Abstract
The δ-globin gene (HBD) of eutherian mammals exhibits a propensity for recombinational exchange with the closely linked β-globin gene (HBB) and has been independently converted by the HBB gene in multiple lineages. Here we report the presence of a chimeric β/δ fusion gene in the African elephant (Loxodonta africana) that was created by unequal crossing-over between misaligned HBD and HBB paralogs. The recombinant chromosome that harbors the β/δ fusion gene in elephants is structurally similar to the “anti-Lepore” duplication mutant of humans (the reciprocal exchange product of the hemoglobin Lepore deletion mutant). However, the situation in the African elephant is unique in that the chimeric β/δ fusion gene supplanted the parental HBB gene and is therefore solely responsible for synthesizing the β-chain subunits of adult hemoglobin. A phylogenetic survey of β-like globin genes in afrotherian and xenarthran mammals revealed that the origin of the chimeric β/δ fusion gene and the concomitant inactivation of the HBB gene predated the radiation of “Paenungulata,” a clade of afrotherian mammals that includes three orders: Proboscidea (elephants), Sirenia (dugongs and manatees), and Hyracoidea (hyraxes). The reduced fitness of the human Hb Lepore deletion mutant helps to explain why independently derived β/δ fusion genes (which occur on an anti-Lepore chromosome) have been fixed in a number of mammalian lineages, whereas the reciprocal δ/β fusion gene (which occurs on a Lepore chromosome) has yet to be documented in any nonhuman mammal. This illustrates how the evolutionary fates of chimeric fusion genes can be strongly influenced by their recombinational mode of origin.
Keywords: chimeric fusion gene, gene duplication, gene family evolution, globin genes, hemoglobin, nonallelic homologous recombination
Introduction
The complete duplication of protein-coding genes typically results in a redundancy of function between the two initially identical daughter copies. This functional redundancy may often entail a relaxation of purifying selection that permits the accumulation of degenerative mutations in one or both gene copies (Ohno 1970; Lynch et al. 2001; Zhang 2003; Lynch and Katju 2004; Taylor and Raes 2004). In the majority of cases, one of the two gene copies will eventually be rendered functionless by inactivating mutations. However, in a small minority of cases, the fixation of previously forbidden mutations may lead to the acquisition of novel function. The chances of evolving a novel function may be improved in cases where the duplication produces a chimeric fusion gene that combines distinct functional modules of two separate parent genes (Katju and Lynch 2003, 2006). The products of chimeric duplication events are structurally distinct at their inception, so they may be especially likely to follow distinct evolutionary trajectories. For this reason, it is important to characterize the structural differences between chimeric fusion genes and their progenitor copies, and to identify the genetic and evolutionary mechanisms that have contributed to their retention in the genome (Long and Langley 1993; Begun 1997; Chen et al. 1997; Nurminsky et al. 1998; Thomson et al. 2000; Jones and Begun 2005; Jones et al. 2005; Shih and Jones 2008).
The δ-globin gene (HBD) of eutherian mammals exhibits a propensity for recombinational exchange with the closely linked β-globin gene (HBB) and has been independently converted by the HBB gene in multiple lineages (Martin et al. 1983; Goodman et al. 1984; Hardies et al. 1984; Hardison 1984; Hardison and Margot 1984; Hutchison et al. 1984; Koop et al. 1989; Tagle et al. 1991; Prychitko et al. 2005; Hoffmann et al. 2008a; Opazo et al. 2008a). The 5′-HBD–HBB-3′gene pair originated via duplication of a proto β-globin gene in the common ancestor of eutherian mammals after the divergence from marsupials (Goodman et al. 1984; Hardison 1984; Opazo et al. 2008a, 2008b). The HBD–HBB gene pair delimits the 3′ end of the β-globin gene cluster, and expression of both paralogs is restricted to fetal and adult erythroid cells. The remaining β-like globin genes at the 5′ end of the gene cluster are typically expressed in embryonic erythroid cells derived from the yolk sac (Hardison 2001). During the course of eutherian evolution, the ontogenetically later-expressed HBD and HBB genes have experienced very different fates. In the majority of species that have been studied to date, the β-chain subunits of fetal and adult hemoglobin (Hb) are exclusively encoded by one or more copies of the HBB gene. The HBD gene has been deleted or inactivated in most eutherian lineages (e.g., rodents, lagomorphs, and many primates; Opazo et al. 2008a). In the few species that have retained a transcriptionally active copy of the HBD gene, it is typically expressed at a much lower level than HBB. For example, HBD is not transcribed in Old World monkeys (Boyer et al. 1971; Barnicot and Hewett-Emmett 1972; Martin et al. 1980; Martin et al. 1983; Vincent and Wilson 1989), but in other primates, the fraction of adult Hb tetramers that incorporate β-chain products of the HBD gene spans a wide range: 1% in hominoids (Boyer et al. 1971), 6% in New World monkeys (Spritz and Giebel 1988), 18% in the Philippine tarsier (Tarsius syrichta; Koop et al. 1989), and 40% in the galago (Otolemur crassicaudatus; Tagle et al. 1991).
In species that synthesize a detectable fraction of HBD-derived β-chain Hb isoforms, the transcriptional activity of HBD always appears to be attributable to the acquisition of HBB-like promoter sequence via gene conversion or unequal crossing-over. As stated by Hardies et al. (1984: 3755), the HBD genes “…can be described as vestigial or partially inactivated genes whose structural integrity may be maintained only by gene conversion.” For example, the galago synthesizes two adult β-chain Hb isoforms, β1 and β2, which are distinguished by a single amino acid substitution (Watanabe et al. 1985; Tagle et al. 1988). The β2 chain is the product of an HBD gene that has been almost completely overwritten by gene conversion from the downstream HBB gene (Tagle et al. 1991). The HBD and HBB genes of the galago share a homology block that extends from 800 bp upstream of the proximal CCAAT element to the 3′ end of the third (and last) exon. After this initial recombination event, recurrent gene conversion between the two paralogs continued to homogenize sequence variation across the entire gene region with the exception of intron 2 and the more distal 5′ flanking region. This evolutionary pattern appears to be fairly general, as the 5′ end of HBD (including exons 1 and 2) has undergone numerous independent recombinational exchanges with the downstream HBB paralog in multiple lineages of eutherian mammals (reviewed by Goodman et al. 1984; Hardison and Margot 1984; Opazo et al. 2008a).
Here we report the presence of a functional chimeric β/δ fusion gene in the African elephant (Loxodonta africana) that originated via unequal crossing-over between misaligned HBD and HBB paralogs. The recombinant chromosome that harbors the β/δ fusion gene in elephants is structurally similar to the “anti-Lepore” duplication mutant of humans (the reciprocal exchange product of the Hb Lepore deletion mutant; Forget 2001) in that the 5′ portion is HBB-like and the 3′ portion is HBD-like. To gain insight into the evolutionary origin of this chimeric fusion gene, we investigated the genomic structure of the β-globin gene cluster in representatives of two basal lineages of eutherian mammals: Afrotheria (which includes elephants, hyraxes, dugongs, manatees, aardvarks, tenrecs, and elephant shrews) and Xenarthra (which includes sloths, armadillos, and anteaters). Results of our comparative survey of β-like globin genes in basal eutherians revealed that the origin of the chimeric β/δ fusion gene and the concomitant inactivation of the HBB gene predated the radiation of “Paenungulata” (Simpson 1945), a clade that contains a morphologically disparate group of afrotherian mammals from three orders: Proboscidea (elephants), Sirenia, (sea cows, including dugongs and manatees), and Hyracoidea (the rodent-like hyraxes).
Materials and Methods
Genomic Sequence Data and Annotation
We obtained genomic DNA sequence spanning the β-globin gene cluster of the African elephant (L. africana) from the High Throughput Genomic Sequences database (accession numbers: AC169166, AC167954) and from the super_3994 contig of the Broad Institute 2× elephant assembly loxAfr1 (May 2005; http://genome.ucsc.edu/). Recent phylogenomic studies have demonstrated that a superordinal clade (Atlantogenata) composed of Afrotheria and Xenarthra is the sister group of all remaining members of the eutherian crown group (Boreoeutheria) (Hallstrom et al. 2007; Wildman et al. 2007). Thus, for the purpose of making comparisons with the elephant β-globin gene cluster, we obtained genomic contigs that spanned the entire β-globin gene cluster of seven other mammalian species, including one additional afrotherian (lesser hedgehog tenrec, Echinops telfairi [AC187664]), two xenarthrans (nine-banded armadillo, Dasypus novemcinctus [AC151518] and Hoffmann's two-toed sloth, Choloepus didactylus [AC231564]), two representative boreoeutherian species (human, Homo sapiens [AC104389] and European rabbit, Oryctalagus cuniculus [AC166202]), and two marsupial species (the gray short-tailed opossum, Monodelphis domestica [352230764–352330641] and the North American opossum, Didelphis virginiana [AC140955]). The genomic sequences analyzed in this study were in phase 2, meaning that the order and orientation of the constituent sequence contigs were firmly established. For each species, we identified β-like globin genes in the unannotated genomic sequence by using the program Genscan (Burge and Karlin 1997) and by comparing known exon sequences with genomic contigs using the program Blast2, version 2.2 (Tatusova and Madden 1999).
Inferring Orthologous Relationships
The genomic structure of the β-globin gene clusters of atlantogenatan mammals was investigated by conducting pairwise comparisons of sequence similarity with the human gene cluster. The comparisons of sequence similarity were conducted with the program Pipmaker (Schwartz et al. 2000). Orthologous relationships among β-like globin genes were inferred from phylogeny reconstructions of flanking sequence and intron 2 sequence (Storz et al. 2007; Hoffmann et al. 2008a, 2008b; Opazo et al. 2008a, 2008b). We conducted phylogeny reconstructions on three separate sequence alignments: 0.8 kb of flanking sequence immediately upstream of the initiation codon, 5,028 bp of intron 2, and 1 kb of flanking sequence immediately downstream of the termination codon. Because previous studies have documented that ectopic recombinational exchanges are typically restricted to the 5′ end of the HBD gene (including exons 1 and 2) and the upstream flanking region (Hardies et al. 1984; Hardison 1984; Hardison and Margot 1984; Prychitko et al. 2005; Hoffmann et al. 2008a; Opazo et al. 2008a), sequence variation in intron 2 and the downstream flanking region should be especially useful for the purpose of assigning orthologous relationships. For the phylogenetic analysis, we included β-like globin gene sequences from six eutherian species (African elephant, tenrec, armadillo, sloth, human, and rabbit) and two marsupial species (the North American opossum and the gray short-tailed opossum). Sequence alignments were carried out using the program MUSCLE (Edgar 2004), as implemented in the European Bioinformatics Institute web server (http://www.ebi.ac.uk). Phylogenetic relationships were inferred in a maximum likelihood framework using Treefinder, version October 2008 (Jobb et al. 2004). For each data partition, we used the “propose model” tool of Treefinder to select the best-fitting model of nucleotide substitution. Model selection was based on the Akaike information criterion with correction for small sample size. For the upstream flanking sequence, intron 2 sequence, and downstream flanking sequence, we used the GTR + Γ5 (Rodriguez et al. 1990), HKY + Γ5 (Hasegawa et al. 1985), and J3 + Γ5 models, respectively. The J3 + Γ5 model accounts for variation in transversion rates in which TA = CG and TG = CA, and rate variation among sites follows a discrete gamma distribution with five categories (Jobb et al. 2004). We evaluated support for the nodes of each tree with 1,000 bootstrap pseudoreplicates.
Phylogenetic Survey of β-Like Globin Genes in Atlantogenata
In addition to the β-like globin gene sequences that we retrieved from the full genome assemblies, we also cloned and sequenced the adult β-like globin genes from nine additional afrotherian species (Asian elephant, Elephas maximus; West Indian manatee, Trichechus manatus; Dugong, Dugong dugon; western tree hyrax, Dendrohyrax dorsalis; yellow-spotted hyrax, Heterohyrax brucei; rock hyrax, Procavia capensis; cape golden mole, Chyrosochloris asiatica; bushveld elephant shrew, Elephantulus intufi; and aardvark, Orycteropus afer) and one additional xenarthran species (pale-throated three-toed sloth, Bradypus tridactylus). We also retrieved sequences of the rock hyrax β-like globin genes from Whole Genome Sequence files on GenBank (HBD: ABRQ01367090, ABRQ01559481; HBB/D: ABRQ01298789, ABRQ01298790; exon 3 of HBBps: ABRQ01298787).
For each of the atlantogenatan species, genomic DNA was extracted from whole blood or liver tissue using DNeasy kits (Qiagen, Valencia, CA). We designed polymerase chain reaction (PCR) primers for the HBD and HBB genes using an alignment of orthologous sequences from the available atlantogenatan species as well as more distantly related eutherian mammals. PCRs were conducted on 100 ng of template DNA (2 μl) in 0.2-ml tubes containing 48 μl of reaction mixture (1.0 μl of each dNTP (2.5 mM), 5.0 μl 10× PCR Reaction Buffer, 3.0–4.5 μl of 50 mM MgCl2, 5.0 μl of each primer (10.0 pmol/μl), 0.5 μl Taq polymerase (5 U/μl), and 24.0–25.5 μl ddH2O), using PTC-0220 DNA Engine Dyad (GMI, Ramsey, MN) and Mastercycler Gradient (Eppendorf, Hamburg, Germany) thermocyclers. The β-like globin genes were amplified using a 30-cycle PCR protocol (94 °C for 30 s; 50 °C for 15 s; and 72 °C for 30–75 s) followed by a 10-min final extension at 72 °C. PCR products were isolated using Montage PCR Centrifugal Filter Devices (Millipore, Billerica, MA). After obtaining DNA sequences of the resulting amplicons, we used a genome-walking protocol to obtain sequences of the upstream and downstream flanking regions (APAgene Genome Walking Kit; Bio S&T Inc., Montreal, Canada). All PCR primer combinations used in this study are listed in supplementary table S1, Supplementary Material online, and sequences for all primers are provided in supplementary table S2 (Supplementary Material online).
Target bands were excised and purified using a MinElute Gel Extraction Kit (Qiagen), and the extracts were ligated using a Qiagen PCR Cloningplus Kit. Transformation products were incubated on agar plates containing 0.1% v/v ampicillin (100 mg/ml), X-GAL (40 mg/ml), and Isopropyl β-D-1-thiogalactopyranoside (100 mM) for 18 h at 37 °C. Plasmid DNA was extracted from positive clones using a QIAprep Spin Miniprep Kit. Plasmids containing globin gene inserts were sequenced in both directions using M-13(F)-40 and M-13(R) sequencing primers, and samples were run on a 3739 ABI PRISM Genetic Analyzer.
Multiple sequence alignments and maximum likelihood phylogeny reconstructions were carried out using methods described above. Phylogeny reconstructions of intron 2 sequences (5,395 bp) were conducted using a J1 + Γ5 model of nucleotide substitution. This model accounts for variation in transversion rates in which TA = TG and CA = CG, and rate variation among sites follows a discrete gamma distribution with five categories (Jobb, von Haeseler, and Strimmer 2007).
Results
Genomic Structure of the β-Globin Gene Cluster in Atlantogenata
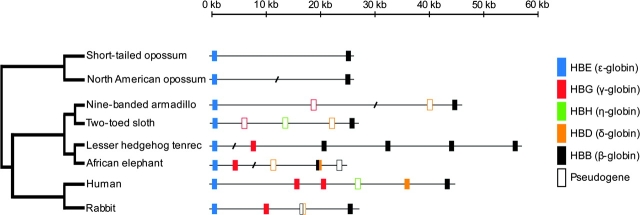
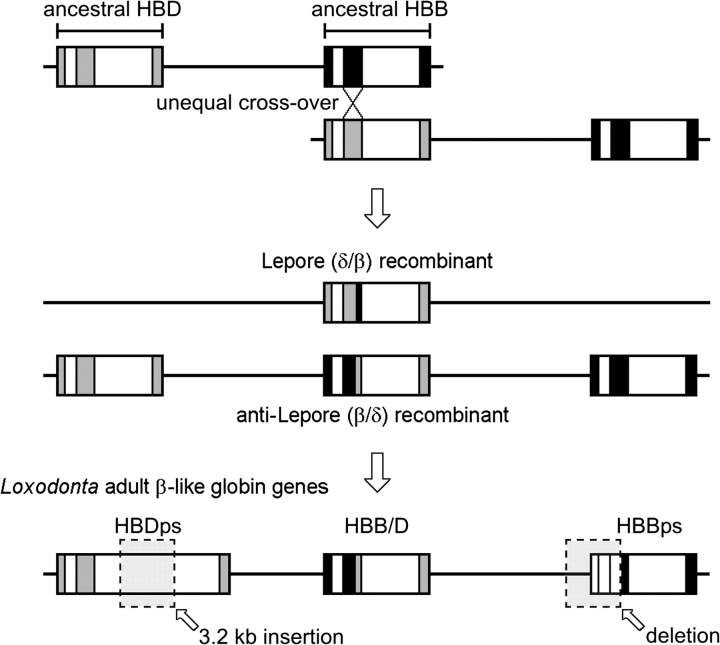
In the African elephant, the adult portion of the β-globin gene cluster contains three genes (from 5′ to 3′): 1 δ-like pseudogene (HBDps), 1 chimeric β/δ fusion gene (HBB/D), and 1 β-globin pseudogene (HBBps; fig. 1). Following the nomenclature of Aguileta et al. (2006), the “ps”suffix indicates that HBDps and HBBps are pseudogenes. A dot plot of pairwise sequence similarity between the β-globin gene clusters of African elephant and human clearly demonstrates that the 5′ flanking sequence and 5′ end of the elephant HBB/D fusion gene is HBB-like, whereas the 3′ flanking sequence and 3′ end of the gene are HBD-like (fig. 2). This pattern of pairwise sequence matches indicates that the fusion gene was created by unequal crossing-over between a misaligned HBD–HBB gene pair (fig. 3).
FIG. 1.—
Genomic structure of the β-globin gene cluster in afrotherians (African elephant and lesser hedgehog tenrec), xenarthrans (nine-banded armadillo and two-toed sloth), two representative boreoeutherian species (human and European rabbit), and two marsupial outgroups (gray short-tailed opossum and North American opossum).
FIG. 2.—

Dot plot of pairwise sequence similarity between the 3′ ends of the β-globin gene clusters of African elephant, Loxodonta africana, and human.
FIG. 3.—
Diagram showing the inferred unequal crossing-over event that produced the 5′-HBD, HBB/D, HBBps-3′ anti-Lepore chromosome in the evolutionary ancestry of elephants.
In African elephants, the HBB/D fusion gene appears to be the only β-like globin gene expressed during adulthood, as a conceptual translation of the coding sequence precisely matches the reported β-chain amino acid sequence of purified Hb from this species (Braunitzer et al. 1984; Kleinschmidt et al. 1986). The HBDps and HBB/D sequences are distinguished from one another by 42 amino acid substitutions and a number of insertion/deletion differences (including a ∼3.2 kb insertion in intron 2 of HBDps). HBDps and HBBps are clearly not functional, as the initiation codon of HBDps was eliminated by a single nucleotide substitution (ATG→ATC) and the entire 5′ portion of HBBps has been deleted (including exon 1 and roughly half of exon 2).
Because the tenrec has four putatively functional HBB genes and no HBD gene (fig. 1), the HBB/D fusion gene of elephant does not appear to be shared by all members of the afrotherian clade. In the case of the xenarthrans, both armadillo (Dasypus) and sloth (Choloepus) possess single copies of the HBD and HBB genes. As a result of inactivations of the embryonic γ-globin (HBG) and η-globin (HBH) genes and the ontogenetically later-expressed HBD gene, the β-globin gene cluster of xenarthans has reverted back to the ancestral 5′-HBE-HBB-3′ structure that is characteristic of marsupials (fig. 1).
Identification of the Crossover Breakpoint in the Elephant HBB/D Fusion Gene
To localize the crossover breakpoint in the elephant HBB/D fusion gene, we searched for abrupt spatial transitions in levels of pairwise sequence identity between the fusion gene and the human HBD and HBB genes. We reasoned that the portion of the elephant HBB/D fusion gene that is located upstream of the breakpoint should be more similar to human HBB than to human HBD, whereas the portion downstream of the breakpoint should be more similar to HBD than to HBB. In comparisons of exon 1 and exon 2 sequences, the elephant HBB/D fusion gene exhibited slightly lower levels of sequence identity to the human HBD gene than to the HBB gene (exon 1: 78.3% and 79.4%, respectively; exon 2: 81.2% and 84.3%, respectively). By contrast, in comparisons of exon 3 sequences, the elephant HBB/D fusion gene exhibited a higher level of sequence identity to the human HBD gene (89.2%) than to the human HBB gene (84.5%). Thus, exon 3 of the elephant HBB/D fusion gene appears to have retained a distinctive HBD-like character that distinguishes it from paralogous HBB-like sequence. The similar level of pairwise sequence identity between the elephant HBB/D fusion gene and the human HBD and HBB genes in the first 2 exons may be attributable to the fact that the HBD gene has been independently converted by the HBB gene in that same gene region in both species (Martin et al. 1983; Goodman et al. 1984). By contrast, exon 3 of HBD typically lies outside of HBB-derived conversion tracts (Martin et al. 1983; Hardison and Margot 1984; Prychitko et al. 2005; Hoffmann et al. 2008a). Based on alignments of nucleotide and amino acid sequences of the HBD and HBB genes (fig. 4), it appears that the crossover breakpoint of the elephant HBB/D fusion gene is located between residue positions 78 and 87 of the polypeptide (encoded by exon 2). Studies of human Hb Lepore and anti-Lepore mutants indicate that the crossover breakpoints are typically located between residue positions 69 and 104. The inferred crossover breakpoint in the elephant HBB/D fusion gene closely coincides with those of three well-characterized Hb Lepore mutants in humans: Hb Baltimore, Hb Boston, and Hb Hollandia (Mavilio et al. 1983; Metzenberg et al. 1991; fig. 4).
FIG. 4.—
An alignment of β-chain amino acid sequences from human, rabbit, and African elephant. The crossover breakpoint that produced the chimeric HBB/D gene of the African elephant is located within the shaded interval. The boxes with dotted lines denote the breakpoint intervals associated with three different Hb Lepore deletion mutants in humans.
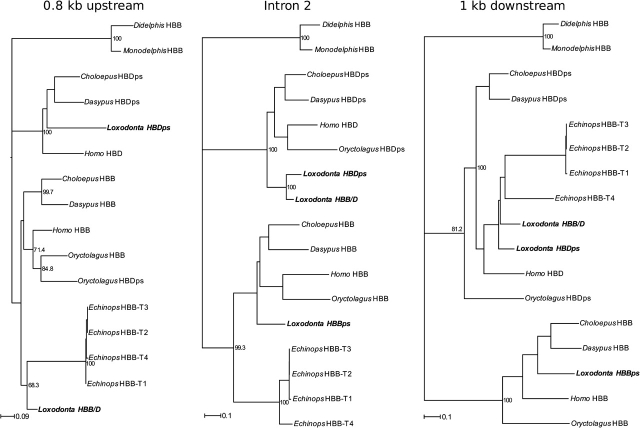
Inferring Orthologous Relationships among β-Like Globin Genes
Although phylogeny reconstructions based on coding sequence indicate that orthologous relationships among HBD and HBB genes have been obscured by a history of concerted evolution (Czelusniak et al. 1982; Opazo et al. 2008a, 2008b), the true history of gene duplication and species divergence is revealed by sequence variation in intron 2 and in the flanking regions. Phylogeny reconstructions based on intron 2 and 3′ flanking sequence provide support for orthologous relationships of the elephant HBD and HBB/D genes with the tenrec, armadillo, sloth, human, and rabbit HBD genes (fig. 5). The phylogeny reconstructions of intron 2 and 3′ flanking sequence also provide support for orthologous relationships among the HBB genes of these same species, including the HBBps pseudogene of the African elephant (fig. 5). Ambiguities in the alignments of noncoding sequence were largely attributable to the inclusion of highly divergent marsupial outgroups. However, these ambiguities do not appear to have had a major effect on the phylogeny reconstructions, as similar sets of relationships among eutherian HBD and HBB sequences were recovered in reconstructions that excluded the marsupial outgroup sequences (supplementary fig. S1, Supplementary Material online). In each of the three trees shown in figure 5, monophyly of the four HBB paralogs of the tenrec indicates that this set of genes originated via successive rounds of lineage-specific gene duplication. Although the four HBB paralogs of the tenrec have 5′ flanking sequences and intron 2 sequences that cluster with the HBB genes of other species, the downstream flanking sequences exhibit strong affinities to those of HBD genes (fig. 5). These results are consistent with those of a previous comparative genomic analysis, which demonstrated that the 5′ flanking sequence and coding sequence of the four tenrec HBB paralogs are HBB-like in character, and that the 3′ flanking sequence is HBD-like in character (Opazo et al. 2008a). This anomalous pattern could be explained by a history of interparalog gene conversion in which the coding region and 5′ flanking region of an ancestral, single-copy HBD gene were completely converted by an HBB donor gene that has since been deleted in the tenrec lineage. Three subsequent rounds of duplication could have then produced a total of four paralogous gene copies that have each retained 5′ flanking sequence and coding sequence that are HBB-like in character.
FIG. 5.—
Maximum likelihood phylograms depicting relationships among adult β-like globin genes based on 0.8 kb of 5′ flanking sequence, 5,028 bp of intron 2, and 1 kb of 3′ flanking sequence. The analysis included data from two afrotherians (African elephant, Loxodonta africana, and the lesser hedgehog tenrec, Echinops telfairi), two xenarthrans (nine-banded armadillo, Dasypus novemcinctus, and Hoffmann's two-toed sloth, Choloepus didactylus), two representative boreoeutherian species (human, Homo sapiens, and European rabbit, Oryctalagus cuniculus), and two marsupial species (the gray short-tailed opossum, Monodelphis domestica, and the North American opossum, Didelphis virginiana). All HBB and HBD sequences were retrieved from full genomic contigs (see text for details). The 5′ flanking sequence of the elephant HBBps pseudogene was not included in the analysis due to missing data (i.e., the upstream region, together with exon 1, intron 1, and part of exon 2 was deleted in the Loxodonta genome; see fig. 3). Because the β-like globin genes of mammals have undergone multiple rounds of duplication that have resulted in tandemly repeated sets of paralogous gene copies, we index each paralog with the symbol—T followed by a number that corresponds to the linkage order in the 5′ to 3′ orientation (Aguileta et al. 2006).
The three β-like globin gene sequences that we retrieved from the 2× assembly of the rock hyrax genome appear to be orthologous to the HBDps, HBB/D, and HBBps genes of the African elephant based on pairwise dot plots and phylogenetic analysis of flanking sequence (data not shown). Our phylogenetic survey of intron 2 sequences from additional atlantogenatan taxa revealed that HBD and HBB sequences form two reciprocally monophyletic clades with high bootstrap support (fig. 6). In contrast to phylogenies based on coding sequence, which primarily reflect a history of concerted evolution, the phylogeny based on intron 2 sequence more accurately reflects the true set of orthologous and paralogous relationships. The inferred tree topology is therefore consistent with previous reports that intron 2 of the HBD gene is largely unaffected by gene conversion from upstream HBB paralogs (Hardison and Margot 1984; Hoffmann et al. 2008a; Opazo et al. 2008a). However, the tree does show evidence of gene conversion between the HBD and HBB/D intron 2 sequences of African elephant (Loxodonta), yellow-spotted hyrax (Heterohyrax), and rock hyrax (Procavia), as these paralogous sequences group together to the exclusion of orthologous sequences in other paenungulates (fig. 6).
FIG. 6.—
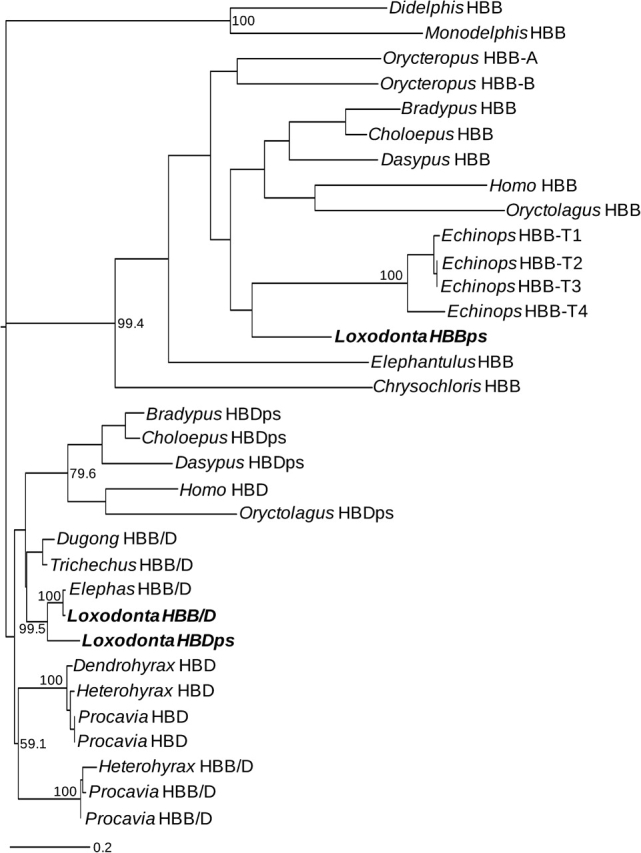
Maximum likelihood phylogram depicting relationships among adult β-like globin genes based on 5,510 bp of intron 2 sequence. The analysis included sequence data from 15 atlantogenatan species in addition to two boreoeutherian outgroups (Oryctolagus and Homo) and two marsupial outgroups (Monodelphis and Didelphis).
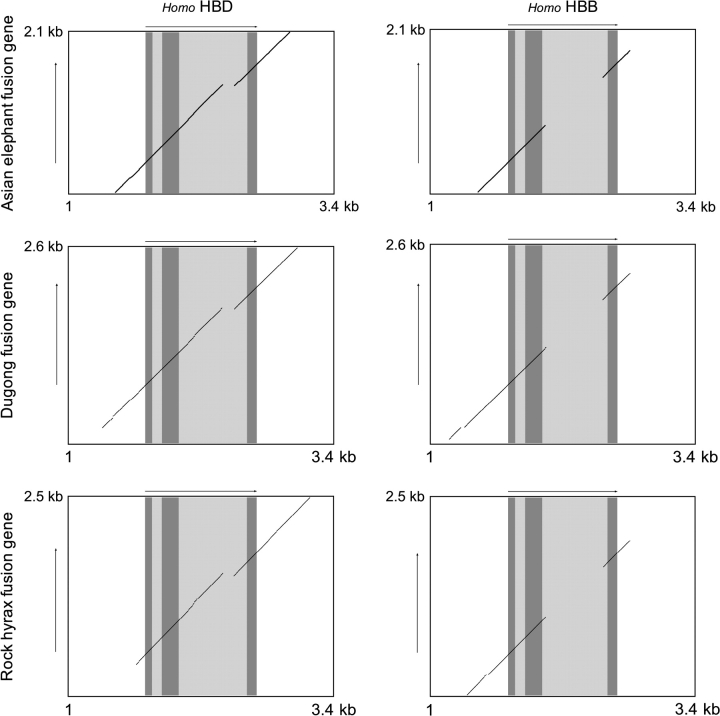
Phylogenetic inferences regarding the orthology of HBB/D fusion genes among paenungulate mammals are corroborated by pairwise dot plots that demonstrate that the fusion genes of Asian elephant, West Indian manatee, and rock hyrax are all characterized by an HBB-like 5′ portion and an HBD-like 3′ portion (fig. 7). Moreover, an alignment of upstream flanking sequences confirms that the chimeric β/δ fusion genes of paenungulate mammals have HBB-like promoters (supplementary fig. S2, Supplementary Material online). It thus seems clear that the origin of the chimeric HBB/D fusion gene and the concomitant inactivation of the HBB gene predated the radiation of paenungulate mammals.
FIG. 7.—
Dot plots of sequence similarity between the chimeric HBB/D (β/δ) fusion genes of paenungulate mammals and the HBD and HBB genes of human. Top row: Asian elephant β/δ fusion gene versus human HBD and HBB; Middle row: Dugong β/δ fusion gene versus human HBD and HBB; Bottom row: Rock hyrax β/δ fusion gene versus human HBD and HBB.
Discussion
Our investigation of the β-globin gene cluster in African elephant revealed that unequal crossing-over between misaligned copies of ancestral, single-copy HBD and HBB genes produced a chimeric β/δ fusion gene that is flanked by inactivated copies of the parental HBD gene on the 5′ side and the parental HBB gene on the 3′ side. This 5′-HBDps–HBB/D–HBBps-3′ recombinant chromosome is structurally similar to the “anti-Lepore” duplication mutant in humans (the reciprocal exchange product of the Hb Lepore deletion mutant). Although structurally similar HBB/D fusion genes have been documented in other mammals, they appear to be functionally inconsequential with respect to oxygen transport because the β-chain products of these genes are not incorporated into functional Hb tetramers (Hardies et al. 1984; Hardison and Margot 1984; Hoffmann et al. 2008a).
The situation in the African elephant is unique because the chimeric HBB/D fusion gene completely supplanted the parental HBB gene, and is therefore solely responsible for synthesizing the β-chain subunits of adult Hb. Only two other mammals were previously known to lack a functional ortholog of the HBB gene: the African pygmy hedgehog (Atelerix albiventris) and Eurasian shrew (Sorex araneus), both of which belong to the order Eulipotyphla (Opazo et al. 2008a). Our initial analysis revealed that the African elephant provides another example of a eutherian mammal that does not possess an intact, functional HBB gene. Comparisons of DNA sequence data with previously published amino acid sequences further indicate that the HBB/D fusion gene supplanted the parental HBB gene not just in the African elephant but in all extant members of the paenungulate clade. The HBB/D sequence of Asian elephant is 100% identical to the β-globin amino acid sequence reported from the purified Hb of this species (Braunitzer et al. 1984; Kleinschmidt et al. 1986). Likewise, the HBB/D sequence of West Indian manatee is 100% identical to the β-globin amino acid sequence of the closely related Amazonian manatee (Trichechus inunguis; Kleinschmidt et al. 1986). The HBB/D sequences that we independently obtained from the rock hyrax are 97–100% identical to the β-globin amino acid sequence reported for this species (Kleinschmidt and Braunitzer 1983; Braunitzer et al. 1984; Kleinschmidt et al. 1986), whereas the HBD gene is only 79% identical. It thus appears that, in all paenungulate mammals, the β-chain subunits of the major adult Hb isoform are encoded by orthologous copies of the same HBB/D fusion gene.
The observation that the β-type Hb chains of elephants and other paenungulates are hybrid polypeptides characterized by a β-like N-terminal portion and a δ-like C-terminal portion is especially surprising because previous workers had suggested that the HBD gene was little more than a relic gene that had been evolving under relaxed functional constraints even before the diversification of eutherian mammals. As stated by Hardies et al. (1984: 3755): “The overall poor evolutionary performance of δ-like genes among the mammals suggests that the proto-δ was already destined for disposal prior to the mammalian radiation.” Thus, if the Hbs of paenungulate mammals are characterized by any unusual functional properties, it would be of interest to know whether they are attributable to amino acid substitutions in the C-terminal, HBD-encoded region of the β-type Hb chain that occurred during a prior history of relaxed functional constraint.
One especially noteworthy substitution in the HBD-derived portion of the β-type Hb chain of African elephant, Asian elephant, and dugong is β131Gln→Glu, which is located at a highly conserved α1β1 intersubunit contact surface. The β131Glu mutation has also been described in humans (Hb Camden; Wade-Cohen et al. 1973; Hubbard et al. 1975). Protein engineering experiments have demonstrated that the β131Gln→Glu substitution disrupts hydrogen bonds between α103His and β131Gln at the α1β1-subunit interface, resulting in a slightly reduced oxygen-binding affinity (Chang et al. 2002). The substitution also diminishes the reactivity of the sulfhydryl group of β93Cys, a residue located in close proximity to the α1β2-subunit interface, and increases the proton exchange rates of α1β1-interface histidyl residues on the α-chain (α103His and α122His; Chang et al. 2002).
The Role of Unequal Crossing-Over in Creating Chimeric Fusion Genes
When the breakpoint of an unequal crossover event interrupts the coding regions of a misaligned gene pair, both reciprocal exchange products will contain a chimeric fusion gene (fig. 3). For example, in humans, crossovers between misaligned copies of the closely linked HBD and HBB genes result in a solitary δ/β fusion gene on one recombinant chromosome (the Hb Lepore deletion mutant) and the reciprocal β/δ fusion gene on the other recombinant chromosome (the “anti-Lepore” duplication mutant; Forget 2001). In the former case, the δ/β fusion gene on the Lepore chromosome is solely responsible for the synthesis of the β-chain subunits of adult Hb. In the latter case, the β/δ fusion gene on the anti-Lepore chromosome is flanked by fully functional copies of the parental HBD gene on the 5′ side and the parental HBB gene on the 3′ side. Heterozygotes for the Hb Lepore deletion produce adult red blood cells that contain normal α2β2 Hb tetramers (HbA), as well as lesser quantities of α2(δ/β)2 tetramers (Hb Lepore) that incorporate products of the δ/β fusion gene. The HbA and Hb Lepore isoforms are not present in equal concentrations in circulating red blood cells because the δ/β fusion gene is under the control of a weak HBD promoter, and is therefore transcribed at a much lower rate than the normal HBB gene. The difference in relative isoform concentrations is also partly attributable to reduced stability of the δ/β mRNA transcript (Forget 2001).
Whereas heterozygotes for the Lepore δ/β fusion gene typically suffer a relatively mild form of δ/β thalassemia (a type of hemolytic anemia caused by reduced synthesis of β-globin subunits), homozygotes for the δ/β fusion gene suffer more severe forms of erythrocytic dysfunction, with clinical manifestations equivalent to the “β thalassemia intermedia” or “β thalassemia major” disease phenotypes (Olivieri and Weatherall 2001). In the β thalassemia syndromes, the imbalance of α- and β-globin synthesis leads to an excess of oxidized α-chain monomers that aggregate and precipitate in bone marrow normoblasts and in circulating red blood cells. The insoluble α-chain aggregates and their associated breakdown products (iron, heme, and hemichrome) contribute to oxidative damage of the cytoskeleton membrane proteins, which eventually results in the destruction of the affected erythroid precursor cells in the bone marrow and premature hemolysis of mature red blood cells in the peripheral circulation (Rachmilewitz and Schrier 2001).
In contrast to the complications associated with inheritance of the Hb Lepore deletion chromosome (and the associated δ/β fusion gene), the inheritance of the reciprocal exchange product of the same crossover event—the 5′ HBD–HBB/D–HBB 3′ anti-Lepore chromosome—is not associated with any hematological pathology because the recombinant chromosome retains a transcriptionally active HBB gene in addition to the HBD and HBB/D genes. The difference in fitness between the Hb Lepore deletion mutant and the anti-Lepore duplication mutant may explain why the two reciprocal β/δ and δ/β fusion genes are not equally common among contemporary mammals. Independently derived β/δ fusion genes (which occur on an anti-Lepore chromosome) have been fixed in a number of mammalian lineages, whereas transcriptionally active copies of the reciprocal δ/β fusion gene (which occurs on the Lepore chromosome) have yet to be documented in the β-globin gene cluster of any nonhuman mammal. This illustrates how the evolutionary fates of chimeric fusion genes can be strongly influenced by their recombinational mode of origin.
Supplementary Material
Supplementary figures S1 and S2, supplementary tables S1 and S2, and all sequence alignments associated with this manuscript are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/). Sequence data from this article have been submitted to GenBank under accession nos. DQ091202–DQ091208, DQ091210, DQ091212, DQ091214, and FJ588200–FJ588203.
Acknowledgments
We thank S.V. Edwards and two anonymous reviewers for helpful suggestions on the manuscript and A. Signore for technical assistance in the laboratory. This work was funded by grants to J.F.S. from the National Institutes of Health, the National Science Foundation, and the Nebraska Research Council, and by grants to K.L.C. by the National Sciences and Engineering Research Council of Canada, the Winnipeg Foundation, and the University of Manitoba Research Grant Program. A.M.S. received support from a University of Manitoba Graduate Fellowship, and J.C.O. received support from the Oliver P. Pearson Award from the American Society of Mammalogists.
References
- Aguileta G, Bielawski JP, Yang Z. Evolutionary rate variation among vertebrate β-globin genes: implications for dating gene family duplication events. Gene. 2006;380:21–29. doi: 10.1016/j.gene.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Barnicot NA, Hewett-Emmett D. Red cell and serum proteins of Cercocebus, Presbytis, Colobus and certain other species. Folia Primatol (Basel) 1972;17:442–457. doi: 10.1159/000155461. [DOI] [PubMed] [Google Scholar]
- Begun DJ. Origin and evolution of a new gene descended from alcohol dehydrogenase in Drosophila. Genetics. 1997;145:375–382. doi: 10.1093/genetics/145.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer SH, Crosby EF, Noyes AN, Fuller GF, Leslie SE, Donaldson LJ, Vrablik GR, Schaefer EW, Jr, Thurmon TF. Primate hemoglobins: some sequences and some proposals concerning the character of evolution and mutation. Biochem Genet. 1971;5:405–448. doi: 10.1007/BF00487132. [DOI] [PubMed] [Google Scholar]
- Braunitzer G, Stangl A, Schrank B, Krombach C, Wiesner H. Phosphate–haemoglobin interaction. The primary structure of the haemoglobin of the African elephant (Loxodonta africana, Proboscidea): asparagine in position 2 of the β-chain. Hoppe Seylers Z Physiol Chem. 1984;365:743–749. doi: 10.1515/bchm2.1984.365.2.743. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Chang C-k, Simplaceanu V, Ho C. Effects of amino acid substitutions at B131 on the structure and properties of hemoglobin: evidence for communication between α1β1- and α1β2-subunit interfaces. Biochemistry. 2002;41:5644–5655. doi: 10.1021/bi011919d. [DOI] [PubMed] [Google Scholar]
- Chen L, DeVries AL, Cheng CH. Convergent evolution of antifreeze glycoproteins in Antarctic notothenioid fish and Arctic cod. Proc Natl Acad Sci USA. 1997;94:3811–3816. doi: 10.1073/pnas.94.8.3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czelusniak J, Goodman M, Hewett-Emmett D, Weiss ML, Venta PJ, Tashian RE. Phylogenetic origins and adaptive evolution of avian and mammalian haemoglobin genes. Nature. 1982;298:297–300. doi: 10.1038/298297a0. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget BG. Molecular mechanisms of beta thalassemia. In: Steinberg M, Forget B, Higgs D, Nagel R, editors. Disoders of hemoglobin: genetics, pathophysiology, and clinical management. Cambridge (UK): Cambridge University Press; 2001. pp. 252–276. [Google Scholar]
- Goodman M, Koop BF, Czelusniak J, Weiss ML. The η-globin gene. Its long evolutionary history in the β-globin gene family of mammals. J Mol Biol. 1984;180:803–823. doi: 10.1016/0022-2836(84)90258-4. [DOI] [PubMed] [Google Scholar]
- Hallstrom BM, Kullberg M, Nilsson MA, Janke A. Phylogenomic data analyses provide evidence that Xenarthra and Afrotheria are sister groups. Mol Biol Evol. 2007;24:2059–2068. doi: 10.1093/molbev/msm136. [DOI] [PubMed] [Google Scholar]
- Hardies SC, Edgell MH, Hutchison CA., 3rd Evolution of the mammalian β-globin gene cluster. J Biol Chem. 1984;259:3748–3756. [PubMed] [Google Scholar]
- Hardison R. Organization, evolution, and regulation of the globin genes. In: Steinberg M, Forget B, Higgs D, Nagel R, editors. Disorders of hemoglobin: genetics, pathophysiology, and clinical management. Cambridge (UK): Cambridge University Press; 2001. pp. 95–115. [Google Scholar]
- Hardison RC. Comparison of the β-like globin gene families of rabbits and humans indicates that the gene cluster 5′-ϵ-γ-δ-β-3′ predates the mammalian radiation. Mol Biol Evol. 1984;1:390–410. doi: 10.1093/oxfordjournals.molbev.a040326. [DOI] [PubMed] [Google Scholar]
- Hardison RC, Margot JB. Rabbit globin pseudogene ψβ2 is a hybrid of δ- and β-globin gene sequences. Mol Biol Evol. 1984;1:302–316. doi: 10.1093/oxfordjournals.molbev.a040321. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF. New genes originated via multiple recombinational pathways in the β-globin gene family of rodents. Mol Biol Evol. 2008a;25:2589–2600. doi: 10.1093/molbev/msn200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF. Rapid rates of lineage-specific gene duplication and deletion in the α-globin gene family. Mol Biol Evol. 2008b;25:591–602. doi: 10.1093/molbev/msn004. [DOI] [PubMed] [Google Scholar]
- Hubbard M, Wilson JB, Wrightstone RN, Efremov GD, Huisman TH. Hb Camden and Hb Hope found during routine testing. Acta Haematol. 1975;54:53–58. doi: 10.1159/000208051. [DOI] [PubMed] [Google Scholar]
- Hutchison CAI, Hardies SC, Padgett RW, Weaver S, Edgell MH. The mouse globin pseudogene bh3 is descended from a premammalian δ-globin gene. J Biol Chem. 1984;259:12881–12889. [PubMed] [Google Scholar]
- Jobb G, von Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jones CD, Begun DJ. Parallel evolution of chimeric fusion genes. Proc Natl Acad Sci USA. 2005;102:11373–11378. doi: 10.1073/pnas.0503528102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CD, Custer AW, Begun DJ. Origin and evolution of a chimeric fusion gene in Drosophila subobscura, D. madeirensis, and D. guanche. Genetics. 2005;170:207–219. doi: 10.1534/genetics.104.037283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katju V, Lynch M. On the formation of novel genes by duplication in the Caenorhabditis elegans genome. Mol Biol Evol. 2006;23:1056–1067. doi: 10.1093/molbev/msj114. [DOI] [PubMed] [Google Scholar]
- Katju V, Lynch M. The structure and early evolution of recently arisen gene duplicates in the Caenorhabditis elegans genome. Genetics. 2003;165:1793–1803. doi: 10.1093/genetics/165.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt T, Braunitzer G. The primary structure of hemoglobins of the rock hyrax (Procavia habessinica, Hyracoidea): insertion of glutamine in the α- chains. Hoppe Seylers Z Physiol Chem. 1983;364:1303–1313. [PubMed] [Google Scholar]
- Kleinschmidt T, Czelusniak J, Goodman M, Braunitzer G. Paenungulata: a comparison of the hemoglobin sequences from elephant, hyrax, and manatee. Mol Biol Evol. 1986;3:427–435. doi: 10.1093/oxfordjournals.molbev.a040411. [DOI] [PubMed] [Google Scholar]
- Koop BF, Siemieniak D, Slightom JL, Goodman M, Dunbar J, Wright PC, Simons EL. Tarsius δ- and β-globin genes: conversions, evolution, and systematic implications. J Biol Chem. 1989;264:68–79. [PubMed] [Google Scholar]
- Long M, Langley CH. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science. 1993;260:91–95. doi: 10.1126/science.7682012. [DOI] [PubMed] [Google Scholar]
- Lynch M, Katju V. The altered evolutionary trajectories of gene duplicates. Trends Genet. 2004;20:544–549. doi: 10.1016/j.tig.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Lynch M, O'Hely M, Walsh B, Force A. The probability of preservation of a newly arisen gene duplicate. Genetics. 2001;159:1789–1804. doi: 10.1093/genetics/159.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL, Vincent KA, Wilson AC. Rise and fall of the δ-globin gene. J Mol Biol. 1983;164:513–528. doi: 10.1016/0022-2836(83)90048-7. [DOI] [PubMed] [Google Scholar]
- Martin SL, Zimmer EA, Kan YW, Wilson AC. Silent δ-globin gene in Old World monkeys. Proc Natl Acad Sci USA. 1980;77:3563–3566. doi: 10.1073/pnas.77.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavilio F, Giampaolo A, Care A, Sposi NM, Marinucci M. The δ/β crossover region in Lepore boston hemoglobinopathy is restricted to a 59 base pairs region around the 5′ splice junction of the large globin gene intervening sequence. Blood. 1983;62:230–233. [PubMed] [Google Scholar]
- Metzenberg AB, Wurzer G, Huisman TH, Smithies O. Homology requirements for unequal crossing over in humans. Genetics. 1991;128:143–161. doi: 10.1093/genetics/128.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurminsky DI, Nurminskaya MV, De Aguiar D, Hartl DL. Selective sweep of a newly evolved sperm-specific gene in Drosophila. Nature. 1998;396:572–575. doi: 10.1038/25126. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- Olivieri NF, Weatherall DJ. Clinical aspects of β thalassemia. In: Steinberg MH, Higgs DR, Nagel RL, editors. Disorders of hemoglobin: genetics, pathophysiology, and clinical management. Cambridge (UK): Cambridge University Press; 2001. pp. 277–341. [Google Scholar]
- Opazo JC, Hoffmann FG, Storz JF. Differential loss of embryonic globin genes during the radiation of placental mammals. Proc Natl Acad Sci USA. 2008a;105:12950–12955. doi: 10.1073/pnas.0804392105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Hoffmann FG, Storz JF. Genomic evidence for independent origins of β-like globin genes in monotremes and therian mammals. Proc Natl Acad Sci USA. 2008b;105:1590–1595. doi: 10.1073/pnas.0710531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prychitko T, Johnson RM, Wildman DE, Gumucio D, Goodman M. The phylogenetic history of New World monkey β-globin reveals a platyrrhine β to δ gene conversion in the atelid ancestry. Mol Phylogenet Evol. 2005;35:225–234. doi: 10.1016/j.ympev.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Rachmilewitz E, Schrier S. Pathophysiology of β thalassemia. In: Steinberg MH, Forget FB, Higgs DR, Nagel RL, editors. Disorders of hemoglobin: genetics, pathophysiology, and clinical management. Cambridge (UK): Cambridge University Press; 2001. pp. 233–251. [Google Scholar]
- Rodriguez F, Oliver JL, Marin A, Medina JR. The general stochastic model of nucleotide substitution. J Theor Biol. 1990;142:485–501. doi: 10.1016/s0022-5193(05)80104-3. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker–a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HJ, Jones CD. Patterns of amino acid evolution in the Drosophila ananassae chimeric gene, siren, parallel those of other Adh-derived chimeras. Genetics. 2008;180:1261–1263. doi: 10.1534/genetics.108.090068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G. The principles of classification and a classification of mammals. Bull Am Mus Nat Hist. 1945;85:1–350. [Google Scholar]
- Spritz RA, Giebel LB. The structure and evolution of the spider monkey δ-globin gene. Mol Biol Evol. 1988;5:21–29. doi: 10.1093/oxfordjournals.molbev.a040474. [DOI] [PubMed] [Google Scholar]
- Storz JF, Baze M, Waite JL, Hoffmann FG, Opazo JC, Hayes JP. Complex signatures of selection and gene conversion in the duplicated globin genes of house mice. Genetics. 2007;177:481–500. doi: 10.1534/genetics.107.078550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagle DA, Koop BF, Goodman M, Slightom JL, Hess DL, Jones RT. Embryonic epsilon and gamma globin genes of a prosimian primate (Galago crassicaudatus). Nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J Mol Biol. 1988;203:439–455. doi: 10.1016/0022-2836(88)90011-3. [DOI] [PubMed] [Google Scholar]
- Tagle DA, Slightom JL, Jones RT, Goodman M. Concerted evolution led to high expression of a prosimian primate δ-globin gene locus. J Biol Chem. 1991;266:7469–7480. [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- Thomson TM, Lozano JJ, Loukili N, et al. (13 co-authors) Fusion of the human gene for the polyubiquitination coeffector UEV1 with Kua, a newly identified gene. Gen Res. 2000;10:1743–1756. doi: 10.1101/gr.gr-1405r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent KA, Wilson AC. Evolution and transcription of old world monkey globin genes. J Mol Biol. 1989;207:465–480. doi: 10.1016/0022-2836(89)90457-9. [DOI] [PubMed] [Google Scholar]
- Wade-Cohen PT, Yates A, Bellingham AJ, Huehns ER. Amino-acid Substitution in the α1β1 Intersubunit Contact of Haemoglobin-Camden β131 (H9) Gln→Glu. Nature. 1973;243:467–468. doi: 10.1038/243467a0. [DOI] [PubMed] [Google Scholar]
- Watanabe B, Fujii T, Nakashima Y, Maita T, Matsuda G. Amino-acid sequences of the alpha and beta chains of adult hemoglobins of the grand galago, Galago crassicaudatus. Biol Chem Hoppe Seyler. 1985;366:265–269. doi: 10.1515/bchm3.1985.366.1.265. [DOI] [PubMed] [Google Scholar]
- Wildman DE, Uddin M, Opazo JC, Liu G, Lefort V, Guindon S, Gascuel O, Grossman LI, Romero R, Goodman M. Genomics, biogeography, and the diversification of placental mammals. Proc Natl Acad Sci USA. 2007;104:14395–14400. doi: 10.1073/pnas.0704342104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18:292–298. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.