Abstract
Lung cancer is the most common cancer worldwide. Polymorphisms in genes associated with carcinogen metabolism may modulate risk of disease. Glutathione S-transferase pi (GSTP1) detoxifies polycyclic aromatic hydrocarbons found in cigarette smoke and is the most highly expressed glutathione S-transferase in lung tissue. A polymorphism in the GSTP1 gene, an A-to-G transition in exon 5 (Ile105Val, 313A → 313G), results in lower activity among individuals who carry the valine allele. The authors present a meta- and a pooled analysis of case-control studies that examined the association between this polymorphism in GSTP1 and lung cancer risk (27 studies, 8,322 cases and 8,844 controls and 15 studies, 4,282 cases and 5,032 controls, respectively). Overall, the meta-analysis found no significant association between lung cancer risk and the GSTP1 exon 5 polymorphism. In the pooled analysis, there was an overall association (odds ratio = 1.11, 95% confidence interval: 1.03, 1.21) between lung cancer and carriage of the GSTP1 Val/Val or Ile/Val genotype compared with those carrying the Ile/Ile genotype. Increased risk varied by histologic type in Asians. There appears to be evidence for interaction between amount of smoking, the GSTP1 exon 5 polymorphism, and risk of lung cancer in whites.
Keywords: Asian continental ancestry group, epidemiology, glutathione S-transferase pi, GSTP1, lung neoplasms, smoking
GENE AND GENE VARIANTS
Glutathione S-transferases are a supergene family of phase II enzymes present in many tissues, including lung (1). These enzymes catalyze the detoxification (through conjugation of glutathione) of a variety of reactive electrophilic compounds, including many environmental carcinogens such as benzo[a]-pyrene and polycyclic aromatic hydrocarbons (PAHs) (2). The soluble glutathione S-transferases comprise 4 main gene classes, alpha (α), mu (μ), pi (π), and theta (θ) (3). Polymorphisms in the glutathione S-transferase pi gene, GSTP1, located on chromosome 11q13 in humans, have been associated with a reduction in enzymatic activity toward several substrates, including both chemotherapy agents (such as cisplatin, a common agent used in lung cancer treatment) and carcinogens found in tobacco smoke (4–9). Of the several thousand chemicals found in tobacco smoke, at least 50 are known to be carcinogenic, including PAHs, aromatic amines, and nitroso compounds (10).
GSTP1 detoxifies PAHs and is the most abundant glutathione S-transferase isoform in the lungs (1). Two single nucleotide polymorphisms in GSTP1 that result in a change in amino acids have been identified. A single nucleotide polymorphism in exon 5 (Ile105Val, 313A → 313G), the A-to-G transition that results in an amino acid change from isoleucine to valine, results in significantly lower conjugating activity among individuals who carry one or more copies of the G (guanine) allele (Ile/Val or Val/Val) compared with those who have the A/A (adenine/adenine; Ile/Ile) genotype (11–13). Having at least one copy of the G allele at this locus is also associated with increased levels of hydrophobic adducts in the lung and higher levels of PAH-DNA adducts in human lymphocytes (14). A second single nucleotide polymorphism in exon 6 (Ala114Val, 341C → 341T) results in an amino acid change from alanine to valine, which also appears to confer lower activity (11). Additionally, 3 functional haplotypes have been identified: GSTP1*A (105Ile;114Ala), GSTP1*B (105Val;114Ala), and GSTP1*C (105Val;114Val) (11). A meta-analysis published in 2006 of 5 polymorphisms in glutathione S-transferases found no association with GSTP1 polymorphisms and lung cancer risk in 25 studies published prior to August 2005 (15). The present report includes additional studies published since that time and a pooled analysis examining the association between the GSTP1 Ile105Val polymorphism and risk of lung cancer.
DISEASE
Lung cancer is the most common cancer worldwide and is responsible for 17.2% of all cancer-related deaths (16). In the United States, overall 5-year survival is about 16% for all stages combined (17). Data from the Surveillance, Epidemiology, and End Results Program indicate that if lung cancer is diagnosed in local stages, survival is significantly better, with overall 5-year survival rates of 49.1%, although fewer than 20% of lung cancers are diagnosed at this stage (17). Along with stage at diagnosis, prognosis also depends on histology type. Because of recent advances in technology that allow a more accurate diagnosis, it is difficult to analyze historic trends in histology types; however, adenocarcinomas of the lung have been increasing in proportion over the last 2–3 decades, especially among women (18). Overall, lung cancer survival rates have not significantly improved with advances in surgical, radiation, or chemotherapy treatments (17).
SMOKING
Cigarette smoking is the greatest risk factor associated with lung cancer development. In the United States and the United Kingdom, approximately 90% of all cases of lung cancer are attributable to current or former cigarette smoking, while the population attributable risks appear to be lower in Japanese populations, especially among women (population attributable risk for men = 67.0%, population attributable risk for women = 14.6%) (19, 20). Other Asian populations report similar risk of lung cancer due to smoking (21, 22). Worldwide, smoking rates have been declining for the past several decades in developed countries and increasing significantly in developing countries. If these trends in smoking rates continue, by 2030, developing countries will account for an estimated 80% of the annual 8 million tobacco-related deaths, many of which will be due to lung cancers (23). Since the induction period for lung cancer appears to be decades, lung cancer will continue to be a major public health issue for generations to come. In addition, the negative health effects of cigarette smoking are not limited to current smokers. In a cohort of former smokers in the United States, 10 years after smoking cessation, the risk of lung cancer is 30%–50% lower than the risk for those who continue to smoke, but lifelong risk remains elevated compared with that for never smokers (24). Furthermore, while cigarette smoking remains the most significant modifiable risk factor, exposure to radon and other occupational and environmental risk factors is associated with development of lung cancer (25, 26).
MATERIALS AND METHODS
Associations and interactions
The association between the exon 5 (Ile105Val, 313A → 313G) polymorphism in GSTP1 and lung cancer was examined through a meta-analysis of all published papers and a pooled analysis of selected published studies. A MEDLINE search was performed from January 1988 (when the structure of GSTP1 was first described (27)) until March 31, 2007, using different combinations of “glutathione S-transferase pi,” “GSTP1,” “lung,” and “lung cancer,” restricting the analysis to “human” with no restriction on language. This search was supplemented by examining the reference sections of all selected papers, plus 2 reviews (28, 29) and a pooled analysis of polymorphisms in candidate genes associated with early-onset (<60 years of age at diagnosis) lung cancer (30).
After reviewing all abstracts ascertained from these searches, 34 articles containing information on GSTP1 polymorphisms and lung cancer were identified. Eligible studies included the frequency of GSTP1 genotypes or the crude odds ratio for the GSTP1 exon 5 polymorphism and lung cancer. Both hospital- and population-based case-control studies were included in the analysis. Additionally, 1 study was a nested case-control study from a large cohort of physicians (31). Of the 34 articles selected, 4 were excluded because they were case-only analyses (32–35), 2 because of subject overlap with more recently published studies (36, 37), and 1 because it did not report the genotypes or unadjusted odds ratios (38). Two studies were included in the meta-analysis even though they contained a small number of overlapping subjects (39, 40). Two studies were found in both non-English and, later, English journals; therefore, the data from the English journals were used (41, 42). Only 4 studies reported on the exon 6 (Ala114Val, 341C → 341T) polymorphism, so we restricted the analysis to the exon 5 polymorphism in GSTP1. The final number of studies in the meta-analysis was 27, including 8,322 cases and 8,844 controls (31, 39–64) (Table 1).
Table 1.
Description of the Studies Included in the Meta-analysis by Ethnicity and Year of Publication
| First Author (Reference No.) | Year | No. of Cases | No. of Controls | Country | Mean Age of Cases, Years | Male Cases, % | Histology | Source of Controls | Matching Criteria |
| Asian studies | |||||||||
| Katoh (47) | 1999 | 47 | 122 | Japan | 64.6 (SD, 10.3) | 85 | SqCC = 51.1%, AC = 25.5%, SCC = 19.1%, LCC = 4.3% | Hospital | None |
| Kihara (48) | 1999 | 358 | 257 | Japan | 62.7 (range, 58–67) | 100 | SqCC = 33.3%, SCC = 20.4%, AC = 46.3% | Hospital | None |
| Kiyohara (41) | 2000 | 86 | 88 | Japan | 63.8 (range, 35–86) | 100 | AC = 45.5%, SqCC = 7.9%, SCC = 13.9%, LCC = 4.7%, others = 7.0% | Hospital | None |
| Lin (51) | 2003 | 198 | 332 | Taiwan | 64 (SD, 9) | 72.2 | AC = 53.0%, SCC = 42.0%, others = 5.0% | Hospital | None |
| Wang (60) | 2003 | 112 | 119 | China | 56.5 (range, 37–75; SD, 8.1) | 64.3 | AC = 100% | Healthy | Age and gender (frequency matching) |
| Chan-Yeung (44) | 2004 | 229 | 197 | China | 53.8 (SD, 14.3) | 67.2 | AC = 55.5%, SqCC = 16.6%, NSCLC = 19.2%, others = 8.7% | Healthy | Ethnicity |
| Chan (43) | 2005 | 75 | 162 | China | 63 (no range or SD) | 82 | AC = 58.7%, SqCC = 41.3% | Hospital | Sex and age |
| Liang (42) | 2005 | 227 | 227 | China | 62.5 (range, 31–86) | 74 | SqCC = 41.4%, AC = 58.6% | Hospital | Age, gender, and ethnicity (frequency matching) |
| White studies | |||||||||
| Ryberg (64) | 1997 | 138 | 297 | Norway | 62.3 (SD, 10.3) | 100 | NSCLC = 100% | Healthy | Age, smoking, and ethnicity |
| Harris (45) | 1998 | 178 | 199 | Australia | 66 (range, 38–91; SD, 9.1) | 69 | SqCC = 43.5%, AC = 18.2%, LCC = 7.7%, SCC = 7.1%, NSCLC = 1.9%, others = 21.6% | Healthy | None |
| Jourenkova-Mironova (46) | 1998 | 150 | 172 | France | 58.4 (no range or SD) | 93 | SqCC = 65.3%, SCC = 34.7% | Hospital | Age and gender (frequency matching) |
| Saarikoski (55) | 1998 | 206 | 293 | Finland | 62 (SD, 9) | 79.8 | SqCC = 45.2%, AC = 39.4%, others = 15.4% | Healthy | None |
| To-Figueras (59) | 1999 | 164 | 200 | Spain | 59 (range, 32–87) | 88.4 | SCC = 34.8%, SqCC = 31.7, AC = 25.6%, LCC = 7.9% | Healthy | Gender |
| Risch (63) | 2001 | 388 | 353 | Germany | 60.9 (range, 28–87) | 75.8 | SqCC = 44.0%, AC = 39.0%, LCC = 4.9%, SCC = 2.8%, others = 10.8% | Hospital | Ethnicity |
| Lewis (50) | 2002 | 93 | 151 | United Kingdom | 67.4 (SD, 10.4) | 63.8 | SCC = 16.1%, SqCC = 34.4%, AC = 10.9, others and nonclassified = 38.7% | Hospital | None |
| Stucker (58) | 2002 | 251 | 264 | France | 59.3 (SD, 9.6) | 100 | SqCC = 46.0%, SCC = 19%, AC = 24.0%, others = 11.0% | Hospital | Age, ethnicity, and gender (frequency matching) |
| Reszka (54) | 2003 | 138 | 165 | Poland | 59.7 (no range or SD) | 76.8 | SqCC = 44.2%, SCC = 25.4%, NSCLC = 17.4%, AC = 8.7%, others = 4.3% | Hospital | Age and gender (frequency matching) |
| Wang (61) | 2003 | 362 | 419 | United States | 60.9 (SD, 10.1) | 52.4 | Hospital | Age, gender, ethnicity, and smoking (frequency matching) | |
| Schneider (56) | 2004 | 446 | 622 | Germany | 64.4 (SD, 8.7) | 90.6 | SCC = 15.0%, LCC = 3.6%, AC = 25.1%, SqCC = 41.1%, others = 15.2% | Hospital | None |
| Larsen (49) | 2006 | 1,095 | 626 | Australia | 63.4 (SD, 9.4) | 71.9 | AC = 45.2%, SqCC = 45.1%, others = 9.7% | Hospital | Age and smoking |
| Miller (52) | 2006 | 1,921 | 1,343 | United States | 66 (no range or SD) | 49.7 | AC = 43.7%, SqCC = 21.4%, LCC = 7.3%, SCC = 9.2%, others = 18.4% | Healthy | None |
| Sorensen (57) | 2007 | 429 | 766 | Denmark | No mean age (range, 50–64) | 53 | SCC = 19%, AC = 32%, SqCC = 23%, others = 26% | Healthy | None |
| Other studies | |||||||||
| Perera (31) | 2002 | 85 | 163 | United States | 61.8 (SD, 7.7) | 100 | AC = 36.0%, SCC = 16.9%, SqCC = 21.3%, LCC = 9.0%, others = 16.8% | Healthy | Smoking, age, and duration of follow-up |
| Nazar-Stewart (53) | 2003 | 253 | 487 | United States | No mean age (range, 18–74) | 100 | SqCC = 29.6%, SCC = 19.0%, NSCLC = 12.0%, LCC = 3.3%, AC = 35.0%, others = 1.1% | Healthy | Age and gender (frequency matching) |
| Yang (62) | 2004 | 235 | 233 | United States | SqCC = 13.5%, SCC = 7.6%, NSCLC = 13.1%, AC = 52.3%, LCC = 3.8%, others = 9.7% | Hospital | None | ||
| Cote (39) | 2005 | 317 | 407 | United States | 42.1 (no range or SD) | 50 | SqCC = 11.7%, SCC = 13.2%, AC = 47.7%, LCC = 9.4%, NSCLC = 3.7%, others = 14.3% | Healthy | Race, sex, 5-year age group, and county of residence |
| Wenzlaff (40) | 2005 | 141 | 180 | United States | 62.4 (range, 40–84; SD, 13.9) | 57.8 | SqCC = 15.7%, SCC = 6.6%, AC = 54.2%, LCC = 7.2%, others = 16.3% | Healthy | Race, sex, county of residence, and age (frequency matching) |
Abbreviations: AC, adenocarcinoma; LCC, large-cell carcinoma; NSCLC, non–small cell lung cancer; SCC, small-cell carcinoma; SD, standard deviation; SqCC, squamous-cell carcinoma.
The pooled analysis was performed by using information collected from researchers who submitted information to the Genetic Susceptibility to Environmental Carcinogens (GSEC) database (www.gsec.net). The design of this study is explained in greater detail elsewhere (65). The primary goal of the GSEC project is to examine the associations between various cancers and genetic polymorphisms by using published and unpublished data solicited from collaborating investigators. These data are then cleaned and entered into a main database that is available to interested investigators for analyses related to the overall goals of the study. Each participating center provided information on the study design, source of controls, laboratory methods used for genotyping, source of DNA for genotyping, and response rates for cases and controls.
From the GSEC database, we selected all studies that included information on GSTP1 and lung cancer. Only 3 studies (46, 55, 62) provided information on the exon 6 (Ala114Val, 341C → 341T) polymorphism; thus, as in the meta-analysis, all analyses reported on in this paper focus on the polymorphism in exon 5 only. Investigators who had not initially participated in the GSEC project were contacted and asked to provide their data for the pooled analysis. We were able to obtain data from 14 of the 27 studies (51.9%) included in the meta-analysis (Wenzlaff et al. (40) and Cote et al. (39), 2 studies from the same principal investigator, combined their data into a single data set, referred to as Cote et al. in the pooled analyses). An additional study not included in the meta-analysis was used in the pooled analysis (32). The number of subjects included in the published reports may differ somewhat from the numbers in this pooled analysis because the GSEC data set includes some unpublished data. The total number of subjects included in the pooled analysis was 4,282 cases and 5,032 controls.
Statistical analysis
For the meta-analysis, study-specific crude odds ratios and 95% confidence intervals were calculated to estimate the association between the exon 5 (Ile105Val, 313A → 313G) polymorphism in GSTP1 and lung cancer based on the reported frequencies of Ile/Ile, Ile/Val, and Val/Val genotypes in cases and controls. Odds ratios and 95% confidence intervals were calculated for individuals carrying 1 (Ile/Val) or 2 (Val/Val) valine alleles compared with individuals carrying 2 isoleucine (Ile/Ile) alleles. Homogeneity among studies was tested by using the Breslow-Day test for homogeneity, and, when not statistically significant (based on P > 0.05), a fixed-effects model was used for the meta-analysis (66). Heterogeneity was also quantified by using the I-squared statistic (67). To test for publication bias, both the Begg and Mazumdar adjusted rank correlation test (68) and the Egger et al. regression asymmetry test (69) were performed. Funnel plots were also created to graphically display evidence of publication bias, and sensitivity analyses to examine the influence of each study on the overall estimate were also performed.
Because the frequency of the polymorphism differs by ethnicity, studies were stratified by the reported ethnicity of the subject population, with 14 studies in whites, 8 studies in Asians, and 5 studies in populations comprising a mix of other ethnic groups, including whites, African Americans, and Mexican Americans. Crude odds ratios and 95% confidence intervals were also estimated by source of controls (healthy or hospital) and then by both ethnicity and control source.
Summary odds ratios and 95% confidence intervals were calculated for all studies combined, as well as for each ethnic group (white, Asian, other), control source (healthy or hospital), and then by both ethnicity and control source. All analyses were performed by examining risk associated with carrying at least 1 valine allele compared with Ile/Ile genotypes at GSTP1 exon 5 (Ile105Val, 313A → 313G). All meta-analyses were performed with the STATA software package (Stata Corporation, College Station, Texas).
To reduce the potential for confounding associated with ethnicity, the pooled analyses were performed separately for the 2 ethnic groups with the greatest number of studies and participants (whites and Asians). Chi-squared tests were conducted to test for Hardy-Weinberg equilibrium in the reported genotype frequencies among the controls in the pooled analysis, after stratification by ethnicity. Study-specific crude odds ratios and 95% confidence intervals for lung cancer and GSTP1 genotype were estimated by using unconditional logistic regression models. As with the meta-analysis, odds ratios and 95% confidence intervals were calculated for individuals carrying 1 or 2 105Val alleles compared with individuals carrying 2 Ile alleles. Heterogeneity between studies was tested by using the Breslow-Day test for homogeneity. Crude and adjusted odds ratios were calculated for each ethnic group, as well as stratified by control source (healthy and hospital), smoking status (nonsmoker/ever smoker), and histologic type (adenocarcinoma, squamous cell carcinoma, and small cell carcinoma). Regarding studies that provided information on pack-years of smoking, this paper presents adjusted odds ratios and 95% confidence intervals for nonsmokers and by tertile of numbers of pack-years of smoking. Regression lines were fitted to test for linear trend between the odds ratios and amount smoked. To formally test for interactions between amount smoked and genotype, cross-product terms were created and tested in the logistic model. The cutpoints for the tertiles of amount smoked were calculated from the controls who smoked. All pooled analyses were performed by using SAS version 9.1 software (SAS Institute, Inc., Cary, North Carolina).
RESULTS
The genotype frequencies for the GSTP1 exon 5 polymorphism varied according to ethnicity. When ethnicity-specific individual data from the controls were used in the pooled analysis, 46.9% of whites carried the Ile/Ile genotype, 41.8% carried the Ile/Val genotype, and 11.3% carried the Val/Val genotype (data not shown). In Asians, the respective percentages were 66.8%, 30.2%, and 3.0%. These frequencies are similar to those for the GSTP1 exon 5 polymorphism found in other control populations for different cancer sites (70, 71). Among all controls in the pooled analysis, the genotype frequencies were in Hardy-Weinberg equilibrium for both Asians (P = 0.38) and whites (P = 0.17).
Meta-analysis
For all 27 studies combined, the meta–odds ratio was 1.04 (95% confidence interval (CI): 0.97, 1.10), with no apparent heterogeneity between the studies (P for Q test = 0.27) (data not shown). Thus, a fixed-effects model versus a random-effects model was used. Sensitivity analysis was conducted to examine the influence of each study. Exclusion of the study by Miller et al. (52) resulted in a summary odds ratio of 1.08 (95% CI: 1.00, 1.15) (data not shown).
Two tests were performed to detect publication bias. Publication bias was not identified when Begg's test was performed (P = 0.10), but the Egger et al. (69) regression asymmetry test, which tends to suggest the presence of publication bias more frequently than Begg's test, did suggest that publication bias was present (P = 0.02). To adjust for this bias, a trim and fill method developed by Duval and Tweedie (72) was implemented. Trimming was based on the fixed-effects model, and the adjusted estimate obtained by using a random-effects model was an odds ratio of 0.99 (95% CI: 0.91, 1.07). Thus, the overall conclusion that there is no association between lung cancer and carrying at least 1 valine allele remained unchanged.
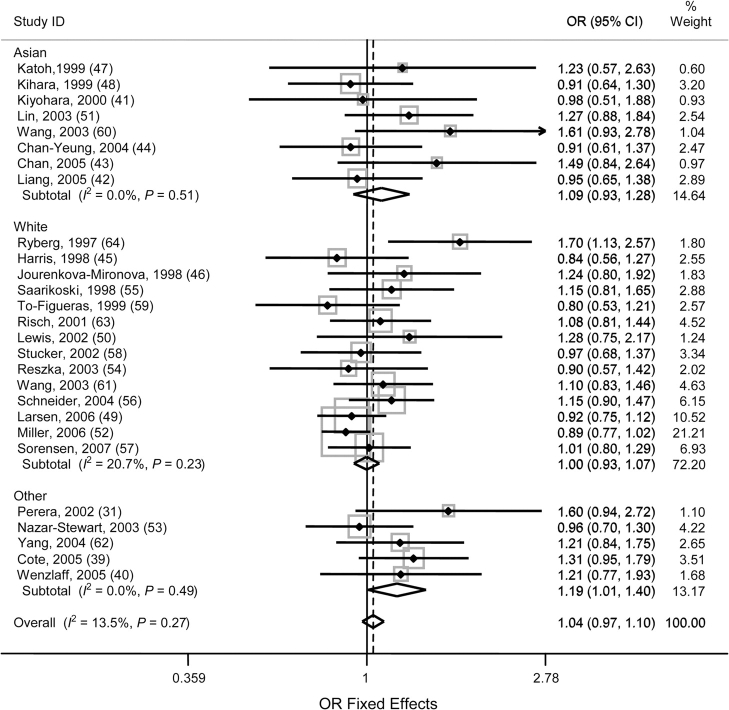
Because of the ethnicity-specific differences in genotype frequency, study-specific crude odds ratios and 95% confidence intervals, stratified by ethnicity, are presented in Figure 1 for individuals carrying at least 1 105Val (Ile/Val and Val/Val vs. Ile/Ile) allele. Among studies of whites, only 1 study (64) reported a statistically significant association (odds ratio (OR) = 1.70, 95% CI: 1.13, 2.57). The remaining 13 studies were clustered around the null effect (6 below 1.0 and 7 above 1.0) and were not statistically significant. None reported a negative association. The overall odds ratio for studies in whites was 1.00 (95% CI: 0.93, 1.07). No heterogeneity was identified between the 14 studies included (P = 0.23, data not shown). Among the 8 studies in Asian populations, none were statistically significant, with 4 slightly below the null and 4 slightly above the null. The overall odds ratio in Asian studies was 1.09 (95% CI: 0.93, 1.28). No heterogeneity was identified between the 8 studies included (P = 0.51, data not shown). In the studies that reported other ethnicities, 4 of the 5 had odds ratio estimates above the null, and 1 was slightly below the null. The overall risk associated with lung cancer and carrying at least 1 valine allele was statistically significant in studies not limited to a single ethnicity, with an odds ratio of 1.19 (95% CI: 1.01, 1.40). No heterogeneity was identified between the 5 studies included (P = 0.49, data not shown).
Figure 1.
Study-specific (first author, year of publication (reference no.)) and meta-log odds ratios (ORs) with 95% confidence intervals (CIs) for a glutathione S-transferase pi gene (GSTP1) exon 5 (Ile105Val, 313A → 313G) polymorphism for individuals carrying at least 1 valine allele (Ile/Val and Val/Val vs. Ile/Ile), by ethnicity. The shading around the point estimate reflects the weight of the study in the meta-analysis. The dashed line indicates the overall OR. ID, an assigned study identifier for each study population; Ile, isoleucine.
No statistically significant differences were identified when studies were stratified by control source (healthy or hospital, data not shown). Stratifying by both ethnicity and control source resulted in sparse strata for “Asian and healthy” (2 studies) and “other and hospital” (1 study). For other strata, no statistically significant associations between genotype and lung cancer were identified, nor were there any significant differences between strata (data not shown).
Pooled analysis
Table 2 describes the 15 studies included in the pooled analysis. Crude odds ratios and 95% confidence intervals for the association between carrying at least 1 105Val allele at GSTP1 exon 5 are presented. The overall pooled odds ratio associated with carrying at least 1 valine allele is 1.11 (95% CI: 1.03, 1.21) (Table 2).
Table 2.
Description of Studies Included in the Pooled Analysis: Study-Specific and Overall Crude Odds Ratios and 95% Confidence Intervals for the Association Between GSTP1 Exon 5 (Ile105Val, 313A → 313G) Polymorphisms and Lung Cancer, by Continent, Country, and Year of Publication
| First Author, Year (Reference No.) | No. of Cases | No. of Controls | Country of Study Origin | Source of Controls | Val/Val and Ile/Val vs. Ile/Ile | |
| Crude OR | 95% CI | |||||
| Asia | ||||||
| Wang, 2003 (60) | 112 | 119 | China | Hospital | 1.61 | 0.93, 2.78 |
| Liang, 2005 (42) | 227 | 227 | China | Hospital | 1.22 | 0.84, 1.78 |
| Kiyohara, 2000 (41) | 62 | 80 | Japan | Hospital | 1.18 | 0.58, 2.42 |
| Lin, 2003 (51) | 198 | 332 | Taiwan | Hospital | 1.27 | 0.88, 1.84 |
| Australia | ||||||
| Larsen, 2006 (49) | 1,095 | 626 | Australia | Hospital | 0.92 | 0.75, 1.12 |
| Europe | ||||||
| Sorensen, 2007 (57) | 429 | 765 | Denmark | Healthy | 1.01 | 0.80, 1.28 |
| Saarikoski, 1998 (55) | 199 | 293 | Finland | Healthy | 1.49 | 1.03, 2.16 |
| Jourenkova-Mironova, 1998 (46) | 150 | 172 | France | Hospital | 1.24 | 0.80, 1.92 |
| Schneider, 2004 (56) | 480 | 630 | Germany | Hospital | 1.09 | 0.86, 1.39 |
| Butkiewicz, 1999 (32) | 165 | 326 | Poland | Healthy | 0.94 | 0.65, 1.37 |
| Reszka, 2003 (54) | 217 | 251 | Poland | Hospital | 0.93 | 0.64, 1.33 |
| To-Figueras, 1999 (59) | 173 | 202 | Spain | Healthy | 1.01 | 0.67, 1.51 |
| Lewis, 2002 (50) | 93 | 151 | United Kingdom | Hospital | 1.28 | 0.75, 2.17 |
| North America | ||||||
| Yang, 2004 (62) | 235 | 234 | United States | Healthy | 1.22 | 0.85, 1.76 |
| Cote, 2005 (39) | 447 | 624 | United States | Healthy | 1.31 | 1.02, 1.69 |
| Total | 4,282 | 5,032 | 1.11 | 1.03, 1.21 | ||
Abbreviations: CI, confidence interval; GSTP1, glutathione S-transferase pi gene; Ile, isoleucine; OR, odds ratio; Val, valine.
Table 3 presents ethnicity-specific crude and adjusted odds ratios and 95% confidence intervals for the association between carrying at least 1 105Val allele at GSTP1 exon 5 and lung cancer. In the crude analysis or after adjusting for study ID (an assigned study identifier for each study population), age, sex, or smoking status (ever/never), no increased risk of lung cancer was seen for whites carrying at least 1 valine allele compared with those with the Ile/Ile genotype. We found no differences in estimates based on whether the control source was healthy or hospital based. There were also no statistically significant differences in risk when subjects were stratified by ever smoker or nonsmoker status; after adjusting for study ID, age, and sex among nonsmokers; and after adjusting for study ID, age, sex, and number of pack-years of smoking among participants who had ever smoked (Table 3).
Table 3.
Odds Ratios and 95% Confidence Intervals for the Association Between a GSTP1 Exon 5 (Ile105Val, 313A → 313G) Polymorphism (Ile/Val and Val/Val vs. Ile/Ile) and Lung Cancer in the Pooled Analysis, Stratified by Smoking Status and Ethnicity
| Ethnic Group | No. of Cases | No. of Controls | OR | 95% CI |
| Asian | ||||
| All studies: unadjusted | 603 | 768 | 1.34 | 1.07, 1.67 |
| All studies: adjusteda | 603 | 768 | 1.35 | 1.07, 1.70 |
| Hospital controls: unadjusted | 599 | 758 | 1.32 | 1.06, 1.65 |
| Hospital controls: adjusteda | 599 | 758 | 1.32 | 1.05, 1.68 |
| Smoker: unadjusted | 287 | 298 | 1.23 | 0.88, 1.72 |
| Smoker: adjustedb | 287 | 298 | 1.17 | 0.83, 1.66 |
| Smoker: adjustedc | 48 | 49 | 1.06 | 0.41, 2.75 |
| Nonsmoker: unadjusted | 254 | 390 | 1.49 | 1.08, 2.07 |
| Nonsmoker: adjustedb | 254 | 390 | 1.52 | 1.09, 2.13 |
| White | ||||
| All studies: unadjusted | 3,538 | 4,098 | 1.07 | 0.98, 1.17 |
| All studies: adjusteda | 3,490 | 4,077 | 1.05 | 0.95, 1.16 |
| Healthy controls: unadjusted | 1,503 | 2,268 | 1.12 | 0.98, 1.27 |
| Healthy controls: adjusteda | 1,497 | 2,254 | 1.15 | 0.99, 1.33 |
| Hospital controls: unadjusted | 2,035 | 1,830 | 1.02 | 0.90, 1.16 |
| Hospital controls: adjusteda | 1,993 | 1,823 | 1.00 | 0.87, 1.14 |
| Smoker: unadjusted | 3,044 | 2,737 | 1.02 | 0.93, 1.14 |
| Smoker: adjustedb | 3,035 | 2,729 | 1.05 | 0.95, 1.17 |
| Smoker: adjustedc | 2,809 | 2,181 | 1.00 | 0.89, 1.13 |
| Nonsmoker: unadjusted | 465 | 1,352 | 1.18 | 0.95, 1.45 |
| Nonsmoker: adjustedb | 455 | 1,348 | 1.17 | 0.92, 1.49 |
Abbreviations: CI, confidence interval; GSTP1, glutathione S-transferase pi gene; Ile, isoleucine; OR, odds ratio; Val, valine.
Adjusted for study ID (an assigned study identifier for each study population), age, sex, and smoking status (ever/never).
Adjusted for study ID, age, and sex.
Adjusted for study ID, age, sex, and pack-years of smoking.
Regarding Asian study subjects, those who carried at least 1 GSTP1 105Val allele compared with those with the Ile/Ile genotype were shown to be at increased risk of lung cancer in both crude analysis (OR = 1.34, 95% CI: 1.07, 1.67) and after adjustment for study ID, age, sex, and smoking status (OR = 1.35, 95% CI: 1.07, 1.70) (Table 3). With the exception of 4 cases and 10 controls, these findings were all based on studies with hospital-recruited controls; thus, in this paper, these data are not presented for healthy controls. After we stratified by ever/never smoking status, the association between genotype and lung cancer was significant among only Asian nonsmokers (OR = 1.52, 95% CI: 1.09, 2.13) after we adjusted for study ID, age, and sex (Table 3).
Risk of lung cancer and GSTP1 exon 5 polymorphism by amount smoked.
The large sample of whites for whom we had individual pack-year information (92.3% of the cases and 79.7% of the controls) allowed us to perform an analysis stratified by number of pack-years of cigarette smoking. Table 4 shows the relation among amount smoked, genotype, and lung cancer risk. For nonsmokers, no statistically significant association was seen between carrying at least 1 105Val allele and lung cancer risk after we adjusted for study ID, sex, and age (OR = 1.17, 95% CI: 0.92, 1.50). For participants who smoked for 1–28 pack-years, the odds of having lung cancer were 1.23-fold higher among those carrying at least 1 valine allele compared with those with the Ile/Ile genotype (95% CI: 1.00, 1.52) after we adjusted for study ID, sex, and age. Among moderate smokers who smoked for 28.01–48 pack-years, risk was not increased (OR = 1.01, 95% CI: 0.82, 1.24), and, among those with the heaviest reported pack-years of smoking (≥48.01), carrying at least 1 105Val allele was associated with a decreased risk of lung cancer compared with that for those with Ile/Ile genotypes after adjustment for study ID, age, and sex (OR = 0.83, 95% CI: 0.67, 1.03). We found no statistically significant linear trend between risk of lung cancer due to genotype and amount smoked (P = 0.16) when nonsmokers were included. There was a statistically significant trend between amount smoked and risk of lung cancer due to genotype (P = 0.05) when only ever smokers were examined. The overall effect of interactions between amount of smoking and genotype on lung cancer risk was marginally significant (P = 0.08, data not shown). When we used nonsmoking and IIe/IIe as the reference group, the group of heavy smokers with lle/Val or Val/Val had a significantly increased risk (P = 0.03, Table 4).
Table 4.
Odds Ratios and 95% Confidence Intervals for the Association Between a GSTP1 Exon 5 (Ile105Val, 313A → 313G) Polymorphism (Ile/Val and Val/Val vs. Ile/Ile) and Lung Cancer in Whites in the Pooled Analysis, Stratified by Pack-years of Smoking
| Smoking Status | No. of Cases | No. of Controls | ORa | 95% CI | P Trend | P Valueb | ||
| Ile/Ile | Ile/Val or Val/Val | Ile/Ile | Ile/Val or Val/Val | |||||
| Nonsmoker | 204 | 251 | 656 | 692 | 1.17 | 0.92, 1.50 | 0.16c | Reference |
| 1–28 pack-years | 279 | 386 | 459 | 539 | 1.23 | 1.00, 1.52 | 0.09 | |
| 28.01–48 pack-years | 490 | 557 | 292 | 334 | 1.01 | 0.82, 1.24 | 0.57 | |
| ≥48.01 pack-years | 505 | 592 | 228 | 329 | 0.83 | 0.67, 1.03 | 0.05d | 0.03 |
Abbreviations: CI, confidence interval; GSTP1, glutathione S-transferase pi gene; Ile, isoleucine; OR, odds ratio; Val, valine.
Adjusted for study ID (an assigned study identifier for each study population), age, and sex.
P value for nonsmokers and with Ile/Ile as the reference group, adjusted for study ID, age, and sex.
P-value test for trend among all 4 categories.
P-value test for trend among smokers only.
Risk of lung cancer and GSTP1 exon 5 polymorphism by histologic type and ethnicity.
Crude and adjusted odds ratios and 95% confidence intervals for the association between carrying at least 1 valine allele and lung cancer risk are presented stratified by histologic type and ethnicity in Table 5. Among whites, there were no statistically significant differences in risk associated with the GSTP1 gene polymorphism and adenocarcinoma, squamous cell carcinoma, or small cell carcinoma. In Asian populations, individuals carrying at least 1 valine allele were at increased risk of adenocarcinoma (OR = 1.33, 95% CI: 1.02, 1.74) after adjustment for study ID, age, sex, and smoking status. Risk of squamous cell lung cancer was increased for Asians carrying at least 1 valine allele (OR = 1.44, 95% CI: 1.05, 1.98), but the odds ratio was no longer statistically significant after adjusting for study ID, age, sex, and smoking status (OR = 1.36, 95% CI: 0.95, 1.94).
Table 5.
Odds Ratios and 95% Confidence Intervals for the Association Between a GSTP1 Exon 5 (Ile105Val, 313A → 313G) Polymorphism (Ile/Val and Val/Val vs. Ile/Ile) and Lung Cancer in the Pooled Analysis, Stratified by Histologic Type and Ethnicity
| Ethnic Group | Unadjusted | Adjusteda | ||||||
| No. of Cases | No. of Controls | OR | 95% CI | No. of Cases | No. of Controls | OR | 95% CI | |
| White | ||||||||
| Adenocarcinoma | 1,048 | 4,098 | 1.08 | 0.95, 1.24 | 1,043 | 4,077 | 1.041 | 0.90, 1.21 |
| Squamous cell carcinoma | 1,394 | 4,098 | 1.00 | 0.86, 1.09 | 1,377 | 4,077 | 0.994 | 0.87, 1.14 |
| Small cell carcinoma | 399 | 4,098 | 1.11 | 0.90, 1.36 | 393 | 4,077 | 1.103 | 0.91, 1.41 |
| Asian | ||||||||
| Adenocarcinoma | 384 | 768 | 1.32 | 1.02, 1.70 | 384 | 768 | 1.33 | 1.02, 1.74 |
| Squamous cell carcinoma | 206 | 768 | 1.44 | 1.05, 1.98 | 206 | 768 | 1.36 | 0.95, 1.94 |
| Small cell carcinoma | 5 | 768 | 0.50 | 0.06, 4.52 | 5 | 768 | 0.44 | 0.04, 4.56 |
| Total | ||||||||
| Adenocarcinoma | 1,496 | 5,032 | 1.05 | 0.94, 1.18 | 1,491 | 5,009 | 1.11 | 0.98, 1.26 |
| Squamous cell carcinoma | 1,617 | 5,032 | 1.02 | 0.91, 1.14 | 1,600 | 5,009 | 1.03 | 0.91, 1.16 |
| Small cell carcinoma | 412 | 5,032 | 1.22 | 1.00, 1.49 | 406 | 5,009 | 1.12 | 0.90, 1.39 |
Abbreviations: CI, confidence interval; GSTP1, glutathione S-transferase pi gene; Ile, isoleucine; OR, odds ratio; Val, valine.
Adjusted for study ID (an assigned study identifier for each study population), sex, smoking (ever/never), and age.
DISCUSSION
The meta-analysis found no association (OR = 1.04, 95% CI: 0.97, 1.10) between lung cancer risk and carrying 1 or more GSTP1 105Val alleles. This finding is similar to that of another meta-analysis of 25 studies with a combined 6,221 cases and 7,602 controls, which reported an unadjusted odds ratio of 1.04 (95% CI: 0.99, 1.09) for the association between lung cancer and carrying the GSTP1 exon 5 Ile105Val variant (15). After stratifying by the ethnicity of study subjects, studies that included subjects of various ethnic backgrounds (i.e., the study had both African-American and white participants) reported an increase in risk associated with carrying at least 1 105Val allele compared with those with Ile/Ile genotypes.
The pooled analysis did identify an overall statistically significant increase in lung cancer risk associated with carrying at least 1 valine allele, with an odds ratio of 1.11 (95% CI: 1.03, 1.21). When the studies were stratified by subject ethnicity, this association was seen among Asian subjects but not among white subjects. A pooled analysis of whites diagnosed with early-onset lung cancer (under age 60 years) also reported no association between lung cancer risk and the GSTP1 exon 5 genotype (30). Unlike the white control populations, who were recruited through either population-based methods or hospitals, almost all Asian controls were recruited by using hospital-based methods. Among Asians, this association between the Ile/Val and Val/Val genotypes and lung cancer risk was strongest among nonsmokers and those with adenocarcinoma. The higher prevalence of adenocarcinoma of the lung in Asians, particularly among women nonsmokers, was identified decades ago (73) and has received more attention recently with the success of using epidermal growth factor receptor tyrosine kinase inhibitors in these populations (74).
The results from the meta-analyses suggest no association between lung cancer risk and the GSTP1 exon 5 polymorphism, either overall or stratified by race/ethnicity, whereas the results from the pooled analysis suggest risk of carrying at least 1 105Val allele is associated with increased risk of lung cancer overall and also in Asians. Examination of the 95% confidence intervals associated with the risk estimates suggests that these apparently discrepant results are not statistically significant. In addition, the pooled analysis did not contain subjects from all studies included in the meta-analysis, and vice versa, and the pooled analysis allowed for individual adjustment by age, sex, and smoking status. It has been suggested that results from individual subject data that allow for adjustment of confounders, such as the pooled analysis presented here, may best summarize results of multiple studies (75).
We found an interaction between GSTP1 exon 5 genotype and personal smoking history. Among whites, those classified as “light” smokers (1–28.00 pack-years) were at increased risk of lung cancer if they carried the Ile/Val or Val/Val genotype compared with those with the Ile/Ile genotype. Conversely, heavy-smoking (≥48.01 pack-years) whites carrying the Ile/Val or Val/Val genotypes were at decreased risk compared with those with the Ile/Ile genotype. This interaction may explain some of the variability seen between populations with different recruitment criteria (i.e. early-onset cases who likely do not have as extensive smoking histories) and highlights the need to investigate the gene-environment interactions between genotype and amount smoked (i.e., pack-years).
Laboratory evidence suggests that carrying a 105Val allele results in reduced GSTP1 enzymatic activity in the cell (11–13). These characterizations, while important for developing a hypothesis about the biologic mechanisms through which carcinogenesis evolves, do not necessarily represent what is occurring in the environment of the human lung. The seemingly protective effect of the Val/Val or Ile/Val genotype in heavy smokers identified in this pooled analysis does not directly support reduced activity associated with carrying the Val allele because it would be expected that those with reduced GSTP1 activity would be at increased risk of malignant transformation after exposure to carcinogens. The continuous assault from heavy smoking may change cellular activity in ways we are currently unable to assess in the human lung. A recent murine model, using GSTP-null mice, found a significantly higher number of adenomas in null mice compared with wild-type mice after exposure to 3 PAHs (76). When adducts in these mice were examined, there were significant differences in the number of adducts formed depending on the PAH they were exposed to, with 1 PAH resulting in no increase in adducts, suggesting that an alternative protective pathway in response to this specific exposure exists. While this study in mice is the first known in vivo model showing the importance of GSTP1 in lung carcinogenesis, it is also apparent that the role of glutathione S-transferase has yet to be fully elucidated. In this study, we were not able to explore the other major roles that GSTP1 is thought to have in the cell, including resistance to chemotherapy (77), because we did not have information on chemotherapy or other exposures that might help clarify this gene-environment interaction.
The variation in risk associated with lung cancer and GSTP1 exon 5 genotypes between Asians and whites is likely due to a number of factors, including different exposures in the populations. For example, studies in Asian women nonsmokers suggest that exposure to the carcinogens found in cooking oils increases risk of lung cancer (78, 79). Further studies of gene-environment interactions in lung cancer should also include occupational risk factors such as ionizing radiation (through radon exposure), asbestos, chromium, and arsenic (25, 80). The ability to examine only a small number of potential confounding variables is a limitation to both pooled and meta-analysis studies. It is also possible that publication bias accounts for some of the difference in risk seen between the Asian and white populations. It has been shown that a large proportion of Chinese literature does not reach PubMed and that studies that do are more likely than non-Chinese studies to be statistically significant and report larger measures of effect (i.e., odds ratios) (81). Thus, our findings may be a result of this publication bias, and the inability to include the entire collection of literature is a limitation of this analysis.
Other potential limitations include the presence of heterogeneity between studies. We tested for heterogeneity and performed a sensitivity analysis to determine whether a particular study or studies were a source of heterogeneity. Various amounts of data regarding smoking behaviors were collected, so we were unable to examine number of years of smoking or number of cigarettes smoked per day; therefore, our analyses were restricted to use of the 2 most commonly collected variables: pack-years of use or dichotomous classification as never or ever smokers. Additionally, pack-years of smoking was missing for approximately 20% of individuals identified as ever smokers. There were also differences in how the subjects were identified (hospital based or population), the histologic types of lung cancers included in the studies, the types of tissues used to extract DNA, and genotyping methods. When possible, we stratified by source of controls and histologic type of cancer. It was not feasible to control for the variation in pathologic reports of histology type that may occur by region or country.
LABORATORY TESTS
The methods used for determining GSTP1 exon 5 genotypes are described in each article. The majority of the studies included in the analyses used genomic DNA extracted from blood, although 3 studies included DNA extracted from blood or lung tissue (32, 43, 49); 2 studies included DNA extracted from blood, lung tissue, or buccal samples (39, 40); and 1 study used blood and bronchial lavage (50). Polymerase chain reaction–based restriction fragment length polymorphism methods were the most frequently cited technique to determine GSTP1 exon 5 genotypes.
POPULATION TESTING
The evidence to date regarding the polymorphism in GSTP1 exon 5 and lung cancer risk is insufficient to suggest testing at the population level.
CONCLUSIONS AND RECOMMENDATIONS FOR RESEARCH
Overall, the meta-analysis found no significant association between lung cancer and the GSTP1 exon 5 (Ile105Val) polymorphism for individuals carrying at least 1 105Val allele. No association was seen when we stratified by ethnicity in white or Asian populations. In the 5 studies that included more than 1 ethnic group, the meta-analysis suggested that an association between lung cancer and carrying at least 1 valine allele (Ile/Val and Val/Val vs. Ile/Ile) was statistically significant, with an odds ratio of 1.19 (95% CI: 1.01, 1.40), although this finding may be the result of population stratification.
In the pooled analysis, there was a statistically significant, mild overall association (OR = 1.11, 95% CI: 1.03, 1.21) between lung cancer and the GSTP1 exon 5 (Ile105Val) polymorphism for individuals carrying a Val/Val or Ile/Val genotype compared with those carrying the Ile/Ile genotype. After stratification by ethnicity and adjustment for study ID, age, sex, and smoking status, increased risk associated with the Val/Val or Ile/Val genotypes and lung cancer was seen in Asian populations only. Among Asians, this risk was highest for nonsmokers and those with adenocarcinoma of the lung.
There is evidence for interaction among amount of smoking (i.e., pack-years), the GSTP1 exon 5 (Ile105Val) polymorphism, and risk of lung cancer in whites. The odds of lung cancer associated with carrying at least 1 valine allele appear to decrease as the amount of pack-years of smoking increases, with heavy smokers who carry a Ile/Val or Val/Val genotype at decreased risk of lung cancer compared with their heavy-smoking counterparts with the Ile/Ile genotype. This finding highlights the importance of context when studying gene-environment interactions and clearly shows the need to collect detailed exposure information on all study participants, because gene expression, and risk of lung cancer, may differ by the environmental exposure.
Acknowledgments
Author affiliations: Population Studies and Prevention, Karmanos Cancer Institute, Internal Medicine, Wayne State University School of Medicine, Detroit, Michigan (Michele L. Cote, Ann G. Schwartz, Angela S. Wenzlaff); Biostatistics Core, Karmanos Cancer Institute, Internal Medicine, Wayne State University School of Medicine, Detroit, Michigan (Wei Chen); Barbara Ann Karmanos Cancer Institute, Detroit, Michigan (Daryn W. Smith); Unit of Cancer Epidemiology (INSERM U351), Paris, France, and CNRS FRE2939, Institut Gustave Roussy, Villejuif, France (Simone Benhamou); Cancer Registry, Geneva, Switzerland (Christine Bouchardy); Department of Tumor Biology, Maria Sklodowska-Curie Memorial Cancer Center and Institute of Oncology, Gliwice, Poland (Dorota Butkiewicz); Department of Thoracic Medicine, The Prince Charles Hospital, Brisbane, Australia (Kwun M. Fong); Public Health Department, University of Barcelona, Barcelona, Spain (Manuel Gené); Finnish Institute of Occupational Health, Helsinki, Finland (Ari Hirvonen); Department of Preventive Medicine, Kyushu University Graduate School of Medical Sciences, Fukuoka, Japan (Chikako Kiyohara); Hamon Center for Therapeutic Oncology Research, University of Texas Southwestern Medical Center, Dallas, Texas (Jill E. Larsen); Institute of Toxicology, Chung-Shan Medical and Dental College, Taichung, Taiwan (Pinpin Lin); Institute of Cancer Epidemiology, Danish Cancer Society, Copenhagen, Denmark (Ole Raaschou-Nielsen); Occupational and Environmental Health Research Group, University of Manchester, Manchester, United Kingdom (Andrew C. Povey); Department of Toxicology and Carcinogenesis, Institute of Occupational Medicine, Lodz, Poland (Edyta Reszka); Department of Toxicology and Cancer Risk Factors, DKFZ-German Cancer Research Center, Heidelberg, Germany (Angela Risch); Institute of Occupational and Social Medicine, University of Giessen, Giessen, Germany (Joachim Schneider); Danish Cancer Society, Institute of Cancer Epidemiology, Copenhagen, Denmark (Mette Sorensen); Hospital Clinic Provincial Toxicology Unit, Barcelona, Spain (Jordi To-Figueras); Department of Health Promotion and Preventive Medicine, Nagoya City University Graduate School of Medical Sciences, Mizuho-ku, Nagoya, Japan (Shinkan Tokudome); School of Public Health, Southeast University, Nanjing, China (Yuepu Pu); Division of Epidemiology, Mayo Clinic College of Medicine, Rochester, Minnesota (Ping Yang); Institute of Tumor Biology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany (Harriet Wikman); and University of Pittsburgh Cancer Institute, Pittsburgh, Pennsylvania (Emanuela Taioli).
This study was supported by National Institutes of Health/National Cancer Institute grant P50 CA090440-06 (E. T.). M. L. C. was supported by a Career Development grant from the National Lung Cancer Partnership and the LUNGevity Foundation.
The authors thank Barbara Stadterman for overall project management and Shawnita Sealy-Jefferson for data management.
Conflict of interest: none declared.
Glossary
Abbreviations
- CI
confidence interval
- GSEC
Genetic Susceptibility to Environmental Carcinogens
- GSTP1
glutathione S-transferase pi
- OR
odds ratio
- PAH
polycyclic aromatic hydrocarbon
References
- 1.Anttila S, Hirvonen A, Vainio H, et al. Immunohistochemical localization of glutathione S-transferases in human lung. Cancer Res. 1993;53(23):5643–5648. [PubMed] [Google Scholar]
- 2.Ketterer B, Harris JM, Talaska G, et al. The human glutathione S-transferase supergene family, its polymorphism, and its effects on susceptibility to lung cancer. Environ Health Perspect. 1992;98:87–94. doi: 10.1289/ehp.929887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coles BF, Kadlubar FF. Detoxification of electrophilic compounds by glutathione S-transferase catalysis: determinants of individual response to chemical carcinogens and chemotherapeutic drugs? Biofactors. 2003;17(1–4):115–130. doi: 10.1002/biof.5520170112. [DOI] [PubMed] [Google Scholar]
- 4.Goto S, Iida T, Cho S, et al. Overexpression of glutathione S-transferase pi enhances the adduct formation of cisplatin with glutathione in human cancer cells. Free Radic Res. 1999;31(6):549–558. doi: 10.1080/10715769900301121. [DOI] [PubMed] [Google Scholar]
- 5.Nakagawa K, Saijo N, Tsuchida S, et al. Glutathione-S-transferase pi as a determinant of drug resistance in transfectant cell lines. J Biol Chem. 1990;265(8):4296–4301. [PubMed] [Google Scholar]
- 6.Strange RC, Jones PW, Fryer AA. Glutathione S-transferase: genetics and role in toxicology. Toxicol Lett. 2000;112–113:357–363. doi: 10.1016/s0378-4274(99)00230-1. [DOI] [PubMed] [Google Scholar]
- 7.Ali-Osman F, Brunner JM, Kutluk TM, et al. Prognostic significance of glutathione S-transferase pi expression and subcellular localization in human gliomas. Clin Cancer Res. 1997;3(12 pt 1):2253–2261. [PubMed] [Google Scholar]
- 8.Armstrong RN. Glutathione S-transferases: reaction mechanism, structure, and function. Chem Res Toxicol. 1991;4(2):131–140. doi: 10.1021/tx00020a001. [DOI] [PubMed] [Google Scholar]
- 9.Strange RC, Faulder CG, Davis BA, et al. The human glutathione S-transferases: studies on the tissue distribution and genetic variation of the GST1, GST2 and GST3 isozymes. Ann Hum Genet. 1984;48(pt 1):11–20. doi: 10.1111/j.1469-1809.1984.tb00829.x. [DOI] [PubMed] [Google Scholar]
- 10.Kiyohara C, Shirakawa T, Hopkin JM. Genetic polymorphism of enzymes involved in xenobiotic metabolism and the risk of lung cancer. Environ Health Prev Med. 2002;7:47–59. doi: 10.1007/BF02897330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ali-Osman F, Akande O, Antoun G, et al. Molecular cloning, characterization, and expression in Escherichia coli of full-length cDNAs of three human glutathione S-transferase Pi gene variants. Evidence for differential catalytic activity of the encoded proteins. J Biol Chem. 1997;272(15):10004–10012. doi: 10.1074/jbc.272.15.10004. [DOI] [PubMed] [Google Scholar]
- 12.Hu X, Ji X, Srivastava SK, et al. Mechanism of differential catalytic efficiency of two polymorphic forms of human glutathione S-transferase P1-1 in the glutathione conjugation of carcinogenic diol epoxide of chrysene. Arch Biochem Biophys. 1997;345(1):32–38. doi: 10.1006/abbi.1997.0269. [DOI] [PubMed] [Google Scholar]
- 13.Sundberg K, Johansson AS, Stenberg G, et al. Differences in the catalytic efficiencies of allelic variants of glutathione transferase P1-1 towards carcinogenic diol epoxides of polycyclic aromatic hydrocarbons. Carcinogenesis. 1998;19(3):433–436. doi: 10.1093/carcin/19.3.433. [DOI] [PubMed] [Google Scholar]
- 14.Butkiewicz D, Grzybowska E, Phillips DH, et al. Polymorphisms of the GSTP1 and GSTM1 genes and PAH-DNA adducts in human mononuclear white blood cells. Environ Mol Mutagen. 2000;35(2):99–105. [PubMed] [Google Scholar]
- 15.Ye Z, Song H, Higgins JP, et al. Five glutathione S-transferase gene variants in 23,452 cases of lung cancer and 30,397 controls: meta-analysis of 130 studies [electronic article] PLoS Med. 2006;3(4):e91. doi: 10.1371/journal.pmed.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shibuya K, Mathers CD, Boschi-Pinto C, et al. Global and regional estimates of cancer mortality and incidence by site: II. Results for the global burden of disease 2000 [electronic article] BMC Cancer. 2002;2:37. doi: 10.1186/1471-2407-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ries LA, Eisner MP, Kosary CL, et al., editors. SEER Cancer Statistics Review, 1975–2000. Bethesda, MD: National Cancer Institute; 2003. [Google Scholar]
- 18.Wu AH, Henderson BE, Thomas DC, et al. Secular trends in histologic types of lung cancer. J Natl Cancer Inst. 1986;77(1):53–56. [PubMed] [Google Scholar]
- 19.Alberg AJ, Samet JM. Epidemiology of lung cancer. Chest. 2003;123(1 suppl):21S–49S. doi: 10.1378/chest.123.1_suppl.21s. [DOI] [PubMed] [Google Scholar]
- 20.Ando M, Wakai K, Seki N, et al. Attributable and absolute risk of lung cancer death by smoking status: findings from the Japan Collaborative Cohort Study. Int J Cancer. 2003;105(2):249–254. doi: 10.1002/ijc.11043. [DOI] [PubMed] [Google Scholar]
- 21.Chan-Yeung M, Koo LC, Ho JC, et al. Risk factors associated with lung cancer in Hong Kong. Lung Cancer. 2003;40(2):131–140. doi: 10.1016/s0169-5002(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 22.Shankar A, Yuan JM, Koh WP, et al. Morbidity and mortality in relation to smoking among women and men of Chinese ethnicity: the Singapore Chinese Health Study. Eur J Cancer. 2008;44(1):100–109. doi: 10.1016/j.ejca.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.WHO Report on the Global Tobacco Epidemic, 2008: The MPOWER Package. Geneva, Switzerland: World Health Organization; 2008. [Google Scholar]
- 24.Alavanja MC, Brownson RC, Benichou J, et al. Attributable risk of lung cancer in lifetime nonsmokers and long-term ex-smokers (Missouri, United States) Cancer Causes Control. 1995;6(3):209–216. doi: 10.1007/BF00051792. [DOI] [PubMed] [Google Scholar]
- 25.Boffetta P. Epidemiology of environmental and occupational cancer. Oncogene. 2004;23(38):6392–6403. doi: 10.1038/sj.onc.1207715. [DOI] [PubMed] [Google Scholar]
- 26.Krewski D, Lubin JH, Zielinski JM, et al. A combined analysis of North American case-control studies of residential radon and lung cancer. J Toxicol Environ Health. 2006;69(7):533–597. doi: 10.1080/15287390500260945. [DOI] [PubMed] [Google Scholar]
- 27.Cowell IG, Dixon KH, Pemble SE, et al. The structure of the human glutathione S-transferase pi gene. Biochem J. 1988;255(1):79–83. doi: 10.1042/bj2550079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiyohara C, Otsu A, Shirakawa T, et al. Genetic polymorphisms and lung cancer susceptibility: a review. Lung Cancer. 2002;37(3):241–256. doi: 10.1016/s0169-5002(02)00107-1. [DOI] [PubMed] [Google Scholar]
- 29.Mohr LC, Rodgers JK, Silvestri GA. Glutathione S-transferase M1 polymorphism and the risk of lung cancer. Anticancer Res. 2003;23(3A):2111–2124. [PubMed] [Google Scholar]
- 30.Skuladottir H, Autrup H, Autrup J, et al. Polymorphisms in genes involved in xenobiotic metabolism and lung cancer risk under the age of 60 years. A pooled study of lung cancer patients in Denmark and Norway. Lung Cancer. 2005;48(2):187–199. doi: 10.1016/j.lungcan.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 31.Perera FP, Mooney LA, Stampfer M, et al. Associations between carcinogen-DNA damage, glutathione S-transferase genotypes, and risk of lung cancer in the prospective Physicians’ Health Cohort Study. Carcinogenesis. 2002;23(10):1641–1646. doi: 10.1093/carcin/23.10.1641. [DOI] [PubMed] [Google Scholar]
- 32.Butkiewicz D, Cole KJ, Phillips DH, et al. GSTM1, GSTP1, CYP1A1 and CYP2D6 polymorphisms in lung cancer patients from an environmentally polluted region of Poland: correlation with lung DNA adduct levels. Eur J Cancer Prev. 1999;8(4):315–323. doi: 10.1097/00008469-199908000-00008. [DOI] [PubMed] [Google Scholar]
- 33.Sweeney C, Nazar-Stewart V, Stapleton PL, et al. Glutathione S-transferase M1, T1, and P1 polymorphisms and survival among lung cancer patients. Cancer Epidemiol Biomarkers Prev. 2003;12(6):527–533. [PubMed] [Google Scholar]
- 34.Yang P, Yokomizo A, Tazelaar HD, et al. Genetic determinants of lung cancer short-term survival: the role of glutathione-related genes. Lung Cancer. 2002;35(3):221–229. doi: 10.1016/s0169-5002(01)00426-3. [DOI] [PubMed] [Google Scholar]
- 35.Lu C, Spitz MR, Zhao H, et al. Association between glutathione S-transferase pi polymorphisms and survival in patients with advanced nonsmall cell lung carcinoma. Cancer. 2006;106(2):441–447. doi: 10.1002/cncr.21619. [DOI] [PubMed] [Google Scholar]
- 36.Miller DP, Neuberg D, de Vivo I, et al. Smoking and the risk of lung cancer: susceptibility with GSTP1 polymorphisms. Epidemiology. 2003;14(5):545–551. doi: 10.1097/01.ede.0000073120.46981.24. [DOI] [PubMed] [Google Scholar]
- 37.Sørensen M, Autrup H, Tjonneland A, et al. Glutathione S-transferase T1 null-genotype is associated with an increased risk of lung cancer. Int J Cancer. 2004;110(2):219–224. doi: 10.1002/ijc.20075. [DOI] [PubMed] [Google Scholar]
- 38.Tsai YY, McGlynn KA, Hu Y, et al. Genetic susceptibility and dietary patterns in lung cancer. Lung Cancer. 2003;41(3):269–281. doi: 10.1016/s0169-5002(03)00238-1. [DOI] [PubMed] [Google Scholar]
- 39.Cote ML, Kardia SL, Wenzlaff AS, et al. Combinations of glutathione S-transferase genotypes and risk of early-onset lung cancer in Caucasians and African Americans: a population-based study. Carcinogenesis. 2005;26(4):811–819. doi: 10.1093/carcin/bgi023. [DOI] [PubMed] [Google Scholar]
- 40.Wenzlaff AS, Cote ML, Bock CH, et al. GSTM1, GSTT1 and GSTP1 polymorphisms, environmental tobacco smoke exposure and risk of lung cancer among never smokers: a population-based study. Carcinogenesis. 2005;26(2):395–401. doi: 10.1093/carcin/bgh326. [DOI] [PubMed] [Google Scholar]
- 41.Kiyohara C, Yamamura KI, Nakanishi Y, et al. Polymorphism in GSTM1, GSTT1, and GSTP1 and susceptibility to lung cancer in a Japanese population. Asian Pac J Cancer Prev. 2000;1(4):293–298. [PubMed] [Google Scholar]
- 42.Liang G, Pu Y, Yin L. Rapid detection of single nucleotide polymorphisms related with lung cancer susceptibility of Chinese population. Cancer Lett. 2005;223(2):265–274. doi: 10.1016/j.canlet.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 43.Chan EC, Lam SY, Fu KH, et al. Polymorphisms of the GSTM1, GSTP1, MPO, XRCC1, and NQO1 genes in Chinese patients with non-small cell lung cancers: relationship with aberrant promoter methylation of the CDKN2A and RARB genes. Cancer Genet Cytogenet. 2005;162(1):10–20. doi: 10.1016/j.cancergencyto.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 44.Chan-Yeung M, Tan-Un KC, Ip MS, et al. Lung cancer susceptibility and polymorphisms of glutathione-S-transferase genes in Hong Kong. Lung Cancer. 2004;45(2):155–160. doi: 10.1016/j.lungcan.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 45.Harris MJ, Coggan M, Langton L, et al. Polymorphism of the Pi class glutathione S-transferase in normal populations and cancer patients. Pharmacogenetics. 1998;8(1):27–31. doi: 10.1097/00008571-199802000-00004. [DOI] [PubMed] [Google Scholar]
- 46.Jourenkova-Mironova N, Wikman H, Bouchardy C, et al. Role of glutathione S-transferase GSTM1, GSTM3, GSTP1 and GSTT1 genotypes in modulating susceptibility to smoking-related lung cancer. Pharmacogenetics. 1998;8(6):495–502. doi: 10.1097/00008571-199812000-00006. [DOI] [PubMed] [Google Scholar]
- 47.Katoh T, Kaneko S, Takasawa S, et al. Human glutathione S-transferase P1 polymorphism and susceptibility to smoking related epithelial cancer; oral, lung, gastric, colorectal and urothelial cancer. Pharmacogenetics. 1999;9(2):165–169. [PubMed] [Google Scholar]
- 48.Kihara M, Kihara M, Noda K. Lung cancer risk of the GSTM1 null genotype is enhanced in the presence of the GSTP1 mutated genotype in male Japanese smokers. Cancer Lett. 1999;137(1):53–60. doi: 10.1016/s0304-3835(98)00337-1. [DOI] [PubMed] [Google Scholar]
- 49.Larsen JE, Colosimo ML, Yang IA, et al. CYP1A1 Ile462Val and MPO G-463A interact to increase risk of adenocarcinoma but not squamous cell carcinoma of the lung. Carcinogenesis. 2006;27(3):525–532. doi: 10.1093/carcin/bgi227. [DOI] [PubMed] [Google Scholar]
- 50.Lewis SJ, Cherry NM, Niven RM, et al. GSTM1, GSTT1 and GSTP1 polymorphisms and lung cancer risk. Cancer Lett. 2002;180(2):165–171. doi: 10.1016/s0304-3835(02)00028-9. [DOI] [PubMed] [Google Scholar]
- 51.Lin P, Hsueh YM, Ko JL, et al. Analysis of NQO1, GSTP1, and MnSOD genetic polymorphisms on lung cancer risk in Taiwan. Lung Cancer. 2003;40(2):123–129. doi: 10.1016/s0169-5002(03)00027-8. [DOI] [PubMed] [Google Scholar]
- 52.Miller DP, Asomaning K, Liu G, et al. An association between glutathione S-transferase P1 gene polymorphism and younger age at onset of lung carcinoma. Cancer. 2006;107(7):1570–1577. doi: 10.1002/cncr.22124. [DOI] [PubMed] [Google Scholar]
- 53.Nazar-Stewart V, Vaughan TL, Stapleton P, et al. A population-based study of glutathione S-transferase M1, T1 and P1 genotypes and risk for lung cancer. Lung Cancer. 2003;40(3):247–258. doi: 10.1016/s0169-5002(03)00076-x. [DOI] [PubMed] [Google Scholar]
- 54.Reszka E, Wasowicz W, Rydzynski K, et al. Glutathione S-transferase M1 and P1 metabolic polymorphism and lung cancer predisposition. Neoplasma. 2003;50(5):357–362. [PubMed] [Google Scholar]
- 55.Saarikoski ST, Voho A, Reinikainen M, et al. Combined effect of polymorphic GST genes on individual susceptibility to lung cancer. Int J Cancer. 1998;77(4):516–521. doi: 10.1002/(sici)1097-0215(19980812)77:4<516::aid-ijc7>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 56.Schneider J, Bernges U, Philipp M, et al. GSTM1, GSTT1, and GSTP1 polymorphism and lung cancer risk in relation to tobacco smoking. Cancer Lett. 2004;208(1):65–74. doi: 10.1016/j.canlet.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 57.Sorensen M, Raaschou-Nielsen O, Brasch-Andersen C, et al. Interactions between GSTM1, GSTT1 and GSTP1 polymorphisms and smoking and intake of fruit and vegetables in relation to lung cancer. Lung Cancer. 2007;55(2):137–144. doi: 10.1016/j.lungcan.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 58.Stücker I, Hirvonen A, de Waziers I, et al. Genetic polymorphisms of glutathione S-transferases as modulators of lung cancer susceptibility. Carcinogenesis. 2002;23(9):1475–1481. doi: 10.1093/carcin/23.9.1475. [DOI] [PubMed] [Google Scholar]
- 59.To-Figueras J, Gené M, Gómez-Catalán J, et al. Genetic polymorphism of glutathione S-transferase P1 gene and lung cancer risk. Cancer Causes Control. 1999;10(1):65–70. doi: 10.1023/a:1008811824890. [DOI] [PubMed] [Google Scholar]
- 60.Wang J, Deng Y, Cheng J, et al. GST genetic polymorphisms and lung adenocarcinoma susceptibility in a Chinese population. Cancer Lett. 2003;201(2):185–193. doi: 10.1016/s0304-3835(03)00480-4. [DOI] [PubMed] [Google Scholar]
- 61.Wang Y, Spitz MR, Schabath MB, et al. Association between glutathione S-transferase p1 polymorphisms and lung cancer risk in Caucasians: a case-control study. Lung Cancer. 2003;40(1):25–32. doi: 10.1016/s0169-5002(02)00537-8. [DOI] [PubMed] [Google Scholar]
- 62.Yang P, Bamlet WR, Ebbert JO, et al. Glutathione pathway genes and lung cancer risk in young and old populations. Carcinogenesis. 2004;25(10):1935–1944. doi: 10.1093/carcin/bgh203. [DOI] [PubMed] [Google Scholar]
- 63.Risch A, Wikman H, Thiel S, et al. Glutathione-S-transferase M1, M3, T1 and P1 polymorphisms and susceptibility to non-small-cell lung cancer subtypes and hamartomas. Pharmacogenetics. 2001;11(9):757–764. doi: 10.1097/00008571-200112000-00003. [DOI] [PubMed] [Google Scholar]
- 64.Ryberg D, Skaug V, Hewer A, et al. Genotypes of glutathione transferase M1 and P1 and their significance for lung DNA adduct levels and cancer risk. Carcinogenesis. 1997;18(7):1285–1289. doi: 10.1093/carcin/18.7.1285. [DOI] [PubMed] [Google Scholar]
- 65.Taioli E. International collaborative study on genetic susceptibility to environmental carcinogens. Cancer Epidemiol Biomarkers Prev. 1999;8(8):727–728. [PubMed] [Google Scholar]
- 66.Egger M, Davey Smith G, Altman DG. Systematic Reviews in Health Care: Meta-analysis in Context. London, United Kingdom: BMJ Books; 2001. [Google Scholar]
- 67.Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. [PubMed] [Google Scholar]
- 69.Egger M, Smith GD, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kellen E, Hemelt M, Broberg K, et al. Pooled analysis and meta-analysis of the glutathione S-transferase P1 Ile 105Val polymorphism and bladder cancer: a HuGE-GSEC review. Am J Epidemiol. 2007;165(11):1221–1230. doi: 10.1093/aje/kwm003. [DOI] [PubMed] [Google Scholar]
- 71.Ye Z, Song H. Glutathione S-transferase polymorphisms (GSTM1, GSTP1 and GSTT1) and the risk of acute leukaemia: a systematic review and meta-analysis. Eur J Cancer. 2005;41(7):980–989. doi: 10.1016/j.ejca.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 72.Duval S, Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56(2):455–463. doi: 10.1111/j.0006-341x.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- 73.Green JP, Brophy P. Carcinoma of the lung in nonsmoking Chinese women. West J Med. 1982;136(4):291–294. [PMC free article] [PubMed] [Google Scholar]
- 74.Engelman JA, Jänne PA. Factors predicting response to EGFR tyrosine kinase inhibitors. Semin Respir Crit Care Med. 2005;26(3):314–322. doi: 10.1055/s-2005-871990. [DOI] [PubMed] [Google Scholar]
- 75.Lyman GH, Kuderer NM. The strengths and limitations of meta-analyses based on aggregate data. BMC Med Res Methodol. 2005;5(1):14–20. doi: 10.1186/1471-2288-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ritchie KJ, Henderson CJ, Wang XJ, et al. Glutathione transferase pi plays a critical role in the development of lung carcinogenesis following exposure to tobacco-related carcinogens and urethane. Cancer Res. 2007;67(19):9248–9257. doi: 10.1158/0008-5472.CAN-07-1764. [DOI] [PubMed] [Google Scholar]
- 77.Inoue T, Ishida T, Sugio K, et al. Glutathione S transferase Pi is a powerful indicator in chemotherapy of human lung squamous-cell carcinoma. Respiration. 1995;62(4):223–227. doi: 10.1159/000196451. [DOI] [PubMed] [Google Scholar]
- 78.Chiang TA, Wu PF, Ko YC. Identification of carcinogens in cooking oil fumes. Environ Res. 1999;81(1):18–22. doi: 10.1006/enrs.1998.3876. [DOI] [PubMed] [Google Scholar]
- 79.Gao YT, Blot WJ, Zheng W, et al. Lung cancer among Chinese women. Int J Cancer. 1987;40(5):604–609. doi: 10.1002/ijc.2910400505. [DOI] [PubMed] [Google Scholar]
- 80.Pohlabeln H, Boffetta P, Ahrens W, et al. Occupational risks for lung cancer among nonsmokers. Epidemiology. 2000;11(5):532–538. doi: 10.1097/00001648-200009000-00008. [DOI] [PubMed] [Google Scholar]
- 81.Pan Z, Trikalinos TA, Kavvoura FK, et al. Local literature bias in genetic epidemiology: an empirical evaluation of the Chinese literature [electronic article] PLoS Med. 2005;2(12):e334. doi: 10.1371/journal.pmed.0020334. [DOI] [PMC free article] [PubMed] [Google Scholar]