Abstract
Horizontal gene transfer is surprisingly common among plant mitochondrial genomes. The first well-established case involves a homing group I intron in the mitochondrial cox1 gene shown to have been frequently acquired via horizontal transfer in angiosperms. Here, we report extensive additional sampling of angiosperms, including 85 newly sequenced introns from 30 families. Analysis of all available data leads us to conclude that, among the 640 angiosperms (from 212 families) whose cox1 intron status has been characterized thus far, the intron has been acquired via roughly 70 separate horizontal transfer events. We propose that the intron was originally seeded into angiosperms by a single transfer from fungi, with all subsequent inferred transfers occurring from one angiosperm to another. The pattern of angiosperm-to-angiosperm transfer is biased toward exchanges between plants belonging to the same family. Illegitimate pollination is proposed as one potential factor responsible for this pattern, given that aberrant, cross-species pollination is more likely between close relatives. Other potential factors include shared vectoring agents or common geographic locations. We report the first apparent cases of loss of the cox1 intron; losses are accompanied by retention of the exonic coconversion tract, which is located immediately downstream of the intron and which is a product of the intron's self-insertion mechanism. We discuss the many reasons why the cox1 intron is so frequently and detectably transferred, and rarely lost, and conclude that it should be regarded as the “canary in the coal mine” with respect to horizontal transfer in angiosperm mitochondria.
Keywords: cox1, group I intron, horizontal transfer, angiosperm, homing endonuclease
Introduction
Until recently, horizontal transfer was virtually unknown to occur between multicellular eukaryotes, except for the special case of mobile genetic elements (Robertson 2002). In the last few years, however, horizontal gene transfer (HGT) from one land plant to another has been shown to be surprisingly common (reviewed in Richardson and Palmer 2007). Almost all cases involve genes transferred from one plant mitochondrial genome to, most likely, that of another plant (Bergthorsson et al. 2003, 2004; Won and Renner 2003; Davis and Wurdack 2004; Mower et al. 2004). In contrast, no convincing evidence of HGT has been found in land plant chloroplast genomes despite far greater sampling (Rice and Palmer 2006), and only 2 cases of horizontally transferred nuclear genes or mobile genetic elements have been described in plants (Diao et al. 2006; Ghatnekar et al. 2006).
The most extensive case of HGT known among flowering plants involves the sole group I intron found thus far in angiosperm mitochondrial genomes. Multiple lines of evidence indicate that this intron, located in the gene encoding cytochrome oxidase subunit 1 (cox1), has been horizontally acquired numerous times during angiosperm evolution, with at least the first transfer having occurred from a fungal donor (Vaughn et al. 1995; Adams et al. 1998; Cho et al. 1998; Cho and Palmer 1999; Barkman et al. 2007). Like many other group I introns (Lambowitz and Belfort 1993; Goddard and Burt 1999), the angiosperm cox1 intron is an invasive genetic element, thanks to its encoded DNA “homing” endonuclease, whose lengthy target site is typically present only once per genome (Lambowitz and Belfort 1993; Lambowitz et al. 1999, supplementary fig. S1, Supplementary Material online). Cleavage by the endonuclease at the target site of an intronless allele results in insertion of the intron (together with its open reading frame [ORF]) via the double-strand break repair pathway (Cech 1990; Lambowitz and Belfort 1993; Lambowitz et al. 1999).
To gain new insights into patterns and mechanisms of cox1 intron transfer and evolution in angiosperms, we have surveyed 197 angiosperms to determine the phylogenetic distribution of the cox1 intron. Almost all the newly discovered intron-containing cox1 genes were sequenced, which led to further targeted sequencing of intron-bearing as well as intron-lacking genes occupying pivotal phylogenetic positions. Our findings more than triple the number of available cox1 intron sequences in angiosperms and double the number of inferred acquisitions of this intron via HGT among angiosperms. This increased sampling also reveals the first known cases of loss of this intron. We conclude that all cox1 intron transfers except for an initial “seed transfer” from fungi occurred by plant-to-plant transfer and that transfer often occurs between closely related species (e.g., between members of the same family). We propose that illegitimate pollination or shared vectoring agents might be important mechanisms of horizontal transfer in angiosperms.
Materials and Methods
Plant DNAs were either prepared using a cetyl trimethyl ammonium bromide DNA extraction protocol (Doyle JJ and Doyle JL 1987) or obtained from other sources (see Acknowledgments). Plant accession numbers are listed in Supplementary table S1 (Supplementary Material online).
The cox1 gene was amplified by polymerase chain reaction (PCR) using 2 sets of primers. Primers cox1-1 (5′-AYGAMAAATCYGGTYGATGG-3′) and cox1-4 (5′-ACCGRATCCAGGCAGAATGRG-3′) amplify exon 1 (and the intron when present), yielding products that were 750 nt long (or 1,735 nt). Primers cox1-3 (5′-CATCTCTTTYTGTTCTTCGGT-3′) and cox1-6 (5′-AGCTGGAAGTTCTCCAAAAGT-3′) amplify exon 2 (and the intron when present), yielding products of 800 nt (or 1.8 kb). PCR products were purified using ExoSAP-IT (United States Biochemical, Cleveland, OH) and sequenced using an ABI 3730 (Applied Biosystems, Foster City, CA). Sequencing primers include PCR primers and 2 other primers located in the intron: cox1-10 (5′-TGACTACTATCAAAGTAGA-3′) and cox1-8 (5′-GTAGAGTCTTATAAGGTAGT-3′). GenBank accession numbers from sequences reported here are listed in Supplementary table S1 (Supplementary Material online).
Sequences were aligned manually with MacClade 4.0 (Maddison W and Maddison P 2000). Phylogenetic analyses were performed on a data set of almost all available cox1 intron sequences from angiosperms and on a data set of cox1 exon sequences that included most intron-containing angiosperms. Supplementary table S1 (Supplementary Material online) includes GenBank accession numbers for all sequences included in phylogenetic analyses.
Maximum likelihood analyses of the intron and exon data sets were performed with Garli 0.951 (Zwickl 2006) under the general time reversible model with parameters for invariable sites and gamma-distributed rate heterogeneity (GTR + I + Γ4; 4 rate categories). This substitution model was supported by hierarchical likelihood ratio tests done using Modeltest v.3.5 (Posada and Crandall 1998). Ten independent runs were conducted using the automated stopping criterion or for up to 5,000,000 generations to ensure convergence to a similar topology and likelihood score. Five hundred bootstrap replicates were performed. The data matrices and optimal trees are available in TreeBase (http://www.treebase.org/treebase/), accession number S1907.
The CONSEL package (Shimodaira and Hasegawa 2001) was used to calculate the approximately unbiased (AU) P values for unconstrained and constrained trees. Using reduced intron data sets including only taxa from Rhamnaceae or Apocynaceae, we compared the unconstrained intron tree topology with the topology constrained according to organismal phylogeny (i.e., with strictly vertical inheritance of the intron). The most likely tree under each constraint was determined by searching for the best tree compatible with that constraint using PAUP* (Swofford et al. 2002). The site likelihoods for this tree and for the best tree in the unconstrained analysis were exported from PAUP*, and the AU P values were calculated from these data.
We scored the presence/absence of the cox1 intron (unordered character) for all taxa included in figure 1 and reconstructed the ancestral states based on the maximum parsimony (MP) criteria on using MacClade (Maddison W and Maddison P 2000). Reconstructions were performed on the organismal cladogram shown in figure 1. Polytomies were resolved in 3 different ways: randomly and such that the minimum and maximum number of steps were necessary. Ambiguous state reconstructions were resolved using both ACCTRAN (accelerates changes toward the root and favors loss over independent gains of character states) and DELTRAN (delays changes away from the root and favors independent gains over losses). In addition, reconstructions based on Dollo parsimony were performed (independent intron gains not allowed, i.e., a single intron acquisition and any number of losses).
FIG. 1.—
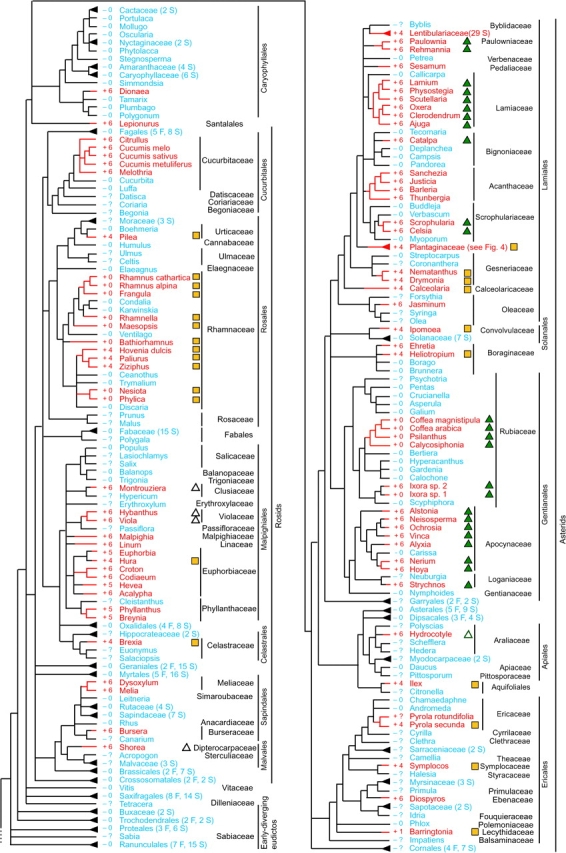

Distribution of the cox1 intron among 640 angiosperms for which cox1 has been sampled. Presence of the intron is shown by red branches on the tree, red taxon names, and plus (+) symbols. Blue lines and names and minus (−) symbols identify taxa lacking the intron. Question marks indicate that exon sequences are not available for that taxon. Blue and red triangles at the tips of branches indicate multiply sampled terminals, with the number of sampled taxa given in parentheses according to the number of families (F) and species (S) sampled. Numbers to the immediate left of plant names give the number of nucleotides changed by exon coconversion (for most collapsed clades, CCT data are available for only a few taxa, whereas full CCT data for the Plantaginaceae are shown in figs. 2 and 3). Open and filled green triangles identify introns belonging to 2 large, nested intron clades so-marked near the top of figure 5. Yellow squares likewise identify introns belonging to the well-supported, so-marked clade at the bottom of figure 5.
We conducted the concentrated changes test as implemented in MacClade (Maddison 1990) to evaluate whether the origins of the coconversion tract (CCT) are concentrated in clades with intron presence. We scored all taxa for which we have information for both intron and CCT presence. The CCT was scored as “present” when all 6 nt differences that define the CCT (i.e., CCT = 6) were observed or a subset of them (CCT = 1–5; see Results and Discussion). Those cases with other differences in this region were scored as CCT = “absent,” for example, in Peperomia and Plantago. Character state reconstructions were performed as described above. Using these reconstructions, we tested for an association between intron presence and CCT presence using the concentrated changes test (Maddison 1990). Because the number of operational taxonomic units was prohibitively large (190), we ran a simulated sample size of 10,000 using the MINSTATE reconstruction option and constraining state 0 to be ancestral.
Results and Discussion
Evidence of HGT from cox1 Intron Distribution
We used PCR to assess the presence/absence of the cox1 intron in 197 angiosperms and sequenced the intron from 85 species belonging to 30 families. Combining these data with cox1 sequences present in GenBank at the time this study was conducted brings the total number of sequenced cox1 introns in angiosperms to 129 out of a total of 640 species of angiosperms examined in this study. Just prior to submission of this manuscript, Barkman et al. (2007) reported an additional 23 cases of the cox1 intron from a diversity of angiosperms, mostly parasitic plants. These additional sequences have not been incorporated in the present study because this would have caused prohibitive delays and because they do not materially affect, but only augment, the conclusions of the current study.
The intron is highly conserved in sequence across angiosperms and is invariably located at the same position near the middle of the cox1 gene, creating 2 exons of typically 726 and 858 nt in length. The presence of a single copy of the cox1 gene and intron (when present) was shown by Southern blots analyses of more than 300 genera (Cho et al. 1998). No evidence for intron presence/absence polymorphism or heteroplasmy was detected in our study.
The phylogenetic distribution of the cox1 intron across angiosperms is shown in figure 1, which includes 162 taxa from 45 families containing the intron. The cladogram shown in figure 1 was compiled from previously published phylogenetic hypotheses (Bremer et al. 2003; Qiu et al. 2005; Soltis et al. 2005; Qiu, Li, Wang, et al. 2006). To keep this figure from being excessively large, many multiply sampled, intron-lacking clades were collapsed to a single, terminal clade (order, family, or genus). These 50 collapsed clades represent 115 sampled families and 318 sampled species and include many of the largest groups of angiosperms, for example, Asterales, Fabales, and Poales, each with more than 20,000 species. In contrast, only 2 intron-containing clades are collapsed: the Lentibulariaceae, because intron sequences were not available for analysis, and the well-sampled Plantaginaceae, which is shown in detail in figures 2 and 3D. The representational bias of figure 1 thus gives an inflated sense of how widespread the cox1 intron is among angiosperms.
FIG. 2.—
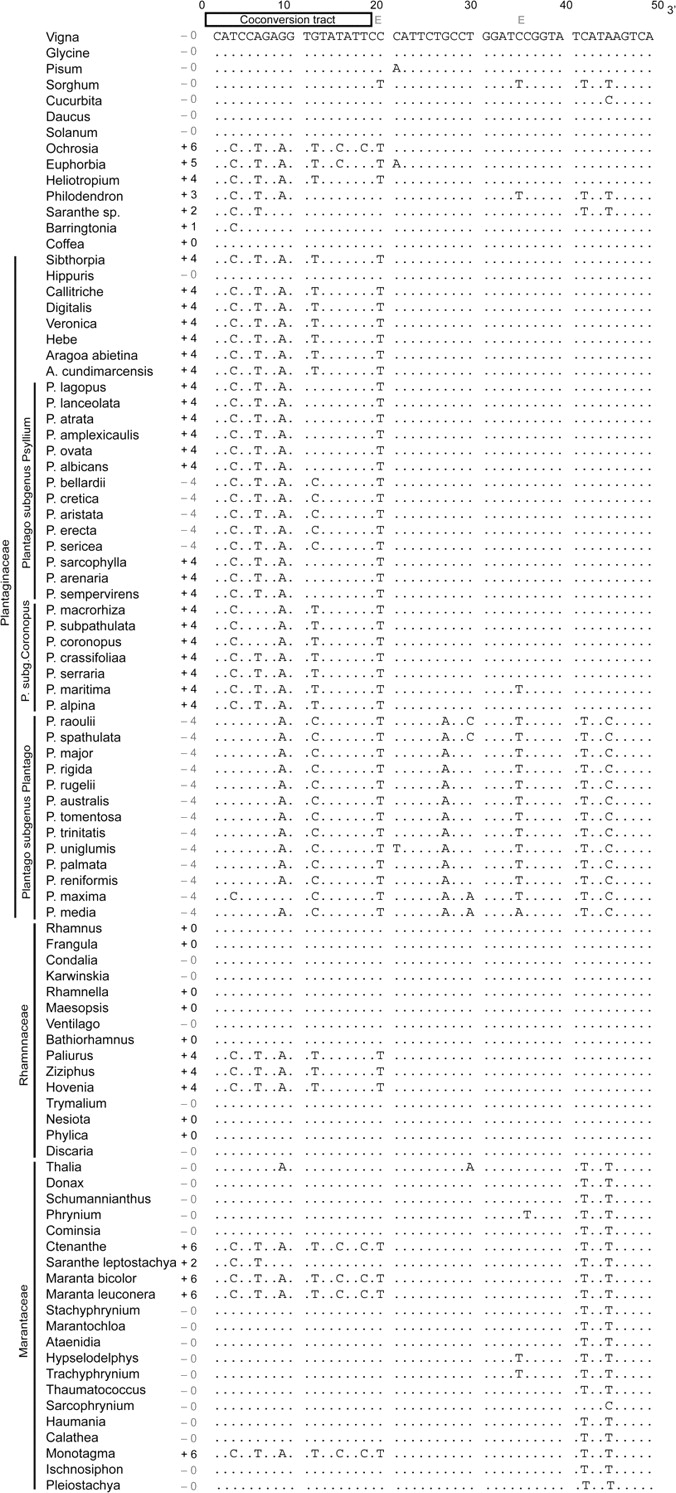
Nucleotide alignment of the 5′ end of cox1 exon 2 from 92 angiosperms, with special reference to Plantaginaceae, Rhamnaceae, and Marantaceae. Dots indicate nucleotide identities to the reference sequence (Vigna) shown at the top of the alignment. Plus (+) and minus (−) symbols indicate intron presence or absence, respectively. The numbers shown next to these symbols indicate the number of nucleotides changed by coconversion. The 2 Es at the top of the figure mark known sites of RNA editing.
FIG. 3.—

Comparison of organismal (left set of trees) and intron (right set) phylogenies for 4 families with complex evolution of the cox1 intron. The presence of the intron is shown by bold plant names and plus (+) symbols, whereas unbolded names and minus (−) symbols identify taxa lacking the intron. Numbers to the immediate left of names give the number of nucleotides “ancestrally” changed by coconversion according to the parsimony criterion (see text and fig. 2 for clarification of the Plantaginaceae situation). The “G” and “L” mark inferred intron gains and losses, respectively. Intron trees and BS values (≥50%) are from figure 5, with subtended intron clades belonging to families other than the one highlighted in a particular tree indicated by a minimal number of dashed lines ending with a star and without any plant names given (fig. 5). The number of species examined for 4 multiply sampled clades of Plantaginaceae is given in parentheses. The rectangle in the Rubiaceae intron tree indicates a shared deletion of 5 nt.
Despite this distortion, the intron clearly has a very patchy distribution across angiosperms, being entirely absent from the early-diverging lineages and sporadically present across more derived groups. From an equal-weighted parsimony standpoint (i.e., with intron gains and losses weighted equally), the phylogenetic distribution of the intron was shaped primarily by numerous independent gain events. MP ancestral state reconstructions suggest 37–50 intron gains and 6–16 losses (totaling 53–59 steps) depending on the resolution of polytomies in the organismal phylogeny and ambiguous character reconstructions. Under Dollo parsimony, a single ancestral intron acquisition and 112–128 intron losses are required, depending on the alternative resolutions of polytomies. In contrast, the phylogenetic distribution of the 22 group II introns in angiosperm mitochondrial genomes (the cox1 intron is the only group I intron known in these genomes) is strikingly different. These group II introns are widely present across angiosperms, clearly ancestrally so (Malek and Knoop 1998; Chaw et al. 2008), with only occasional absences readily interpreted as cases of intron loss (Gugerli et al. 2001; Joly et al. 2001; Kudla et al. 2002). The cox1 intron is not expected to be more susceptible to loss than these group II introns. In fact, the intron's homing properties should make it more recalcitrant to loss, so the very different distributions of these 2 classes of introns are again consistent with frequent, horizontal acquisition of the cox1 intron.
Evidence of HGT from CCTs
Additional evidence in support of the hypothesis of frequent horizontal transfer of the cox1 intron comes from analysis of exonic CCTs. Group I homing introns propagate themselves into intron-lacking alleles via homing endonuclease–mediated cleavage of the intron's target (homing) site, followed by exonucleolytic degradation and double-strand break repair (Cech 1990; Lambowitz and Belfort 1993; Lambowitz et al. 1999; Burt and Trivers 2006). During this process, the exonic regions immediately flanking the insertion site are usually degraded to varying lengths, and this damage is repaired using the same intron-containing template. CCTs are the segments of flanking exons in the recipient genome that are converted during this process. These tracts can vary in length from one to hundreds of nucleotides (Mueller et al. 1996; Jurica and Stoddard 1999; Lambowitz et al. 1999; Caprara and Waring 2005). The extent to which exonic coconversion is detectable depends on the number and distribution of nucleotide differences in the exons of donor and recipient alleles (fig. 2 and supplementary fig. S2 [Supplementary Material online]).
There is no evidence in angiosperms of a CCT immediately 5′ to the cox1 intron, whereas a 3′ CCT of 3–18 nt in length is indicated by the presence of 1–6 nt differences, respectively (we refer to these tracts as CCT = 1 through CCT = 6, fig. 2 and supplementary fig. S2 [Supplementary Material online]). About 90% of intron-containing cox1 genes have a clearly identifiable 3′ CCT, whereas the motif is absent in virtually all intronless genes (figs. 1–3). Intronless cox1 genes are instead nearly identical in sequence to each other—and thus to the inferred common ancestor of angiosperms—in the 18-nt region corresponding to the CCT (fig. 2 and supplementary fig. S2 [Supplementary Material online]). Aside from the very rapidly evolving cox1 genes of Plantago (see below), sequences in the CCT are identical across intron-containing angiosperms, with the tract length varying in a polar manner with respect to the intron insertion site (fig. 2 and supplementary fig. S2 [Supplementary Material online]). The concentrated changes test implemented in MacClade indicated that CCT presence is significantly associated with intron presence (P < 0.01). The highly significant correlation (figs. 1–3) between intron presence/absence and CCT presence/absence supports the hypothesis that each phylogenetically disparate group containing the intron probably acquired it by an independent HGT event. If the present-day intron distribution reflects a few ancient transfer events with many subsequent intron losses, we would expect to find a relict CCT in the many cox1 genes that lost the intron (see below). This hypothesis has little support, however, because the only CCT evidence for intron loss is restricted to a single family of angiosperms (Plantaginaceae; figs. 1–3).
Evidence of HGT from Phylogenetic Analysis of the cox1 Intron
Phylogenetic analyses support the hypothesis of widespread HGT of the cox1 intron, with the caveat that the low substitution rates of plant mitochondrial genomes (Wolfe et al. 1987; Palmer and Herbon 1988) sometimes yield trees with poor resolution and/or low statistical support (Qiu, Li, Hendry, et al. 2006). Despite this caveat, it is striking that whereas a phylogenetic tree of cox1 exon sequences (fig. 4) recovers the expected organismal phylogeny reasonably well—with not a single strongly supported conflict indicative of HGT—an intron-based tree (fig. 5) conflicts grossly with the organismal phylogeny. Conflicts are evident from various perspectives. Most globally, taxa color coded (green, red, and blue) according to 3 large groups of well-established monophyly (monocots, rosids, and asterids, respectively) (Jansen et al. 2007; Moore et al. 2007) are interspersed across the intron tree (fig. 5). In contrast, all 3 groups show the expected pattern of monophyly in the cox1 exon tree, consistent with a pattern of strictly vertical inheritance of cox1 exons (fig. 4). More specifically, there are numerous well-supported groupings of closely related introns from disparately related angiosperms, for example, the asterid Sesamum with the rosid Acalypha (97% bootstrap support [BS], AU P = 3.00 × 10−5; see top of fig. 5), the asterid Hydrocotyle with the monocot family Marantaceae (99% BS, AU P = 4.00 × 10−5; near top of figure), and a subset of Apocynaceae (Hoya + Nerium) with Rubiaceae (90% BS, AU P = 0.009; a little lower in the tree). Finally, the 2 large, well-supported intron clades marked with colored squares or triangles in figure 5 represent taxa scattered across many disparate lineages of either rosids, asterids, and monocots (triangles) or just rosids and asterids (squares; figs. 1 and 5).
FIG. 4.—

Maximum likelihood phylogeny of angiosperm cox1 coding regions using the GTR + I + Γ4 model of substitution. The data set includes 126 taxa, most of which have the cox1 intron (fig. 5) and 1,256 aligned exonic positions. The CCT was excluded from the character matrix, as were sites of known RNA editing. Taxa in red are rosids; blue, asterids; and green, monocots. Numbers on branches correspond to BS values ≥50% from 500 bootstrap replicates. In stark contrast to the cox1 intron phylogeny (fig. 5), this coding region phylogeny agrees very well with organismal phylogeny. The only notable conflict involves Siphonochilus, the basal-most member sampled from the Zingiberaceae, which groups instead with the Musaceae (Musa + Musella). However, the latter relationship is not well supported and is probably the result of long-branch attraction (the Siphonochilus and Musaceae branches are both very long).
FIG. 5.—
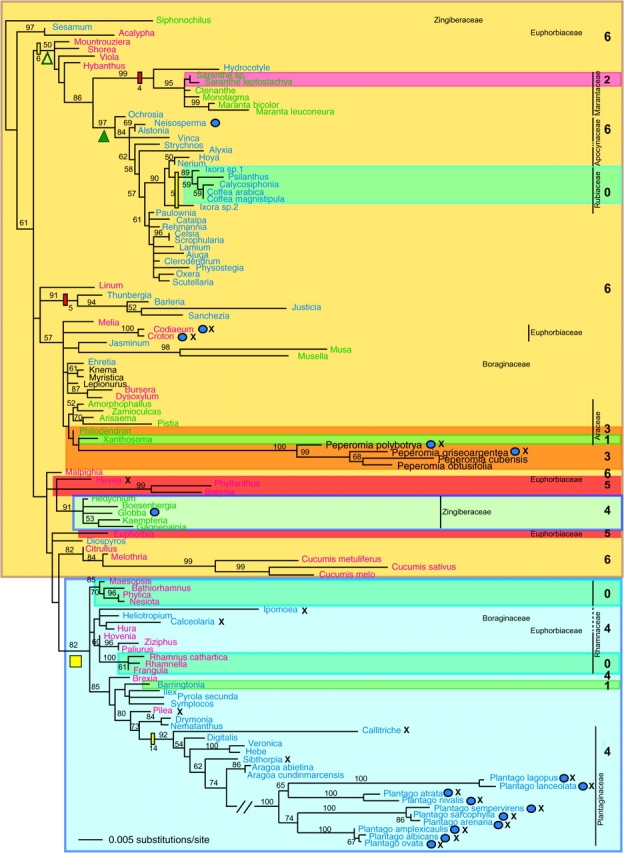
Maximum likelihood phylogeny of the angiosperm cox1 intron estimated using the GTR + I + Γ4 model of nucleotide substitution. The data set includes 121 taxa and 1,179 aligned intronic positions. Numbers on branches correspond to support values ≥50% from 500 bootstrap replicates. Taxa in red are rosids; blue, asterids; and green, monocots. Selected indels of phylogenetic importance are shown as red rectangles (insertions) and yellow rectangles (deletions), with the length of the indel in nucleotides given. Crosses and blue ovals mark taxa whose intronic ORFs are disrupted by out-of-frame indels or are out-of-frame with exon 1, respectively. Shaded boxes and numbers at far right give the number of nucleotides in exon 2 changed by coconversion. The yellow square and open and filled green triangles mark large, well-supported intron clades whose distributions are so-marked on the organismal phylogeny of figure 1. The root of the angiosperm intron tree probably lies among those introns with the longest CCT (CCT = 6, see text). Taxa belonging to the 9 families with multiple inferred intron gains are marked according to their family names to the right of the tree.
Despite recent discovery of a cox1 intron in the zygomycete fungus Rhizopus that is more closely related in sequence to the clade of angiosperm introns than are any other known introns (Seif et al. 2005), we were unable to root the angiosperm clade with confidence. Further sampling within fungi might reveal still more closely related fungal introns that would serve as appropriate outgroups, but owing to relatively high rates of mitochondrial sequence evolution in fungi relative to angiosperms, we are not optimistic that the angiosperm intron clade will ever be rooted. Despite lack of a proper outgroup, the root chosen for the cox1 intron phylogeny as shown in figure 5 is by no means arbitrary, as on 2 grounds, we can exclude the root from most parts of the angiosperm intron tree. First, we expect the root to lie among those introns with the longest, putatively ancestral CCT (CCT = 6); this excludes about half of the intron phylogeny. Second, the region of the 2 amino acid gap in the intron ORF shared by all members of the large, CCT = 6 clade (Montrouziera through Scutellaria) located near the top of figure 5 can be readily aligned with fungal ORFs from Rhizopus and Monoblepharella (data not shown; Seif et al. 2005), and the gap can confidently be interpreted as a deletional synapomorphy for this large group of introns (clade marked with yellow rectangle in fig. 5). On this basis, the root can also be excluded from that clade, which includes more than half of the taxa possessing the ancestral CCT.
Revisiting the Evidence for Horizontal versus Vertical Transmission of the cox1 Intron
In a recent study on the angiosperm cox1 intron, Cusimano et al. (2008) reached entirely opposite conclusions compared with this and previous studies (Cho et al. 1998; Cho and Palmer 1999; Barkman et al. 2007), as stated in the concluding sentence of their abstract: “Together, these results suggest that the cox1 intron entered angiosperms once, has since largely or entirely been transmitted vertically, and has been lost numerous times, with CCT footprints providing unreliable signal of former intron presence.” As explained in detail in the 3 above sections and elaborated on in previous studies (Cho et al. 1998; Cho and Palmer 1999), 3 lines of evidence support HGT of the cox1 intron among angiosperms: 1) the highly sporadic distribution of the intron, 2) significant incongruence between intron and organismal phylogenies based on AU tests and BS values, and 3) significant association of intron and CCT presence based on the concentrated changes test. With respect to point 2, we believe that Cusimano et al. have seriously downplayed the extent and magnitude of the phylogenetic incongruence. In addition, no plausible mechanism exists for replacement of the CCT with the ancestral exonic sequence “during the intron excision process,” as suggested by Cusimano et al. (2008, p. 275) to explain the absence of the CCT in more than 400 intron-lacking angiosperms (ca. 200 cox1 sequences are available from those angiosperms), which according to their model must have lost the intron via some 112–128 putative intron-loss events after a single ancestral intron acquisition. Thus, in the absence of statistical support for their model and with no plausible mechanism to account for loss of the CCT (Cusimano et al. 2008), we see no reason whatsoever to favor the intron loss hypothesis over the HGT hypothesis.
Similar Introns with Different Length CCTs
The most challenging intron patterns to interpret involve 3 small clades of entirely intron-containing taxa (Ixora sp1 and Ixora sp2 from the Rubiaceae; Saranthe, Ctenanthe, and Maranta from the Marantaceae; and Amorphophallus and Xanthosoma from the Araceae) for which the introns within each group are similar and phylogenetically consistent with a single shared gain but which nonetheless possess different length CCTs (fig. 5). At least 4 hypotheses could explain such patterns; the first 2 postulate a single horizontal transfer in each group, and the latter 2 postulate multiple transfers. Hypothesis 1 posits that the differences in CCT length are unrelated to intron transfer and reflect sequence drift via 4–6 single-nucleotide substitutions (reversions) within the 18-nt long CCT region. Hypothesis 2 posits that one of the homing events during the presumably brief period when the intron went to fixation vis-a-vis the population of ancestrally intronless alleles happened to generate a shorter CCT (CCT = 0–2, depending on the group) than the CCT = 6 form of the intron initially introduced by HGT and that the 2 forms of the intron were then fixed within descendant subgroups via lineage sorting. Hypothesis 3 posits independent gains of each of the 2 different length CCT introns in each group (plus 2 separate gains of the CCT = 6 intron in the Marantaceae group, i.e., separate intron gains in each Marantaceae genus listed above; see fig. 1). Hypothesis 4, like 1 and 2, posits intron gain in the ancestor of a given group, followed in this case by replacement of this intron-containing gene in one subgroup by a foreign, intron-containing cox1 gene with the other length CCT. Under both hypotheses 3 and 4, the donor for the additional horizontal transfers—of the intron or whole gene, respectively—is a close relative, probably a member of the same family (fig. 5).
Hypothesis 1 is extremely unlikely given how otherwise conserved each pair of cox1 genes is (fig. 4): the 2 Ixora cox1 coding sequences are identical across the 684 nt sequenced outside of the CCT yet differ at 6 of 18 sites within the CCT, Saranthe and Ctenanthe are also identical apart from 4 CCT differences, and Amorphophallus and Xanthosoma differ at only 4 sites apart from 5 CCT differences. Note also that all 6 CCT differences are at silent third codon positions and therefore unlikely to be under significant selective pressure, that codon usage bias is minimal in plant mitochondrial genomes, and that the 3′–5′ polarity of the CCT differences in the latter 2 pairs of taxa is inconsistent with the occurrence of independent, single-nucleotide substitutions. In addition, we did not find evidence for coevolution of this exonic region and the intron sequence. The putative interaction of the intron and the 3′ flanking exon involves only the first 2 nt of cox1 exon 2 (Vaughn et al. 1995), which are not variable in angiosperms (regardless of whether they contain the intron).
Based on current sampling, hypothesis 4 is regarded as substantially less likely than hypothesis 3 in the case of the Rubiaceae and Araceae. This is because although the 2 hypotheses involve the same number of horizontal transfers, the probability of active, homing endonuclease–mediated intronic HGT is much higher than that of passive, “genic” HGT (see final section). For the Marantaceae, hypothesis 4 cannot be so readily dismissed vis-a-vis hypothesis 3 because the latter postulates 2 additional HGTs versus only 1 for the former. Hypotheses 2 and 4 invoke the at least transitory coexistence within the mitochondrion of virtually identical or at least highly similar, respectively, intron-containing forms of the cox1 gene. Because repeated sequences of this length (ca. 2.6 kb) found thus far in angiosperm mitochondrial genomes are in general kept identical via frequent concerted evolution (Ogihara et al. 2005), such highly similar cox1 genes may not escape homogenization before they can sort out and be fixed in different plants. If so, then hypothesis 3 should be regarded as most likely.
Further sampling within each of the 3 groups should help in evaluating hypotheses 2 through 4. Hypotheses 2 (CCT reduction and lineage sorting) and 4 (genic, replacement HGT) predict that all members of a clade will contain a cox1 intron (of one CCT length form or the other), whereas hypothesis 3 (separate intron transfers) predicts the likely existence of intron- and CCT-less taxa that are sister to each subgroup with a different length CCT. Because lineage sorting is expected to occur relatively rapidly, hypothesis 2 is most likely in cases where the 2 intron types trace back to the base of a group (e.g., Ixora sp1 and sp2), whereas hypothesis 4 is more likely where 1 intron type is deeply nested within a paraphyletic grade of the other type (e.g., Marantaceae). Hypotheses 3 and 4, unlike number 2, predict the existence of intron-containing, donor plants related to but outside the group in question, a not unlikely scenario considering the findings described in the following sections.
On balance, although hypotheses 2 and 4 are certainly quite viable, hypothesis 3 seems to be the most likely explanation for these patterns. This is for the reasons advanced 2 paragraphs above, as well as the fact that the major prediction made by this hypothesis—that increased taxon sampling will lead to an ever finer interspersion of intron-containing and intron- and CCT-lacking taxa—fits with what has been seen thus far (Cho et al. 1998; Cho and Palmer 1999; this study). Accordingly, we have based figures 1 and 3 on hypothesis 3 for the 3 cases discussed in this section.
Families with Multiple Gains of the cox1 Intron
Of the 70 intron transfers inferred under hypothesis 3, fully 37 are restricted to just 9 of 212 families of angiosperms examined (these families are marked on fig. 5). Moreover, most of these transfers appear to result from phylogenetically local, within-family transfers. Because this last conclusion has important mechanistic implications and because interpretation of intron history is complicated for some of these families, the following subsections plus figure 3 provide synopses for 4 of the most complex families in which multiple, within-family intron acquisitions are inferred. Synopses for the other 5 families for which multiple transfers are inferred are provided in supplementary text (Supplementary Material online).
Rhamnaceae
The cox1 intron is present in 11 disparate members of the 17 Rhamnaceae examined (figs. 1 and 3A). We infer fully 6 separate transfers in the family, all but the first occurring by intrafamilial transfer, based on the intron's patchy distribution within Rhamnaceae and on conflict between intron and organismal phylogenies (fig. 3A). Overall evidence for this conflict comes from the AU test, which rejected (P < 0.01) the topology in which all 11 Rhamnaceae introns were constrained according to the hypothesis of strictly vertical transmission. In particular, the intron phylogeny provides strong support (85% BS) for a clade comprising the phylogenetically disparate lineages Maesopsis, Bathiorhamnus, and Nesiota + Phylica (fig. 3A). The existence of this clade raises the possibility of multiple horizontal transfers between members of this family. The strongly supported (100% BS) clustering of the Rhamnella intron with those of Rhamnus and Frangula suggests another HGT event between members of the family under the assumption that the sole intervening, intron-lacking clade (Condalia + Karwinskia) never possessed the intron. This assumption seems safe because the homing endonuclease ORF is intact and therefore probably still functional in this family, which would make intron loss extremely unlikely (see below). Additional within-family transfers are suggested by the existence of the otherwise very rare, CCT = 0 condition in 5 of the 6 putative intron-acquisition clades within the family (the poor resolution of all these Rhamnaceae taxa in the intron phylogeny of fig. 5 is not inconsistent with this hypothesis).
Apocynaceae
We provisionally infer 6 separate intron acquisitions, all but the first from Apocynaceae-like donors, to account for the intron's presence in 7 of 8 Apocynaceae examined (figs. 1 and 3B). We do so for 2 sets of reasons despite the overall phylogenetic clustering of these 7 introns (fig. 5). First, Carissa, the only examined member of the family that lacks the intron, occupies a relatively derived phylogenetic position within the family and also lacks the CCT (fig. 3B). Carissa also groups with the intron-containing Apocynaceae in cox1 exon phylogeny (data not shown); that is, there is no evidence that Carissa either acquired its cox1 gene by HGT or lost its cox1 intron and CCT by HGT. Second, intron and organismal phylogenies are in conflict for the family: a tree in which all 7 Apocynaceae introns were constrained according to the organismal phylogeny was rejected by the AU test (P < 0.05). Most notably, the intron from Ochrosia branches with good support (84% BS) at the base of an overall well-supported intron clade that contains all other Apocynaceae introns (fig. 3B). Conversely, the intron from the early-diverging Alstonia branches instead with Neisosperma, with 69% BS (fig. 3B).
Rubiaceae
Another within-family transfer, in the Rubiaceae, is suggested by a strongly supported (89% BS) clade of 5 introns, all with CCT = 0, comprising Ixora sp1 and a clade of 4 taxa that is separated from Ixora sp1 by multiple intron-lacking clades (figs. 2 and 3C). In addition, although poorly resolved in the intron phylogeny (fig. 3C), the intron from Ixora sp2 shares a 5-nt deletion with these other 5 Rubiaceae introns. As discussed at length above, their different CCT lengths suggest that Ixora sp1 (CCT = 0) and Ixora sp2 (CCT = 6) may have acquired their introns independently. Based on current sampling, one plausible scenario for the Rubiaceae is that the intron was first introduced (probably from an Apocynaceae donor) into the Ixora sp2 lineage, with CCT = 6, and then twice transferred within the family, to the 2 groups with CCT = 0. Because reductions in CCT length are relatively rare (see fig. 5 and below), it is likely that only the first of these intrafamilial transfers occurred from an Ixora sp2 donor, in an event accompanied by an unusually short 3′ coconversion event such that the characteristic CCT was effectively lost and that the second transfer occurred between the 2 CCT = 0 clades of Rubiaceae. However, as discussed above, alternative hypotheses can by no means be ruled out.
Plantaginaceae
Most clades within the family contain an intron with the same length CCT and whose relationships, with one important exception, are congruent with the organismal phylogeny (fig. 3D). However, 3 clades of intronless taxa are interspersed with intron-containing clades of the family. For 2 of the 3 intronless clades, we invoke intron loss rather than multiple gains to explain the distribution of the intron. Both cases involve members of the large and well-sampled genus Plantago. All 5 species sampled from the clade containing Plantago bellardii through Plantago sericea (all Plantago subgenus Psyllium) lack the intron but contain the same diagnostic CCT as intron-containing members of subgenus Psyllium (figs. 2 and 3D). Consistent with the exceptionally high mutation rates in Plantago (Cho et al. 2004), this is the only group for which we infer base substitutional divergence of the CCT following intron acquisition (fig. 2). A separate loss is inferred in Plantago subgenus Plantago, all of whose members lack the intron. This inference is weaker because only 1 of the 3 CCT differences inferred at the base of the subgenus is present (fig. 2). However, note that P. subgenus Plantago contains the very highest mutation rates (e.g., see fig. 2), with extensive sequence divergence accumulated at the base of the subgenus (Cho et al. 2004). This greatly increases the plausibility of substitutional reversion at 2 of the CCT sites. Hippuris, the third intronless member of the family, entirely lacks the CCT. For this reason, because Hippuris cox1 is not notably divergent (fig. 4), and because of the conflicted placement (fig. 3D) of the cox1 intron from its relative Sibthorpia, we infer that Hippuris never possessed the intron. If this inference is correct (see supplementary text, Supplementary Material online, for alternative explanations deemed less likely), then the intron must have been acquired 3 times within the family (fig. 3D), with 2 of the donors probably being other members of the family (fig. 5).
Overall Estimate of Intron Transfers and CCT Reductions
Based on all the above considerations, plus those presented in supplementary text (Supplementary Material online), we estimate 70 horizontal transfers of the cox1 intron to account for its presence in 162 diverse taxa among the 640 angiosperms sampled. The increased sampling of cox1 genes in this study has more than tripled the number of previously sequenced cox1 introns in angiosperms (from 37 to 129) and has doubled the number of inferred gains of the intron by horizontal transfer (from 35 to 70). Considering that the vast majority of the >250,000 species and >13,500 genera of angiosperms have yet to be examined, we are confident that many more horizontal transfers of this remarkable intron await discovery. Indeed, Barkman et al. (2007) recently reported 23 new cases of the cox1 intron from a diverse set of mostly parasitic angiosperms. They concluded that their estimate of cox1 intron phylogeny is “highly discordant with angiosperm phylogeny as indicated by a significant S–H test” and stated that their results “corroborate earlier findings (Cho et al. 1998) of widespread HGT of this mobile sequence.” Based on the distribution of these 23 new introns across angiosperm phylogeny, we estimate at least 10 additional intron transfers among flowering plants, bringing the total estimate of intron gains/transfers to 80.
We may have overestimated the number of transfers in some groups (e.g., Apocynaceae, see above) or underestimated in others (e.g., Meliaceae, where we provisionally infer a single common transfer in Melia and Dysoxylum despite their weakly supported separation in the cox1 intron phylogeny and the strong grouping of Dysoxylum with Bursera, a member of a different family). In both situations, increased taxon sampling is the key to gaining a better understanding of the intron's evolutionary history. For example, our interpretation for the Apocynaceae predicts the likely existence of intron- and CCT-lacking Apocynaceae clades sister to each of the 6 examined clades inferred to have separately acquired the intron. The predicted pattern is similar to that observed in the Rhamnaceae, for which 6 separate, mostly intrafamilial transfers were also inferred, but with greater confidence (figs. 1 and 3). With more than 400 genera and 4,500 species, there is plenty of opportunity for further sampling and illumination within the Apocynaceae.
Absent substantially more comprehensive taxon sampling and incorporation of divergence time estimates, we are unable to estimate with confidence the direction and absolute timing of cox1 intron transfers. However, with respect to taxonomic rank, all cox1 intron transfers are relatively recent events. Within the limits of current sampling, none of the transfers is shared by even sister families, and most are restricted to a single subfamilial clade. In many cases, the intron is present in the only examined member of one genus among 2 or more genera sampled in a given family, and thus, some or many of these transfers may have occurred intragenerically. These cases present potentially fruitful material for more comprehensive follow-up sampling to identify especially recent cases of intron gain.
We conservatively estimate that at least 8 of the 70 inferred transfers were accompanied by reductions in length of the 3′ CCT (fig. 5). Length reduction probably occurred either by an unusually short intron invasion event with minimal 3′ exon degradation and repair or by partial 3′ exon conversion with a native, intronless cox1 allele (with this conversion erasing part or all the CCT with a 3′–5′ polarity). Conversely, therefore, most of the transfers must have involved CCTs at least as long, and probably longer, than the 18-nt region that encompasses the 6 CCT differences. How much longer and whether coconversion also occurs 5′ to the intron is difficult to know given the high level of conservation of the cox1 coding region. Careful scrutiny of the cox1 alignment among closely related introns from disparately related plants failed to reveal convincing evidence for coconversion outside of the 18-nt 3′ tract (fig. 2). This scrutiny did, however, reveal intriguing but complex patterns of variation at the RNA editing site located just 2 nt downstream of the CCT at position 20 (fig. 2). The complexities of these patterns, which we hypothesize reflect selection for efficient editing at this position in ways that affect the CCT, are discussed in supplementary text (Supplementary Material online).
Horizontal Intron Transfer Is Biased toward Phylogenetically Local Transfers
Even with 300 angiosperms examined, the density of taxon sampling in Cho et al. (1998) was insufficient to reveal any clear phylogenetic patterning with respect to intron transfer. Increased sampling of the Araceae (Cho and Palmer 1999) first suggested that the cox1 intron might tend to transfer preferentially between closely related plants. The much greater sampling in this study greatly strengthens this conclusion. Fully 37 of the 70 acquisitions of the cox1 intron inferred under hypothesis 3 are restricted to just 9 of the 212 angiosperm families examined. Of these 37 inferred transfers, 22 are likely to have occurred from one family member to another (although none of the 7 transfers inferred for the Euphorbiaceae and Boraginaceae appear to be within-family events [see supplementary text, Supplementary Material online], all 30 transfers involving the other 7 families (see above and fig. 5) appear to be within-family events with the exception of the initial transfer into each family, plus the transfer differentiating the Siphonochilus intron from the rest of the Zingiberaceae introns). Furthermore, our results also suggest a tendency for intron transfer to occur within orders of angiosperms. Note, for instance, the phylogenetic grouping of introns inferred to represent at least 4 different transfers within the Lamiales, the grouping of all introns belonging to 3 families within the Gentianales, and the grouping of introns from 3 disparate clades of the Ericales (figs. 1 and 5).
Ironically, the extraordinary promiscuity of the cox1 intron also limits our ability to infer with complete confidence the donor group for any particular transfer. Even if unequivocal phylogenetic evidence was obtained nesting a recipient intron clade “C” within a thereby-paraphyletic clade (A) of putative donor taxa showing vertical transmission, we could never rule out that some unsampled plant lineage (B), be it extant or extinct, served as a phylogenetic intermediate in the apparent transfer from A to C, that is, that A actually transferred the intron to B, which then gave it to C. With this general caveat in mind, the patterns found in this study are nonetheless most straightforwardly interpreted as reflecting a tendency of the cox1 intron to make phylogenetically local jumps, within orders, families, and, in the case of Ixora, possibly genera.
To the extent that intron transfer is driven by phylogeny, stochastic founder effects will come into play. The earlier in its evolution that the intron is introduced into a particular family, the greater the opportunity for it to spread widely within the family, both because there is more time for transfer to occur and because there will be a relatively large clade of potential familial donors. All things being equal, this stochastic factor should lead to certain families (orders, etc.) possessing relatively many and deeply arising intron clades, whereas others will contain relatively few and shallow intron clades. Greatly increased sampling of large families that already show evidence of either the former pattern (e.g., Apocynaceae, Rhamnaceae, and Rubiaceae) or the latter (e.g., Asteraceae, Fabaceae, Poaceae, and Solanaceae) should therefore be illuminating.
Forces Favoring Phylogenetically Local Intron Transfer
Any bias toward phylogenetically local transfer probably involves multiple forces acting on the introduction and, possibly, fixation of the intron. At the level of intron introduction, common ancestry could in principle cause related plants to share a similar set of potential vectoring agents (e.g., pollinators, sapsuckers, pathogens, and mycorrhizal fungi), occupy similar habitats, or be more likely to take up foreign mitochondrial DNA via mitochondrial fusion (Arimura et al. 2004; Sheahan et al. 2005) or transformation (Koulintchenko et al. 2003). It could also make them more likely to engage in “illegitimate pollination,” that is, the rare (but potentially common on an evolutionary timescale) breach of the normal forces that prevent germination and growth of pollen from other species. Keep in mind that although angiosperm sperm cells usually lack mitochondria, the pollen tube (cell) itself is loaded with mitochondria and may occasionally contribute one or more of them to the egg cell (Yu and Russell 1994; Svab and Maliga 2007). Given the invasive properties of the homing cox1 intron and the propensity of plant mitochondria to fuse (Arimura et al. 2004; Sheahan et al. 2005), it is easy to imagine the occasional successful introduction of the intron via illegitimate pollination (Vervaeke et al. 2001; Faivre 2002). If direct evidence is obtained for a role of illegitimate pollination in plant mitochondrial HGT, this would make it a second important demonstrated mechanism for the delivery of DNA from one plant to another, the other involving direct contact between parasitic plants and their host plants (Davis and Wurdack 2004; Mower et al. 2004; Nickrent et al. 2004; Davis et al. 2005; Barkman et al. 2007).
Forces disfavoring fixation of introns acquired from distantly related plants could in principle involve molecular incompatibilities, for example, at the level of COX1 protein function or editing of cox1 RNA, resulting from the introduction of foreign cox1 coding sequences via exonic coconversion. However, considering the high level of conservation of the cox1 gene among angiosperms, and especially the concomitant difficulty in detecting any evidence of coconversion and chimerism apart from the 18-nt 3′ CCT, it seems unlikely that such incompatibilities have played an important role in the intron's evolutionary history.
Intron Loss and Intronic ORF Endonuclease and Maturase Function
This study reports the first 2 cases, both in Plantago (fig. 5), in which loss of the angiosperm cox1 intron can be inferred with confidence. In both cases, the intron-lacking taxa retained part or all the CCT and were nested well within a clade that otherwise showed vertical transmission of the intron. In addition, there are a few other cases, discussed above (e.g., Hippuris, Carissa, and the Condalia/Karwinskia clade), for which loss is not implausible, although deemed less likely than other explanations (either because there is no sign of a CCT in these intronless genes and/or because the introns in related taxa probably encode active homing endonucleases; see below). Group II intron loss has been reported to occur occasionally for the 22 mitochondrial group II introns that all predate the origin of angiosperms (Malek and Knoop 1998; Chaw et al. 2008). In general, intron loss is thought to occur via retroprocessing, the recombination of a reverse-transcribed mRNA with its encoding gene. There is good evidence for this mechanism—in the form of phylogenetically coincident loss of RNA editing at positions flanking the site of intron loss—in some (Geiss et al. 1994; Lu et al. 1998; Itchoda et al. 2002; Lopez et al. 2007) but not all (Kudla et al. 2002) cases of plant mitochondrial intron loss. For one case of cox1 intron loss in Plantago, intron loss is indeed phylogenetically coincident with loss of RNA editing, indicating a retroprocessing event that affected much of the gene, whereas in the other case, editing is unaffected (data not shown). The editing status of crucial intron-lacking genes, for example, from Hippuris, Carissa, and Condalia/Karwinskia, is consistent with the hypothesis advanced above that they never contained the intron.
A prerequisite for loss of homing group I introns is the loss of the homing ability itself; otherwise the intron would be expected to swiftly reoccupy alleles that occasionally lose the intron via retroprocessing and thus the intronless alleles would never go to fixation. Consistent with this hypothesis, all sequenced intronic ORFs from Plantago have frameshift mutations that almost certainly render the ORF nonfunctional. Several other intronic ORFs, scattered across the cox1 intron phylogeny (crosses in fig. 5), are also predicted to be pseudogenes. Two other intronic ORFs are out of frame with respect to cox1 exon 1 (marked by filled ovals in fig. 5), as are most pseudogenized ORFs. These 2 ORFs may also be nonfunctional because in fungi this intron's ORF is cotranscribed and cotranslated with exon 1 and then proteolytically processed (Lambowitz et al. 1999; Caprara and Waring 2005). Because the angiosperm endonuclease ORF has repeatedly become an obvious pseudogene, it is highly unlikely that it also serves as a splicing maturase, as is the case for certain homing endonucleases (Caprara and Waring 2005). No experimental work has been published in flowering plants on the ORF located in the cox1 group I intron; thus, whether it acts as an endonuclease, maturase, both, or neither, remains a matter of speculation.
Loss of homing endonuclease function is expected under the cyclical model of homing intron gain–loss (Goddard and Burt 1999; Haugen et al. 2005; Gogarten and Hilario 2006) for those homing introns whose ORFs function solely as homing endonucleases, as is evidently the case for the angiosperm cox1 intron. Once these introns go to fixation in a population, selection on the ORFs should be relaxed, and they should decay as pseudogenes. The existence of intact ORFs with both LAGLIDADG motifs conserved in most angiosperm cox1 introns (fig. 5) probably reflects a combination of generally very low substitution rates in plant mitochondria and, in most cases, the recent arrival of the intron. Most intron ORFs may not have had sufficient time to accumulate even a single obvious pseudogenization mutation, and many probably still encode active homing endonucleases. This would make loss of this particular intron exceptionally difficult because any cox1 allele that happens to lose its intron via retroprocessing and which still encodes an active endonuclease should quickly reacquire the intron via intramitochondrial homing before it could go to fixation.
A Single Fungal Seed of the cox1 Intron in Angiosperms
When this homing group I intron was first discovered in the angiosperm Peperomia (Vaughn et al. 1995), it was shown that its closest relatives were to be found within the fungi and therefore hypothesized that it had probably been acquired from fungi by horizontal transfer. When the intron was then found to have been acquired on many different occasions within angiosperm evolution (Cho et al. 1998), the question arose as to what extent its transfer history involved multiple fungal-to-plant transfers versus mostly (potentially even all but one) plant-to-plant transfers. Subsequent evidence leads us to now strongly favor the extreme model according to which angiosperms acquired the intron but once from fungi, followed by numerous, exclusively angiosperm-to-angiosperm transfers. First, there is now abundant evidence (Bergthorsson et al. 2003, 2004; Davis and Wurdack 2004; Mower et al. 2004; Richardson and Palmer 2007) that mitochondrial genes are, on an evolutionary timescale, relatively frequently transferred between angiosperms, whereas there is no evidence for even a second fungal-to-plant transfer. Second, if fungal-to-plant transfer was at all common, then we would expect to find other fungal-derived group I introns in plants as many fungal mitochondrial genomes are now known to contain multiple homing group I introns (Seif et al. 2005; Lang et al. 2007).
cox1 Intron Still Stands Out
Although it is now clear that mitochondrial sequences other than the cox1 intron are horizontally transferred at some frequency during angiosperm evolution, there are good reasons to regard this intron as a very special case with an elevated rate of transfer, a depressed rate of loss, and a high likelihood of being detected. Three of these reasons are a function of the intron's endonuclease-specified homing ability. First, assuming that horizontal transfer introduces not only just the cox1 intron but also the endonuclease, the intron should have a higher probability of insertion into a recipient mitochondrial genome than any random piece of DNA. Second, once introduced into one mitochondrial genome and expressed, the intron's probability of spread and fixation throughout the population of mitochondrial genomes within the panmictic population of mitochondria in a plant cell (Arimura et al. 2004; Sheahan et al. 2005) should be higher, again owing to its invasive homing properties. Third, as described in the section above on intron loss, any cox1 intron carrying a still functional endonuclease gene should be much more refractory to loss than other angiosperm mitochondrial introns, none of which are homing introns.
Other factors relate to the cox1 intron's generic properties as an intron. As previously noted (Cho et al. 1998), introns as a class are essentially immune to normal forces of genomic deletion (as opposed to the cDNA-mediated force of retroprocessing), and subsequent analyses have shown that introns are indeed lost much less readily during angiosperm mitochondrial evolution than is spacer DNA (Adams et al. 2000). In addition, because the cox1 intron is a relatively large and readily scored segment of foreign DNA—inserted in the middle of an essential, invariantly mitochondrial, and frequently sequenced gene—its probability of detection is higher than for most pieces of acquired DNA. For all these reasons, the cox1 intron stands out as the most frequently acquired, least often lost, and overall most notable segment of horizontally transferred DNA in angiosperm mitochondrial evolution.
Although it is currently impossible to assign a meaningful rate of cox1 intron transfer (or loss), one further consideration highlights its extraordinary propensity to be horizontally transferred during angiosperm evolution. A total of 40 protein genes and 22 group II introns were present in the ancestral angiosperm mitochondrial genome, and with a few exceptions, most of these are present throughout most if not all the hundreds of diverse angiosperms examined to date (Adams et al. 2002). In contrast, the cox1 intron was initially introduced into angiosperms relatively late in their evolution, and despite frequent subsequent horizontal spread, it is nonetheless present in only a minority of “extant” angiosperms. Most genes and introns were thus present in donor genomes and therefore potentially transferable in virtually all HGT events across the history of angiosperms, whereas the cox1 intron has been much more rarely present in donor genomes and, in some unknown fraction of these cases, its homing ORF is defunct. That despite this, the cox1 intron has been so often transferred further emphasizes its extraordinary mobility. Over time, as the intron continues to spread horizontally—and in an autocatalytic manner (the more it spreads, the more potential donors there are)—a state of near saturation should eventually be reached, balanced only by occasional loss of the intron in lineages with a defunct homing endonuclease. Overall, then, this intron behaves as a truly remarkably invasive and persistent genetic element in angiosperm mitochondria, an element that should be regarded as the “canary in the coal mine” with respect to mitochondrial HGT in angiosperms.
Supplementary Material
Supplementary text, table S1, and figures S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Han Ong for carrying out PCR assays for cox1 intron presence in 29 Poales, Yin-Long Qiu and John Kress for providing DNAs, Julie Gummow for technical contributions, and Greg Young for comments on RNA editing. This work was supported by National Institutes of Health (NIH) grant 1F32GM080079 to A.J.A. and by NIH research grant RO1-GM-70612 and the METACyt Initiative of Indiana University, funded in part through a major grant from the Lilly Endowment, Inc. to J.D.P.
References
- Adams KL, Clements MJ, Vaughn JC. The Peperomia mitochondrial cox1 group I intron: timing of horizontal transfer and subsequent evolution of the intron. J Mol Evol. 1998;46:689–696. doi: 10.1007/pl00006349. [DOI] [PubMed] [Google Scholar]
- Adams KL, Daley DO, Qiu Y-L, Whelan J, Palmer JD. Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature. 2000;408:354–357. doi: 10.1038/35042567. [DOI] [PubMed] [Google Scholar]
- Adams KL, Qiu Y-L, Stoutemyer M, Palmer JD. Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci USA. 2002;99:9905–9912. doi: 10.1073/pnas.042694899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura S, Yamamoto J, Aida G, Nakazono M, Tsutsumi N. Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc Natl Acad Sci USA. 2004;101:7805–7808. doi: 10.1073/pnas.0401077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkman TJ, McNeal JR, Lim SH, Coat G, Croom HB, Young ND, Depamphilis CW. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evol Biol. 2007;7:248. doi: 10.1186/1471-2148-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U, Adams KL, Thomason B, Palmer JD. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature. 2003;424:197–201. doi: 10.1038/nature01743. [DOI] [PubMed] [Google Scholar]
- Bergthorsson U, Richardson A, Young G, Goertzen L, Palmer JD. Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella. Proc Natl Acad Sci USA. 2004;101:17747–17752. doi: 10.1073/pnas.0408336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer B, Bremer K, Chase MW, et al. (24 co-authors) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc. 2003;141:399–436. [Google Scholar]
- Burt A, Trivers R. Genes in conflict. The biology of selfish genetic elements. Cambridge (MA): The Belknap Press of Harvard University Press; 2006. [Google Scholar]
- Caprara M, Waring R. Group I introns and their maturases: uninvited, but welcome guests. In: Belfort M, Stoddard B, Wood D, Derbyshire V, editors. Homing endonucleases and inteins. Heidelberg (Germany): Springer; 2005. [Google Scholar]
- Cech TR. Self-splicing of group-I introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- Chaw SM, Chun-Chieh Shih A, Wu YW, Liu SM, Chou TY, Wang D. The mitochondrial genome of the gymnosperm Cycas taitungensis contains a novel family of short interspersed elements, Bpu sequences, and abundant RNA editing sites. Mol Biol Evol. Forthcoming 2008;25:603–615. doi: 10.1093/molbev/msn009. [DOI] [PubMed] [Google Scholar]
- Cho Y, Mower J, Qiu Y-L, Palmer JD. Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. Proc Natl Acad Sci USA. 2004;101:17741–17746. doi: 10.1073/pnas.0408302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Qiu YL, Kuhlman P, Palmer JD. Explosive invasion of plant mitochondria by a group I intron. Proc Natl Acad Sci USA. 1998;95:14244–14249. doi: 10.1073/pnas.95.24.14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YR, Palmer JD. Multiple acquisitions via horizontal transfer of a group I intron in the mitochondrial cox1 gene during evolution of the Araceae family. Mol Biol Evol. 1999;16:1155–1165. doi: 10.1093/oxfordjournals.molbev.a026206. [DOI] [PubMed] [Google Scholar]
- Cusimano N, Zhang LB, Renner SS. Reevaluation of the cox1 group I intron in Araceae and angiosperms indicates a history dominated by loss rather than horizontal transfer. Mol Biol Evol. 2008;25:265–276. doi: 10.1093/molbev/msm241. [DOI] [PubMed] [Google Scholar]
- Davis C, Andersen W, Wurdack K. Gene transfer from a parasitic flowering plant to a fern. Proc R Soc Lond B Biol Sci. 2005;272:2237–2242. doi: 10.1098/rspb.2005.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CC, Wurdack KJ. Host-to-parasite gene transfer in flowering plants: phylogenetic evidence from Malpighiales. Science. 2004;305:676–678. doi: 10.1126/science.1100671. [DOI] [PubMed] [Google Scholar]
- Diao X, Freeling M, Lisch D. Horizontal transfer of a plant transposon. PLoS Biology. 2006;4:e5. doi: 10.1371/journal.pbio.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid isolation procedure for small quantities of fresh leaf tissues. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Faivre AE. Variation in pollen tube inhibition sites within and among three heterostylous species of Rubiaceae. Int J Plant Sci. 2002;163:783–794. [Google Scholar]
- Geiss K, Abbas G, Makaroff C. Intron loss from the NADH dehydrogenase subunit 4 gene of lettuce mitochondrial DNA: evidence for homologous recombination of a cDNA intermediate. Mol Gen Genet. 1994;243:97–105. doi: 10.1007/BF00283881. [DOI] [PubMed] [Google Scholar]
- Ghatnekar L, Jaarola M, Bengtsson BO. The introgression of a functional nuclear gene from Poa to Festuca ovina. Proc R Soc Lond B Biol Sci. 2006;273:395–399. doi: 10.1098/rspb.2005.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard MR, Burt A. Recurrent invasion and extinction of a selfish gene. Proc Natl Acad Sci USA. 1999;96:13880–13885. doi: 10.1073/pnas.96.24.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten JP, Hilario E. Inteins, introns, and homing endonucleases: recent revelations about the life cycle of parasitic genetic elements. BMC Evol Biol. 2006;6:94. doi: 10.1186/1471-2148-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugerli F, Sperisen C, Buchler U, Brunner I, Brodbeck S, Palmer JD, Qiu YL. The evolutionary split of Pinaceae from other conifers: evidence from an intron loss and a multigene phylogeny. Mol Phylogenet Evol. 2001;21:167–175. doi: 10.1006/mpev.2001.1004. [DOI] [PubMed] [Google Scholar]
- Haugen P, Simon D, Bhattacharya D. The natural history of group I introns. Trends Genet. 2005;21:111–119. doi: 10.1016/j.tig.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Itchoda N, Nishizawa S, Nagano H, Kubo T, Mikami T. The sugar beet mitochondrial nad4 gene: an intron loss and its phylogenetic implication in the Caryophyllales. Theor Appl Genet. 2002;104:209–213. doi: 10.1007/s001220100744. [DOI] [PubMed] [Google Scholar]
- Jansen RK, Cai Z, Raubeson LA, et al. (13 co-authors) Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly S, Brouillet L, Bruneau A. Phylogenetic implications of the multiple losses of the mitochondrial coxII.i3 intron in the angiosperms. Int J Plant Sci. 2001;162:359–373. [Google Scholar]
- Jurica M, Stoddard B. Homing endonucleases: structure, function and evolution. Cell Mol Life Sci. 1999;55:1304–1326. doi: 10.1007/s000180050372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulintchenko M, Konstantinov Y, Dietrich A. Plant mitochondria actively import DNA via the permeability transition pore complex. Embo J. 2003;22:1245–1254. doi: 10.1093/emboj/cdg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Albertazzi F, Blazevic D, Hermann M. Loss of the mitochondrial cox2 intron 1 in a family of monocotyledonous plants and utilization of mitochondrial intron sequences for the construction of a nuclear intron. Mol Genet Genomics. 2002;267:223–230. doi: 10.1007/s00438-002-0657-6. [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Belfort M. Introns as mobile genetic elements. Annu Rev Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Caprara M, Zimmerly S, Perlman P. Group I and group II ribozymes as RNPs: clues to the past and guides to the future. In: Gesteland R, Cech TR, Atkins J, editors. The RNA world. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1999. pp. 451–485. [Google Scholar]
- Lang BF, Laforest MJ, Burger G. Mitochondrial introns: a critical view. Trends Genet. 2007;23:119–125. doi: 10.1016/j.tig.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Lopez L, Picardi E, Quagtiariello C. RNA editing has been lost in the mitochondrial cox3 and rps13 mRNAs in Asparagales. Biochimie. 2007;89:159–167. doi: 10.1016/j.biochi.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Lu M, Szmidt A, Wang X. RNA editing in gymnosperms and its impact on the evolution of the mitochondrial cox1 gene. Plant Mol Biol. 1998;37:225–234. doi: 10.1023/a:1005972513322. [DOI] [PubMed] [Google Scholar]
- Maddison W. A method for testing the correlated evolution of two binary characters: are gains or losses concentrated on certain branches of a phylogenetic tree? Evolution. 1990;44:539–557. doi: 10.1111/j.1558-5646.1990.tb05937.x. [DOI] [PubMed] [Google Scholar]
- Maddison W, Maddison P. Sunderland (MA): Sinauer Associates; 2000. MacClade version 4: analysis of phylogeny and character evolution. [Google Scholar]
- Malek O, Knoop V. Trans-splicing group II introns in plant mitochondria: the complete set of cis-arranged homologs in ferns, fern allies, and a hornwort. RNA. 1998;4:1599–1609. doi: 10.1017/s1355838298981262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Bell CD, Soltis PS, Soltis DE. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci USA. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower J, Stefanovic S, Young GJ, Palmer JD. Plant genetics: gene transfer from parasitic to host plants. Nature. 2004;432:165–166. doi: 10.1038/432165b. [DOI] [PubMed] [Google Scholar]
- Mueller J, Smith D, Belfort M. Exon coconversion biases accompanying intron homing: battle of the nucleases. Genes Dev. 1996;10:2158–2166. doi: 10.1101/gad.10.17.2158. [DOI] [PubMed] [Google Scholar]
- Nickrent DL, Blarer A, Qiu YL, Vidal-Russell R, Anderson FE. Phylogenetic inference in Rafflesiales: the influence of rate heterogeneity and horizontal gene transfer. BMC Evol Biol. 2004;4:40. doi: 10.1186/1471-2148-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara Y, Yamazaki Y, Murai K, et al. (11 co-authors) Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res. 2005;33:6235–6250. doi: 10.1093/nar/gki925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD, Herbon LA. Plant mitochondrial-DNA evolves rapidly in structure, but slowly in sequence. J Mol Evol. 1988;28:87–97. doi: 10.1007/BF02143500. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall K. ModelTest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Qiu YL, Dombrovska O, Lee J, et al. (17 co-authors) Phylogenetic analyses of basal angiosperms based on nine plastid, mitochondrial, and nuclear genes. Int J Plant Sci. 2005;166:815–842. [Google Scholar]
- Qiu YL, Li L, Hendry T, Li R, Taylor D, Issa M, Ronen A, Vekaria M, White A. Reconstructing the basal angiosperm phylogeny: evaluating information content of mitochondrial genes. Taxon. 2006;55:837–856. [Google Scholar]
- Qiu YL, Li L, Wang B, et al. (18 co-authors) The deepest divergences in land plants inferred from phylogenomic evidence. Proc Nat Acad Sci USA. 2006;103:15511–15516. doi: 10.1073/pnas.0603335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice D, Palmer JD. An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol. 2006;4:31. doi: 10.1186/1741-7007-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AO, Palmer JD. Horizontal gene transfer in plants. J Exp Bot. 2007;58:1–9. doi: 10.1093/jxb/erl148. [DOI] [PubMed] [Google Scholar]
- Robertson H. Evolution of DNA transposons in eukaryotes. In: Craig N, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. Washington (DC): ASM Press; 2002. pp. 1093–1110. [Google Scholar]
- Seif E, Leigh J, Liu Y, Roewer I, Forget L, Lang BF. Comparative mitochondrial genomics in zygomycetes: bacteria-like RNase P RNAs, mobile elements and a close source of the group I intron invasion in angiosperms. Nucleic Acids Res. 2005;33:734–744. doi: 10.1093/nar/gki199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan MB, McCurdy DW, Rose RJ. Mitochondria as a connected population: ensuring continuity of the mitochondrial genome during plant cell dedifferentiation through massive mitochondrial fusion. Plant J. 2005;44:744–755. doi: 10.1111/j.1365-313X.2005.02561.x. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Endress P, Chase MW. Phylogeny and evolution of angiosperms. Sunderland (MA): Sinauer Associates, Inc; 2005. [Google Scholar]
- Svab Z, Maliga P. Exceptional transmission of plastids and mitochondria from the transplastomic pollen parent and its impact on transgene containment. Proc Natl Acad Sci USA. 2007;104:7003–7008. doi: 10.1073/pnas.0700063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D, Olsen G, Waddell P, Hillis D. PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland (MA): Sinauer Associates; 2002. [Google Scholar]
- Vaughn JC, Mason MT, Sperwhitis GL, Kuhlman P, Palmer JD. Fungal origin by horizontal transfer of a plant mitochondrial group I intron in the chimeric cox1 gene of Peperomia. J Mol Evol. 1995;41:563–572. doi: 10.1007/BF00175814. [DOI] [PubMed] [Google Scholar]
- Vervaeke I, Parton E, Maene L, Deroose R, De Proft MP. Prefertilization barriers between different Bromeliaceae. Euphytica. 2001;118:91–97. [Google Scholar]
- Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, Renner S. Horizontal gene transfer from flowering plants to Gnetum. Proc Natl Acad Sci USA. 2003;100:10824–10829. doi: 10.1073/pnas.1833775100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Russell S. Occurrence of mitochondria in the nuclei of tobacco sperm cells. Plant Cell. 1994;6:1477–1484. doi: 10.1105/tpc.6.10.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Austin (TX): University of Texas; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


