass fiber filters, followed by three washes with 3 mL of ice-cold 50 mM Tris-HCl, pH 7.4. Bound radioactivity was measured by liquid scintillation spectrophotometry in Liquid Scintillation Analyzer (TRI-CARB 2900TR, PerkimElmer) after overnight extraction in 5 mL liquiscint scintillation fluid (National Diagnostic Inc., Atlanta, GA, USA).
Results
Isolating three new exon 11-associated splice variants from the human OPRM1 gene
The published human genome databases provide a valuable resource for identifying potential alternatively spliced exons and variants that are homologous to those from other species. Searching the human genome databases using the mouse sequences had led us to isolate at least seven splice variants of the human OPRM1 gene (Pan et al. 2003, 2005a). Using a similar strategy, we blasted the human genome databases with the mouse exon 11 sequence to identify a mouse exon 11 homolog. One alignment with moderate homology was mapped to a region located at ∼28 kb upstream of exon 1 in the human OPRM1 gene locus, a distance similar to the ∼30 kb between exons 11 and 1 in the mouse OPRM1 gene, suggesting that the region was a good candidate for the human exon 11 (Fig. 1). We then isolated three partial clones in RT-PCR with a sense primer from the region and anti-sense primers from exon 2. Sequence analysis indicated that all three partial clones contained the sequence defined by the sense primer as the 5′ exon, which was homologous to the mouse exon 11, and shared similar splicing patterns of mMOR-1H, mMOR-1I and mMOR-1G, respectively. These results suggested that the human OPRM1 gene contained the mouse exon 11 homolog, human exon 11, and its associated splice variants. Sequential RT-PCR using primers from exons 11 and 4 enabled us to isolate three full length clones, hMOR-G1, hMOR-1G2 and hMOR-1i (Figs 1 and 2).
Fig. 2.
The partial nucleotide sequence and predicted amino acid sequence of the human variants. Exon-exon boundaries are indicated by arrows. The stop codons are showed by *. The complete cDNA and deduced amino acid sequences of hMOR-1G1, hMOR-1G2, hMOR-1H and hMOR-1i have been deposited in the GenBank database with Accession numbers: EU340242, EU340243, DQ680044 and EU340241, respectively. (Although variants have traditionally be designated using MOR-1 and an uppercase letter, hMOR-1i and hMOR-1o are give lowercase letters to avoid confusion with similar appearing numbers.)
The human exon 11 (> 600 bp) was longer than the mouse exon 11 (∼270 bp) and contained a splice site within it that yielded variants splicing together exons 11a and 1, as seen with the clones hMOR-1G1 and hMOR-1G2. Both hMOR-1G1 and hMOR-1G2 contained exons 2, 3 and 4, but differential usage of the splice sites in exon 11 (Fig. 2). In hMOR-1G2, exon 11b was skipped and exon 11a was spliced directly to exon 2. The predicted protein sequence of hMOR-1G2 from exon 11a was in frame with those from exons 2, 3 and 4 and suggested a 6 TM structure similar to that of mMOR-1G The sixteen amino acids deduced from human exon 11a shared ∼43% homology with that from mouse exon 11.
In hMOR-1G1, both exons 11a and 11b were included by using the downstream splice site of exon 11b. Translation from the translational start codon, AUG, in exon 11a predicted only a short 24 amino acid peptide because of early termination of translation within exon 11b. However, if the first AUG from exon 2 is used to initiate translation, hMOR-1G1 would generate a 6 TM protein.
The exon composition of hMOR-1i included exons 11a/11b, 1c, 1a, 2, 3 and 4, similar to that of mMOR-1I (Fig. 2). Interestingly, a 105 bp sequence (exon 1b) between exons 1c and 1a was spliced out in hMOR-1i, unlike the continuous exon 1 in mMOR-1I, and assigned as exon 1b. Translation from the AUG in exon 11a predicted a short peptide because of early termination, as in hMOR-1G1. However, translation using the AUG from exon 1c, which was 279 bases upstream of the initiation AUG in hMOR-1, predicted a protein with a novel sequence of 93 amino acids at the N-terminus followed by the same sequence as hMOR-1. These initial 93 amino acids did not contain any predicted glycosylation sites or transmembrane domains, implying that they simply led to an elongated extracellular N-terminus for hMOR-1i and a classical 7 TM G-protein coupled receptor structure. We were unable to isolate the full length of hMOR-1H variant despite several attempts with sense primers from exon 11 and anti-sense primers from all the exons downstream from exon 3. This might reflect a low abundance of the mRNA or a different downstream exon.
Regional distribution of the expression of the human exon 11-associated variant mRNAs
We next examined the expression of the variant mRNAs in several human brain regions by using RT-PCR (Fig. 3). hMOR-1 mRNA was detected in all regions with a 35-cycle PCR protocol whereas all the other variants required 45-cycles, suggesting a lower abundance. Unlike hMOR-1, which was expressed in all the regions examined, the expression of the variant mRNAs clearly differed among the brain regions. The most restricted distribution was seen with hMOR-1G1, which was detected only in the pons and cerebellum, followed by hMOR-1i, which was present only in the prefrontal cortex, pons and piriform cortex and not in the temporal cortex, nucleus accumbens, cerebellum and spinal cord. Although hMOR-1G2 was detectable in all the regions, its expression level in the spinal cord and temporal cortex was much lower than the other regions. These results suggested region-specific processing of these exon 11-associated variant mRNAs. The exon 11-associated variants are also present within the human BE(2)-C neuroblastoma cell line (Fig. 3), as determined by RT-PCR.
Fig. 3.
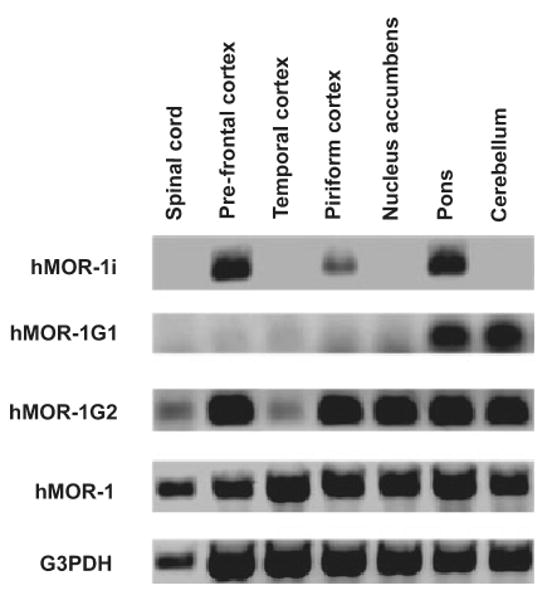
Regional distribution of the mRNAs from the human exon 11-associated variants. RT-PCRs were performed using total RNAs isolated from the indicated brain regions and Be(2)C cells using primers designed for amplifying hMOR-1G1, hMOR-1G2, hMOR-1i and hMOR-1 as described in Materials and Methods. G3PDH was used as RNA loading control.
Expression of human exon 11-associated variant protein
In vitro translation of the three full length clones revealed the predicted molecular weights on sodium dodecyl sulfate– polyacrylamide gel electrophoresis (Fig. 4). hMOR-1 generated a 44 kDa band that was also close to the predicted 45 kDa value from the AUG of exon 1a. hMOR-1G1 showed a band of approximately 33 kDa, corresponding to the 34 kDa predicted from the first AUG of exon 2. A 36 kDa band seen with hMOR-1G2 was similar to the 37 kDa size predicted from the AUG of exon 11a, suggesting that the AUG of exon 11a is preferred in hMOR-1G2. Although the first AUG of exon 2 can be used to start translation of hMOR-1G1 in vitro, it was not used in hMOR-1G2, hMOR-1i and hMOR-1 which upstream AUG codons. In vitro translation of hMOR-1i produced two bands, ∼54 kDa and ∼50 kDa, comparable to the 56 kDa and 51 kDa proteins predicted by using the first and second AUGs of exon 1c, respectively. The similar intensity of the two bands suggests that both AUGs are utilized equally well in this assay. In the mouse, several variants predicted small peptides, although we were not able to detect them on western blots (Pan et al. 2001). Several of the human variants also have the potential of generating similar small proteins. However, we did not observe these with in vitro translation. While this might be because of detection issues, it is not clear whether or not the small peptides can be produced in vitro.
Fig. 4.
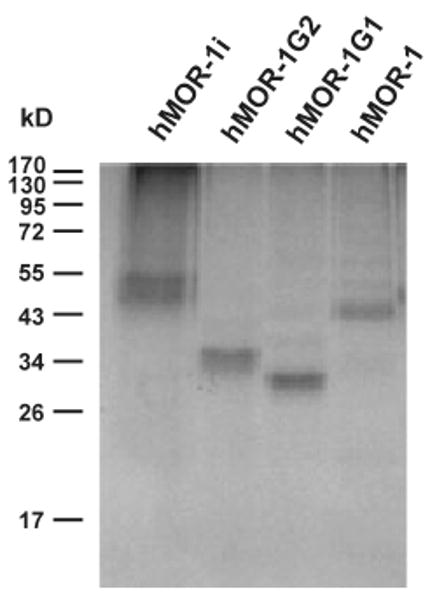
In vitro translation of the human exon 11-associated variants. In vitro transcription coupled translation was performed as described in Materials and Methods.
Pharmacological characterization of hMOR-1i in CHO cells
hMOR-1i was the only full length exon 11 variant isolated. To investigate its pharmacological profile, we established a stably transfected CHO cell line expressing hMOR-1i cDNA under control of a human cytomegalovirus early promoter and examined opioid binding using [3H]DAMGO in membranes isolated from the stable cell line. In saturation studies hMOR-1i bound [3H]DAMGO with high affinity indistinguishable from hMOR-1 (Table 1). Competition studies confirmed the mu selectivity of the binding (Table 2). Mu ligands such as morphine and M6G competed binding with high affinity, whereas the delta-selective ligand, [d-Pen2, d-Pen5]enkephalin (DPDPE), and the kappa1-selective opioid, U50,488H, were ineffective. The partial agonists, nalorphine and buprenorphine, also displayed high affinities for both hMOR-1 and hMOR-1i. The lack of any significant binding affinity differences among the drugs between hMOR-1 and hMOR-1i suggested that the extended N-terminal sequence in hMOR-1i did not influence mu binding characteristics.
Table 1.
Saturation studies with [3H]DAMGO binding
| Clone | KD (nM) | Bmax (pmol/mg protein) |
|---|---|---|
| hMOR-1 | 0.25 ± 0.02 | 0.18 ± 0.02 |
| hMOR-1i | 0.28 ± 0.01 | 0.28 ± 0.01 |
Saturation studies [3H]DAMGO binding was performed in membranes isolated from CHO cells stably transfected with the indicated cDNA clones, as described in Materials and Methods. Results are the mean ± SEM of at least three independent determinations.
Table 2.
Competition of [3H]DAMGO binding in hMOR-1 and hMOR-1i cells membranes
| Ki value (nM) | ||
|---|---|---|
| Ligand | hMOR-1 | hMOR-1i |
| Morphine | 0.8 ± 0.2 | 1.0 ± 0.1 |
| M6G | 2.5 ± 0.3 | 2.7 ± 0.2 |
| DADLE | 6.6 ± 1.1 | 6.2 ± 1.3 |
| DSLET | 4.9 ± 0.7 | 6.6 ± 1.5 |
| β-Endorphin | 1.4 ± 0.2 | 1.6 ± 0.2 |
| Dynorphin A | 9.3 ± 1.1 | 13.1 ± 2.0 |
| Methadone | 7.7 ± 0.2 | 11.2 ± 5.0 |
| Nalorphine | 1.1 ± 0.2 | 1.5 ± 0.3 |
| Buprenorphine | 1.8 ± 0.04 | 2.0 ± 0.2 |
| Naloxone | 0.6 ± 0.1 | 0.7 ± 0.2 |
| U50,488H | > 500 | > 500 |
| DPDPE | > 500 | > 500 |
Competition studies were performed with at least four concentrations of the indicated drugs against [3H]DAMGO binding in membranes from CHO cells stably transfected with the indicated cDNA clones, as described in Materials and Methods. IC50 values were determined by non-linear regression analysis and converted to Ki values (Cheng and Prusoff 1973). The results are the means ± SEM of at least three independent determinations.
C-terminal splicing led to significant functional differences in mu agonist-induced G protein coupling, as determined by opioid stimulated [35S]GTPγS binding (Bolan et al. 2004; Pasternak et al. 2004; Pan et al. 2005a,b). To investigate whether the changes at the N-terminus had a similar effect, we examined the effect of several agonists and partial agonists on stimulating [35S]GTPγS binding in the stable CHO clones expressing hMOR-1i or hMOR-1.
We observed significant differences in the EC50 of the [35S]GTPγS stimulation of a number of the agonists between the two splice variants despite the fact that no significant differences were observed in their binding affinities (Table 3). For example, the M6G and β-endorphin EC50 values differed over four-fold between hMOR-1 and hMOR-1i. The EC50 values for DAMGO and dynorphin A also differed by two- to three-fold.
Table 3.
Opioid-induced stimulation of [35S]GTPγS binding in hMOR-1 and hMOR-1i cells
| hMOR-1 | hMOR-1i | |||||
|---|---|---|---|---|---|---|
| EC50 (nM) | % Increase maximal stimulation | Relative efficacy (%) | EC50 (nM) | % Increase maximal stimulation | Relative efficacy (%) | |
| Morphine | 35 ± 13 | 127 ± 6 | 66 | 27 ± 8 | 207 ± 13 | 67 |
| M6G | 60 ± 9 | 143 ± 2 | 75 | 238 ± 24** | 227 ± 3 | 74 |
| DAMGO | 23 ± 6 | 149 ±116 | 77 | 76 ± 6 | 276 ± 12 | 90 |
| β-Endorphin | 42 ± 6 | 159 ±16 | 83 | 193 ± 32** | 263 ± 5 | 86 |
| Dynorphin A | 109 ± 23 | 192 ± 20 | 100 | 297 ± 35** | 307 ± 10 | 100 |
| Methadone | 81 ± 21 | 181 ± 19 | 94 | 117 ± 45 | 286 ± 10 | 93 |
| Nalorphine | 3.9 ± 2.6 | 22 ± 2 | 11 | 2.6 ± 1.9 | 40 ± 2 | 13 |
| Buprenorphine | 0.005 ± 0.002 | 38 ± 4 | 20 | 0.007 ± 0.005 | 74 ± 4 | 24 |
Membranes were prepared from CHO cells stably transfected with the indicated cDNA clones and [35S]GTPγS binding carried out as described in Materials and Methods. In brief, dose-response curves were determined for each opioid and the EC50 and % maximal stimulation values determined. The % maximal stimulation value represents the increase over basal levels of [35S]GTPγS binding and was calculated as: (Stimulated binding) basal binding)/(basal binding). Results are the means ± SEM of at least three independent determinations. Two-way ANOVA was performed on EC50 values to determine whether there were differences between hMOR-1 and hMOR-1i. The EC50 values for M6G, β-endorphin and dynorphin A were significantly different between hMOR-1 and hMOR-1i (**p < 0.001). Relative efficacy (%) of the drugs for the indicated variant was determined comparing the maximal stimulation of the drug compared to the drug that gave the highest level of stimulation (i.e. 100%).
It is difficult comparing the maximal stimulation of [35S]GTPγS binding between cell lines, particularly when expression levels differ since the maximal stimulation of [35S]GTPγS binding is dependent upon the number of receptors. Thus, the comparison of maximal stimulation levels (i.e. efficacy) is best made among drugs within the same cell line. In our current studies, the relative maximal stimulation among the drugs for hMOR-1 was very similar to that seen with hMOR-1i. Although the maximal stimulation levels with hMOR-1i cells were higher, this likely resulted from its higher expression levels than hMOR-1. Both nalorphine and buprenorphine were particularly noteworthy in that their maximal stimulation was far less than the other opioids, confirming their classification as partial agonists. Although more efficacious, morphine and M6G also were partial agonists, which was consistent with our earlier studies of the C-terminal splice variants (Bolan et al. 2004; Pan et al. 2005a,b).
Discussion
The mouse OPRM1 gene undergoes extensive splicing to generate multiple mu subtypes, confirming their initial suggestion (Wolozin and Pasternak 1981). The functional relevance of the mouse splice variants has been suggested by anti-sense and KO models, including recent studies with the exon 11 KO mice. Disruption of MOR-1 exon 1 eliminated morphine actions, but not those of heroin and morphine-6β-glucurnoide (Schuller et al. 1999). This residual analgesia was not because of cross activation of delta and kappa1 receptors since it was retained in a mu/delta/kappa1 triple KO mouse (M. King, J. Pintar and G.W. Pasternak, in preparation), suggesting a possible role for exon 11-associated variants that were still expressed in these mice (unpublished observation). On the other hand, elimination of the exon 11-associated variants had little effect on morphine and methadone analgesia, but significantly impaired the analgesic activity of heroin and M6G (Y.X. Pan, J. Xu, M. Xu, G. Rossi, J. Matulonis and G.W. Pasternak, submitted), strongly implying a role for exon 11-associated variants in heroin, and not morphine, actions. The dissociation of morphine and heroin actions in the mouse may prove important in understanding both opioid analgesia and addiction.
The primary question in the current study was whether or not a similar 5′ splicing profile existed in humans. Thus, our isolation of a human homolog of the mouse exon 11 along and three human exon 11-associated splice variants in both brain and a human neuroblastoma cell line raises the possibility that a similar pharmacological dissociation between morphine and heroin exists in people. Our findings, in conjunction with additional studies identifying seven homologous exon 11-associated variants in the rat (J. Xu, M. Xu, G.W. Pasternak and Y.X. Pan, in preparation), suggests conservation of exon 11 and its associated splice variants across species. With the new exon 11-associated variants, the human OPRM1 gene now contains at least 10 exons spanning over 200 kb, a size comparable to ∼250 kb of the mouse OPRM1 gene that generates at least 17 splice variants.
Overall, splicing of the human OPRM1 gene corresponds closely with that in the mouse. The exon 11 homologs are located ∼30 kb upstream of exon 1 in the mouse and ∼28 kb in humans. However, the human exon 11 is longer than mouse exon 11 and contains an alternative splice site that divides the human exon 11 into two parts, a splicing pattern not seen present in the mouse exon 11. The mouse contains a separate promoter located just upstream from exon 11 that controls the expression of the nine exon 11-associated splice variants (Pan 2002; Xu et al. 2006). The location of the human exon 11 approximately 28 kb upstream from exon 1 raises the obvious question of whether a similar human exon 11 promoter exists at upstream of exon 11. Preliminary studies have suggested that the 5′ flanking region of the human exon 11 contains promoter activity, as demonstrated in human neuroblastoma cell lines using a secreted alkaline phosphotase reporter assay (J. Xu, M. Xu and X.Y. Pan, in preparation).
The mouse has three exon 11-associated variants that contain exon 1 and predict full length receptor proteins identical to that of mMOR-1 itself. In humans, hMOR-1i contains exon 1 and predicts a full length receptor very similar to hMOR-1. The difference at the protein level between hMOR-1i and hMOR-1 is because of the presence of an additional initial 93 amino acids at the tip of the N-terminus. hMOR-1i was the first splice variant isolated so far from OPRM1 genes that predicts a receptor with additional amino acid sequences extended at the N-terminus of hMOR-1. It was not surprising that the pharmacological binding profiles of hMOR-1i were indistinguishable with those of hMOR-1 since their predicted binding pockets involve identical transmembrane domains. However, differences in agonist-induced G protein coupling assessed with [35S]GTPγS binding assay raises a number of interesting questions. Foremost is how the additional extracellular 93 amino acids at the tip of the N-terminus can alter receptor-G-protein coupling which is presumably more dependent upon the intracellular structures, as well as how can these changes influence transduction without changing opioid binding.
A number of exon 11-associated variants in mice predict truncated variants. Several predict very small (< 10 kDa) proteins that can be generated with in vitro translation, but that have not yet been isolated from brain. However, the 6 TM variants mMOR-1G, mMOR-1M and mMOR-1N are expressed in mouse brain (Pan et al. 2001). Although the functional significance of these truncated variants remains unclear, evidence is mounting for their relevance. [3H]Diprenorphine can label the mouse 6 TM variants (KD ∼10 nM) and the mouse 6 TM variants are able to modulate the expression and function of mMOR-1 when they were co-expressed (Y.X. Pan, J. Xu and G.W. Pasternak, unpublished observation). Furthermore, they can physically associate with, and modulate the activity of, traditional full length MOR-1 variants (X.Y. Pan, J. Xu, M. Xu and G.W. Pasternak, unpublished observations). The functional significance of the mouse exon 11-associated splice variants was further supported by finding in an exon 11 KO mouse. Disrupting exon 11 greatly diminished the analgesic actions of M6G, fentanyl and heroin, while the analgesic activity of morphine and methadone were not affected. Similar 6 TM variants have now been isolated from humans. Like the mouse 6 TM variants, both hMOR-1G1 and hMOR-1G2 produced a 6 TM protein, as illustrated by in vitro translation studies. The conservation of these variants in mice, rats and humans is consistent with a functional significance.
The 6 TM variants lack the first TM encoded by exon 1. This is interesting since there is another human variant comprised only of TM 1 (Du et al. 1997; Pan 2005), the one missing in these new human variants. Preliminary studies with the mouse variants indicate that the single TM and the 6 TM variants can physically associate, raising the question of whether the complexed variants might complement each other to form a complete 7 TM receptor (X.Y. Pan and G.W. Pasternak, unpublished observations). However, the functional effects of these dimers have not yet been documented.
Finally, the regional distribution of the exon 11-associated variants varied markedly among brain regions, implying cell-specific and region-specific mRNA processing. This is similar to the mouse, which also demonstrated differences in their distribution at both the mRNA and protein level (Pan et al. 2001; Abbadie et al. 2004).
In conclusion, the discovery of human exon 11 and its associated splice variants further illustrates the complexity of the human OPRM1 gene and the overall conservation of the splicing patterns among mice, rats and humans. The importance of the exon 11-associated variants in mice in mediating the actions of heroin and M6G suggests that these human exon 11-associated variants may also prove important clinically and may help explain the diverse responses of patients to the various mu opioids.
Acknowledgments
This work was supported, in part, by research grants to Y.-X.P (DA13997), G.W.P (DA02615) and a Senior Scientist Award to G.W.P. (DA00220) and Y.L.H (DA15446) from the National Institute on Drug Abuse, a grant from the National Genetics Foundation to G.W.P. and by a Core Grant to MSKCC from the National Cancer Institute (CA8748).
Abbreviations
- 5-HT
5-hydroxytryptamine
- CHO
Chinese hamster ovary
- DAMGO
[D-Ala2,MePhe4,Gly(ol)5]enkephalin
- KO
knockout
- M6G
morphine-6β-glucuronide
- MOR
mu opioid receptor
- OPRM1
mu opioid receptor gene
- TM
transmembrane
References
- Abbadie C, Gultekin SH, Pasternak GW. Immunohistochemical localization of the carboxy terminus of the novel mu opioid receptor splice variant MOR-1C within the human spinal cord. Neuroreport. 2000a;11:1953–1957. doi: 10.1097/00001756-200006260-00029. [DOI] [PubMed] [Google Scholar]
- Abbadie C, Pan YX, Pasternak GW. Differential distribution in rat brain of mu opioid receptor carboxy terminal splice variants MOR-1C-like and MOR-1-like immunoreactivity: evidence for region-specific processing. J Comp Neurol. 2000b;419:244–256. doi: 10.1002/(sici)1096-9861(20000403)419:2<244::aid-cne8>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Abbadie C, Pan YX, Drake CT, Pasternak GW. Comparative immunhistochemical distributions of carboxy terminus epitopes from the mu opioid receptor splice variants MOR-1D, MOR-1 and MOR-1C in the mouse and rat central nervous systems. Neuroscience. 2000c;100:141–153. doi: 10.1016/s0306-4522(00)00248-7. [DOI] [PubMed] [Google Scholar]
- Abbadie C, Pasternak GW, Aicher SA. Presynaptic localization of the carboxy-terminus epitopes of the mu opioid receptor splice variants MOR-1C and MOR-1D in the superficial laminae of the rat spinal cord. Neuroscience. 2001;106:833–842. doi: 10.1016/s0306-4522(01)00317-7. [DOI] [PubMed] [Google Scholar]
- Abbadie C, Pan YX, Pasternak GW. Immunohistochemical study of the expression of exon11-containing mu opioid receptor variants in mouse brain. Neuroscience. 2004;127:419–430. doi: 10.1016/j.neuroscience.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Bare LA, Mansson E, Yang D. Expression of two variants of the human l opioid receptor mRNA in SK-N-SH cells and human brain. FEBS Lett. 1994;354:213–216. doi: 10.1016/0014-5793(94)01129-x. [DOI] [PubMed] [Google Scholar]
- Bender E, Pindon A, Van Oers I, Zhang YB, Gommeren W, Verhasselt P, Jurzak M, Leysen J, Luyten W. Structure of the human serotonin 5-HT4 receptor gene and cloning of a novel 5-HT4 splice variant. J Neurochem. 2000;74:478–489. doi: 10.1046/j.1471-4159.2000.740478.x. [DOI] [PubMed] [Google Scholar]
- Blondel O, Gastineau M, Dahmoune Y, Langlois M, Fischmeister R. Cloning, expression, and pharmacology of four human 5-hydroxytryptamine 4 receptor isoforms produced by alternative splicing in the carboxyl terminus. J Neurochem. 1998;70:2252–2261. doi: 10.1046/j.1471-4159.1998.70062252.x. [DOI] [PubMed] [Google Scholar]
- Bolan EA, Pan YX, Pasternak GW. Functional analysis of MOR-1 splice variants of the mouse mu opioid receptor gene Oprm. Synapse. 2004;51:11–18. doi: 10.1002/syn.10277. [DOI] [PubMed] [Google Scholar]
- Cadet P, Mantione KJ, Stefano GB. Molecular identification and functional expression of mu 3, a novel alternatively spliced variant of the human mu opiate receptor gene. J Immunol. 2003;170:5118–5123. doi: 10.4049/jimmunol.170.10.5118. [DOI] [PubMed] [Google Scholar]
- Canton H, Emeson RB, Barker EL, Backstrom JR, Lu JT, Chang MS, Sanders-Bush E. Identification, molecular cloning, and distribution of a short variant of the 5-hydroxy-tryptamine2C receptor produced by alternative splicing. Mol Pharmacol. 1996;50:799–807. [PubMed] [Google Scholar]
- Chang A, Emmel DW, Rossi GC, Pasternak GW. Methadone analgesia in morphine-insensitive CXBK mice. Eur J Pharmacol. 1998;351:189–191. doi: 10.1016/s0014-2999(98)00366-5. [DOI] [PubMed] [Google Scholar]
- Chen Y, Mestek A, Liu J, Hurley JA, Yu L. Molecular cloning and functional expression of a μ-opioid receptor from rat brain. Mol Pharmacol. 1993;44:8–12. [PubMed] [Google Scholar]
- Cheng YC, Prusoff WH. Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50 percent inhibiton (IC50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- Cherny N, Ripamonti C, Pereira J, Davis C, Fallon M, McQuay H, Mercadante S, Pasternak G, Ventafridda V. Strategies to manage the adverse effects of oral morphine: an evidence-based report. J Clin Oncol. 2001;19:2542–2554. doi: 10.1200/JCO.2001.19.9.2542. [DOI] [PubMed] [Google Scholar]
- Choi HS, Kim CS, Hwang CK, Song KY, Wang W, Qiu Y, Law PY, Wei LN, Loh HH. The opioid ligand binding of human mu-opioid receptor is modulated by novel splice variants of the receptor. Biochem Biophys Res Commun. 2006;343:1132–1140. doi: 10.1016/j.bbrc.2006.03.084. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Doyle GA, Sheng XR, Lin SS, Press DM, Grice DE, Buono RJ, Ferraro TN, Berrettini WH. Identification of three mouse micro-opioid receptor (MOR) gene (Oprm1) splice variants containing a newly identified alternatively spliced exon. Gene. 2007a;388:135–147. doi: 10.1016/j.gene.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Doyle GA, Sheng XR, Lin SS, Press DM, Grice DE, Buono RJ, Ferraro TN, Berrettini WH. Identification of five mouse mu-opioid receptor (MOR) gene (Oprm1) splice variants containing a newly identified alternatively spliced exon. Gene. 2007b;395:98–107. doi: 10.1016/j.gene.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du YL, Elliot K, Pan YX, Pasternak GW, Inturrisi CE. A splice variant of the mu opioid receptor is present in human SHSY-5Y cells. Soc Neurosci. 1997;23:1206. [Google Scholar]
- Foley KM. Management of cancer pain. In: DeVita VT, Hellman S, Rosenberg SA, editors. Cancer: Principles and Practice of Oncology. Lippincott; New York: 1985. pp. 1940–1961. [Google Scholar]
- Guest PC, Salim K, Skynner HA, George SE, Bresnick JN, McAllister G. Identification and characterization of a truncated variant of the 5-hydroxytryptamine(2A) receptor produced by alternative splicing. Brain Res. 2000;876:238–244. doi: 10.1016/s0006-8993(00)02664-0. [DOI] [PubMed] [Google Scholar]
- Hope AG, Downie DL, Sutherland L, Lambert JJ, Peters JA, Burchell B. Cloning and functional expression of an apparent splice variant of the murine 5-HT3 receptor A subunit. Eur J Pharmacol. 1993;245:187–192. doi: 10.1016/0922-4106(93)90128-v. [DOI] [PubMed] [Google Scholar]
- Jasper JR, Kosaka A, To ZP, Chang DJ, Eglen RM. Cloning, expression and pharmacology of a truncated splice variant of the human 5-HT7 receptor (h5-HT7b) Br J Pharmacol. 1997;122:126–132. doi: 10.1038/sj.bjp.0701336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Ling GSF, MacLeod JM, Lee S, Lockhart SH, Pasternak GW. Separation of morphine analgesia from physical dependence. Science. 1984;226:462–464. doi: 10.1126/science.6541807. [DOI] [PubMed] [Google Scholar]
- Ling GSF, Spiegel K, Lockhart SH, Pasternak GW. Separation of opioid analgesia from respiratory depression: evidence for different receptor mechanisms. J Pharmacol Exp Ther. 1985;232:149–155. [PubMed] [Google Scholar]
- Lord JAH, Waterfield AA, Hughes J, Kosterlitz HW. Endogenous opioid peptides: multiple agonists and receptors. Nature. 1977;267:495–499. doi: 10.1038/267495a0. [DOI] [PubMed] [Google Scholar]
- Martin WR, Eades CG, Thompson JA, Huppler RE, Gilbert PE. The effects of morphine and nalorphine-like drugs in the nondependent and morphine-dependent chronic spinal dog. J Pharmacol Exp Ther. 1976;197:517–532. [PubMed] [Google Scholar]
- Monsma FJ, McVittie LD, Gerfen CR, Mahan LC, Sibley DR. Multiple D2 dopamine receptors produced by alternative RNA splicing. Nature. 1989;342:926–929. doi: 10.1038/342926a0. [DOI] [PubMed] [Google Scholar]
- Moulin DE, Ling GSF, Pasternak GW. Unidirectional analgesic cross tolerance between morphine and levorphanol in the rat. Pain. 1988;33:233–239. doi: 10.1016/0304-3959(88)90095-4. [DOI] [PubMed] [Google Scholar]
- Namba T, Sugimoto Y, Negishi M, Irie A, Ushikubi F, Kakizuka A, Ito S, Ichikawa A, Narumiya S. Alternative splicing of C-terminal tail of prostaglandin E receptor subtype EP3 determines G-protein specificity. Nature. 1993;365:166–170. doi: 10.1038/365166a0. [DOI] [PubMed] [Google Scholar]
- Olsen MA, Nawoschik SP, Schurman BR, Schmitt HL, Burno M, Smith DL, Schechter LE. Identification of a human 5-HT6 receptor variant produced by alternative splicing. Brain Res Mol Brain Res. 1999;64:255–263. doi: 10.1016/s0169-328x(98)00338-6. [DOI] [PubMed] [Google Scholar]
- Pan YX. Identification and characterization of a novel promoter of the mouse mu opioid receptor gene (Oprm) that generates eight splice variants. Gene. 2002;295:97–108. doi: 10.1016/s0378-1119(02)00825-9. [DOI] [PubMed] [Google Scholar]
- Pan YX. Diversity and complexity of the mu opioid receptor gene: alternative pre-mRNA splicing and promoters. DNA Cell Biol. 2005;24:736–750. doi: 10.1089/dna.2005.24.736. [DOI] [PubMed] [Google Scholar]
- Pan YX, Cheng J, Xu J, Rossi G, Jacobson E, RyanMoro J, Brooks AI, Dean GE, Standifer KM, Pasternak GW. Cloning and functional-characterization through antisense mapping of a kappa(3)-related opioid receptor. Mol Pharmacol. 1995;47:1180–1188. [PubMed] [Google Scholar]
- Pan YX, Xu J, RyanMoro J, Mathis J, Hom JSH, Mei JF, Pasternak GW. Dissociation of affinity and efficacy in KOR-3 chimeras. FEBS Lett. 1996;395:207–210. doi: 10.1016/0014-5793(96)01023-x. [DOI] [PubMed] [Google Scholar]
- Pan YX, Xu J, Bolan E, Abbadie C, Chang A, Zuckerman A, Rossi G, Pasternak GW. Identification and characterization of three new alternatively spliced mu-opioid receptor isoforms. Mol Pharmacol. 1999;56:396–403. doi: 10.1124/mol.56.2.396. [DOI] [PubMed] [Google Scholar]
- Pan YX, Xu J, Bolan E, Chang A, Mahurter L, Rossi G, Pasternak GW. Isolation and expression of a novel alternatively spliced mu opioid receptor isoform, MOR-1F. FEBS Lett. 2000;466:337–340. doi: 10.1016/s0014-5793(00)01095-4. [DOI] [PubMed] [Google Scholar]
- Pan YX, Xu J, Mahurter L, Bolan E, Xu M, Pasternak GW. Generation of the mu opioid receptor (MOR-1) protein by three new splice variants of the Oprm gene. Proc Natl Acad Sci USA. 2001;98:14084–14089. doi: 10.1073/pnas.241296098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YX, Xu J, Mahurter L, Xu MM, Gilbert AK, Pasternak GW. Identification and characterization of two new human mu opioid receptor splice variants, hMOR-1O and hMOR-1X. Biochem Biophys Res Commun. 2003;301:1057–1061. doi: 10.1016/s0006-291x(03)00089-5. [DOI] [PubMed] [Google Scholar]
- Pan L, Xu J, Yu R, Xu MM, Pan YX, Pasternak GW. Identification and characterization of six new alternatively spliced variants of the human mu opioid receptor gene, Oprm. Neuroscience. 2005a;133:209–220. doi: 10.1016/j.neuroscience.2004.12.033. [DOI] [PubMed] [Google Scholar]
- Pan YX, Xu J, Bolan E, Moskowitz HS, Xu M, Pasternak GW. Identification of four novel exon 5 splice variants of the mouse mu-opioid receptor gene: functional consequences of C-terminal splicing. Mol Pharmacol. 2005b;68:866–875. doi: 10.1124/mol.105.011858. [DOI] [PubMed] [Google Scholar]
- Pasternak GW. Pharmacological mechanisms of opioid analgesics. Clin Neuropharmacol. 1993;16:1–18. doi: 10.1097/00002826-199302000-00001. [DOI] [PubMed] [Google Scholar]
- Pasternak GW. The pharmacology of mu analgesics: from patients to genes. Neuroscientist. 2001;7:220–231. doi: 10.1177/107385840100700307. [DOI] [PubMed] [Google Scholar]
- Pasternak GW. Multiple opiate receptors: deja vu all over again. Neuropharmacology. 2004;47(Suppl 1):312–323. doi: 10.1016/j.neuropharm.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Pasternak DA, Pan L, Xu J, Yu R, Xu MM, Pasternak GW, Pan YX. Identification of three new alternatively spliced variants of the rat mu opioid receptor gene: dissociation of affinity and efficacy. J Neurochem. 2004;91:881–890. doi: 10.1111/j.1471-4159.2004.02767.x. [DOI] [PubMed] [Google Scholar]
- Payne R, Pasternak GW. Pain. In: Johnston MV, Macdonald RL, Young AB, editors. Principles of Drug Therapy in Neurology. F.A. Davis; Philadelphia: 1992. pp. 268–301. [Google Scholar]
- Portenoy RK. Opioid analgesics. In: Portenoy RK, Kanner RM, editors. Pain Management: Theory and Practice. F.A. Davis; Philadelphia: 1996. pp. 248–276. [Google Scholar]
- Rossi GC, Pan YX, Brown GP, Pasternak GW. Antisense mapping the MOR-1 opioid receptor: evidence for alternative splicing and a novel morphine-6β-glucuronide receptor. FEBS Lett. 1995a;369:192–196. doi: 10.1016/0014-5793(95)00757-z. [DOI] [PubMed] [Google Scholar]
- Rossi GC, Standifer KM, Pasternak GW. Differential blockade of morphine and morphine-6β-glucuronide analgesia by antisense oligodeoxynucleotides directed against MOR-1 and G-protein α subunits in rats. Neurosci Lett. 1995b;198:99–102. doi: 10.1016/0304-3940(95)11977-5. [DOI] [PubMed] [Google Scholar]
- Rossi GC, Leventhal L, Pan YX, Cole J, Su W, Bodnar RJ, Pasternak GW. Antisense mapping of MOR-1 in rats: distinguishing between morphine and morphine-6beta-glucuronide antinociception. J Pharmacol Exp Ther. 1997;281:109–114. [PubMed] [Google Scholar]
- Schnell SA, Wessendorf MW. Expression of MOR1C-like mu-opioid receptor mRNA in rats. J Comp Neurol. 2004;473:213–232. doi: 10.1002/cne.20103. [DOI] [PubMed] [Google Scholar]
- Schuller AG, King MA, Zhang J, et al. Retention of heroin and morphine-6 beta-glucuronide analgesia in a new line of mice lacking exon 1 of MOR-1. Nat Neurosci. 1999;2:151–156. doi: 10.1038/5706. [DOI] [PubMed] [Google Scholar]
- Stamm S, Riethoven JJ, Le Texier V, Gopalakrishnan C, Kumanduri V, Tang Y, Barbosa-Morais NL, Thanaraj TA. ASD: a bioinformatics resource on alternative splicing. Nucleic Acids Res. 2006;34:D46–D55. doi: 10.1093/nar/gkj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Narumiya S. Prostaglandin E receptors. J Biol Chem. 2007;282:11613–11617. doi: 10.1074/jbc.R600038200. [DOI] [PubMed] [Google Scholar]
- Thompson RC, Mansour A, Akil H, Watson SJ. Cloning and pharmacological characterization of a rat μ opioid receptor. Neuron. 1993;11:903–913. doi: 10.1016/0896-6273(93)90120-g. [DOI] [PubMed] [Google Scholar]
- Vanetti M, Kouba M, Wang X, Vogt G, Hollt V. Cloning and expression of a novel mouse somatostatin receptor (SSTR2B) FEBS Lett. 1998;311:290–294. doi: 10.1016/0014-5793(92)81122-3. [DOI] [PubMed] [Google Scholar]
- Vukusic I, Grellscheid SN, Wiehe T. Applying genetic programming to the prediction of alternative mRNA splice variants. Genomics. 2007;89:471–479. doi: 10.1016/j.ygeno.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Wang JB, Imai Y, Eppler CM, Gregor P, Spivak CE, Uhl GR. μ opiate receptor: cDNA cloning and expression. Proc Natl Acad Sci USA. 1993;90:10230–10234. doi: 10.1073/pnas.90.21.10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin BL, Pasternak GW. Classification of multiple morphine and enkephalin binding sites in the central nervous system. Proc Natl Acad Sci USA. 1981;78:6181–6185. doi: 10.1073/pnas.78.10.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Xu M, Pan YX. Characterizing exons 11 and 1 promoters of the mu opioid receptor (Oprm) gene in transgenic mice. BMC Mol Biol. 2006;7:41. doi: 10.1186/1471-2199-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pan YX, Kolesnikov Y, Pasternak GW. Immunohistochemical labeling of the mu opioid receptor carboxy terminal splice variant mMOR-1B4 in the mouse central nervous system. Brain Res. 2006;1099:33–43. doi: 10.1016/j.brainres.2006.04.133. [DOI] [PubMed] [Google Scholar]
- Zimprich A, Simon T, Hollt V. Cloning and expression of an isoform of the rat l opioid receptor (rMOR 1 B) which differs in agonist induced desensitization from rMOR1. FEBS Lett. 1995;359:142–146. doi: 10.1016/0014-5793(95)00028-8. [DOI] [PubMed] [Google Scholar]