Abstract
Proteins involved in reproductive fitness have evolved unusually rapidly across diverse groups of organisms. These reproductive proteins show unusually high rates of amino acid substitutions, suggesting that the proteins have been subject to positive selection. We sought to identify seminal fluid proteins experiencing adaptive evolution because such proteins are often involved in sperm competition, host immunity to pathogens, and manipulation of female reproductive physiology and behavior. We performed an evolutionary screen of the mouse prostate transcriptome for genes with elevated evolutionary rates between mouse and rat. We observed that secreted rodent prostate proteins evolve approximately twice as fast as nonsecreted proteins, remarkably similar to findings in the primate prostate and in the Drosophila male accessory gland. Our screen led us to identify and characterize a group of seminal vesicle secretion (Svs) proteins and to show that the gene Svs7 is evolving very rapidly, with many amino acid sites under positive selection. Another gene in this group, Svs5, showed evidence of branch-specific selection in the rat. We also found that Svs7 is under selection in primates and, by using three-dimensional models, demonstrated that the same regions have been under selection in both groups. Svs7 has been identified as mouse caltrin, a protein involved in sperm capacitation, the process responsible for the timing of changes in sperm activity and behavior, following ejaculation. We propose that the most likely explanation of the adaptive evolution of Svs7 that we have observed in rodents and primates stems from an important function in sperm competition.
Keywords: evolutionary screen, positive selection, adaptive evolution, seminal vesicle proteins, prostate gene expression
Introduction
Rapid changes in a protein's amino acid sequence over evolutionary time have often been attributed to the effects of positive selection, characterized at the molecular level by an excess of nonsynonymous nucleotide substitutions compared with synonymous ones (Nei and Gojobori 1986; Hughes and Nei 1988; Yang and Bielawski 2000; Nielsen 2005; Jensen et al. 2007). Repeated selection for amino acid substitutions is thought to be a form of adaptive evolution, resulting in an increase in the fitness of the organism. Proteins involved in reproductive fitness have been shown to have evolved unusually rapidly across diverse groups of organisms (Swanson and Vacquier 2002; Emes et al. 2003; Bustamante et al. 2005; Clark et al. 2006). In the case of reproductive proteins, coevolutionary cycles involving adaptation and counter adaptation are expected to apply continuous selective pressure, resulting in rapid changes at amino acid sites involved in the function of the protein. These proteins often have roles in sperm competition, host immunity to pathogens, and manipulation of female reproductive physiology and behavior; however, in many other cases, the function of the rapidly evolving protein is unknown.
We sought to determine the extent of positive selection in rodent seminal fluid proteins and to identify new proteins subject to positive selection. Evolutionary screens, that is, “scanning” for elevated dN/dS between genes in a designated group, or in the whole genome, are a powerful way to identify, in a single experiment, many candidate genes undergoing positive selection. Because there is no a priori requirement to know the function of the protein before determining that it has experienced positive selection, those with either unknown or poorly understood functions can be identified for further study and, as a consequence, our knowledge about proteins with important effects on reproductive fitness should grow more rapidly.
We performed an evolutionary screen of rodent proteins and compared the results with similar studies in diverse taxonomic groups including Drosophila and primates (Clark and Swanson 2005; Dean et al. 2007; Ramm et al. 2007). Although most studies focus on one taxonomic group, we are interested in comparing evolutionary pressures on reproductive proteins across a wide taxonomic spectrum. For example, we find that, although several rodent and primate prostate proteins evolved rapidly, their complement of highly expressed genes are largely different. The availability of the mouse (Mus musculus) and rat (Rattus norvegicus) genomes (Waterston et al. 2002; Gibbs et al. 2004) and the mouse prostate-expressed sequence tag (EST) database (Nelson et al. 2002) presented the opportunity to conduct an evolutionary screen of murid rodent taxa. The advantage of this screen is that it provides candidate proteins for studies of rapid evolution in rodent seminal fluid proteins.
In the process of our evolutionary screen of rodents, we identified and characterized a group of candidate seminal vesicle secretion (Svs) proteins under selection and showed that one of them, Svs7, is evolving very rapidly in rodents. Consequently, we also studied Svs7 evolution in primates and found a similar high rate of evolution. We note that the regions of protein structure changing most rapidly are quite similar between rodent and primate Svs7 proteins. Svs7 has been identified as mouse caltrin (for reviews, see Lardy 1985, 2003), a protein involved in sperm capacitation, the process responsible for the timing of changes in sperm activity and behavior, following ejaculation. This is interesting in regard to the strong selection on Svs7 that we report here because Svs7 is only one member of a heterogeneous group of proteins identified by classical biochemical characterizations as having caltrin activity in various mammals (Lardy 1985, 2003). Nonetheless, our data suggest that Svs7 is involved in a competitive aspect of reproductive fitness in rodents and primates, and we present arguments that sperm competition must be considered as a likely competitive mechanism driving the rapid evolution seen in this protein.
Materials and Methods
Evolutionary Screen
Genes transcribed in the mouse prostate were retrieved from the mouse prostate expression database (mPEDB), a compilation of ESTs from several prostate samples (Nelson et al. 2002). To remove low abundance transcripts, we only included a transcript if it was observed three or more times in the compiled transcriptome. Transcript accession numbers were linked to RefSeq genes. Mouse RefSeq exons were matched to the rat genome through Blast, and top hits above a threshold e value of 1 × 10−5 were used to create mouse–rat pairwise alignments (Altschul et al. 1990).
An excess of nonsynonymous substitutions relative to synonymous substitutions, the dN/dS ratio, in a protein's amino acid coding sequence is evidence for positive selection (Jensen et al. 2007). A dN/dS ratio greater than one is indicative of positive selection, whereas a ratio less than one indicates purifying selection. A ratio of one is taken for the absence of selection (neutral evolution). When measured over the entire length of a gene, the dN/dS ratio is a conservative measure of positive selection because the averaging effect of conservation of amino acids at some sites will reduce the ratio in spite of strong positive selection at others. Thus, we consider an elevated dN/dS measured over the entire gene to be suggestive of positive selection acting on a portion of the gene even though the ratio may not reach one (Swanson et al. 2004). A pairwise dN/dS value for each coding sequence was estimated for all 2,077 alignments using CODEML of the phylogenetic analysis by maximum likelihood (PAML) package (Yang 1997, 2007). The presence of a secretion signal was inferred by SignalP (http://www.cbs.dtu.dk/services/SignalP/; Bendtsen et al. 2004) on virtually translated amino acid sequences. To assess the statistical significance of the difference in evolutionary rates between secreted and nonsecreted sets, we compared the observed difference with differences found in 10,000 random permutations.
DNA Amplification and Sequencing of Svs Genes
The coding regions of four Svs genes were sequenced in seven rodent taxa. Purified DNA from Mus musculus domesticus (Zalende/EiJ), Mus musculus musculus (CZECHII/EiJ), Mus musculus castaneus (CAST/EiJ), Mus spicilegus (PANCEVO/EiJ), Mus spretus (SPRET/EiJ), Mus caroli/EiJ, and Mus pahari/EiJ were obtained from Jackson Laboratory, and gene sequences were amplified by polymerase chain reaction (PCR) with primers designed for each coding region of interest. PCR primer sequences and conditions are available from the authors upon request. The PCR products were evaluated on 1% agarose gels and diluted 1:4, and 4 μl aliquots were sequenced using Big Dye v.3.1 (Applied Biosystems Foster City CA). The sequencing products were analyzed on an ABI 3100 DNA sequencer. The rat sequences were obtained from the 2004 Rattus genome release. Sequences for six primate taxa were obtained from their genomes using the method of Young et al. (2007).
Genome Databases
We searched the following trace archives (http://www.ncbi.nlm.nih.gov/Traces/trace.cgi?) for primate Svs7 genes, using the human Svs7 sequence as a search query: lemur: 8.4 × 106 traces, as of 4 February 2008; bush baby: 12.5 × 106 traces, as of 4 February 2008; marmoset: 28.2 × 106 traces, as of 18 January 2007; orangutan: 12.2 × 106 traces, as of 18 January 2007; tarsier: 16.8 × 106 traces, as of 15 June 2007; and tree shrews: 8.6 × 106 traces, as of 12 January 2007. A computer script to search primate genomes was kindly provided by Janet M. Young.
Data Analysis
Sequence traces were edited in the program Chromas 2.3 (http://www.technelysium.com.au). DNA sequence alignment, coding region assembly, and in silico translation were done using the DNAsis Max program (Hitachi). Positive selection was assessed in the program CODEML in the PAML 3.14 package (Yang 1997, 2007). The phylogeny of Chevret et al. (2005) was used for the mouse species for the PAML tests. The three subspecies of M. musculus were treated as an unresolved polytomy.
For each gene, three different comparisons of neutral and selection models gave similar results (M1 vs. M2, M7 vs. M8, and M8A vs. M8; Yang et al. 2000; Bielawski and Yang 2003; Swanson et al. 2003). Model M1 (neutral) allows two classes of codons, one with dN/dS over the interval (0,1) and the other with a dN/dS value of one. Model M2 (selection) is similar to M1 except that it allows an additional class of codons with a freely estimated dN/dS value. Model M7 (neutral) estimates dN/dS with a beta-distribution over the interval (0, 1), whereas model M8 (selection) adds parameters to M7 for an additional class of codons with a freely estimated dN/dS value. M8A (neutral) is a special case of M8 that fixes the additional codon class at a dN/dS value of one. To evaluate variation in selective pressure over a phylogeny, the branch model of CODEML estimated dN/dS values for each branch (Yang et al. 1998, 2005). The branch model is compared with the null hypothesis, model M0, in which all lineages have the same dN/dS value.
The PSI-Blast program on the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) Web site was used to obtain Svs7-like protein sequences from vertebrate genomes. In addition, Svs7-like protein sequences were obtained using ENSEMBL (http://www.ensembl.org/index.html). The protein sequences were aligned with those obtained experimentally in DNAsis Max. Nucleotide sequences corresponding to the Svs7-like proteins were obtained on the NCBI Web site. Conserved gene order was determined with both the BLAT program of the University of California Santa Cruz genome browser (http://genome.ucsc.edu/) and with ENSEMBL. When the gene in question was not identified with either of these programs, the DNA region between flanking genes was analyzed with the program GENESCAN (http://genes.mit.edu/GENSCAN.html) to search for peptides corresponding to those of Svs7 proteins. Three-dimensional structures of the Svs7 proteins were modeled using the PHYRE threading program (http://www.sbg.bio.ic.ac.uk/∼phyre/), and the resulting models were visualized using PYMOL (http://pymol.sourceforge.net/).
Structural Analysis of Positively Selected Sites
Positively selected sites in Svs7 were mapped onto structural models. The clustering of selected sites was measured as the mean distance in angstroms between all pairs of selected sites. Statistical tests compared the observed clustering to 100,000 random sets of surface-exposed sites with the same number of sites as the selected set. We restricted analysis to surface-exposed sites because buried core sites evolve slowly and are rarely among sites inferred under positive selection. Surface exposure per residue was estimated as percent solvent accessibility by GETAREA1.1 (Franzkiewicz and Braun 1998). Sites with 10% or less solvent accessibility were considered buried. The similarity of selected sites between rodent and primate Svs7 proteins was assessed on the rodent structural model. Rodent and primate selected sites were considered to be neighbors if their Van der Waal's surface areas were within 1 Å of each other. The observed number of neighboring residues was compared with 10,000 random sets of the same size. Analysis was restricted to surface-exposed sites.
Results
Evolutionary Screen
To identify candidate seminal fluid proteins evolving under positive selection, we screened the mouse prostate transcriptome for genes with elevated evolutionary rates between mouse and rat. We gathered 2,077 moderate-to-highly expressed transcripts from the mPEDB (http://www.pedb.org/) (Nelson et al. 2002) and aligned their full-length mouse-coding sequences with orthologous sequence from the rat genome. We then estimated the ratio of nonsynonymous to synonymous divergence (dN/dS ratio) between mouse and rat for each coding sequence using the maximum likelihood and the Nei and Gojobori (1986) methods (Yang 1997), both of which gave similar results.
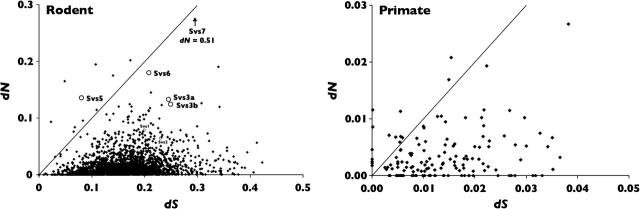
Several genes showed coding sequences with elevated dN/dS ratios (fig. 1A). From all 2,077 coding sequences, we chose to focus on the set of 437 proteins with inferred secretion signal sequences because they are likely secreted from the prostate and hence represent potential seminal fluid proteins. This requirement should greatly enrich for proteins present in seminal fluid although a few known seminal fluid proteins, such as transglutaminase 4, do not have a secretion signal (Pilch and Mann 2006). The secreted set had significantly higher dN/dS values (mean dN/dS = 0.176) compared with the nonsecreted set (mean dN/dS = 0.096; P < 0.0001). Although the elevated dN/dS ratios of secreted proteins could result from frequent positive selection, they could also result from lower selective constraint. Hence, this alone does not constitute evidence for positive selection. However, prostate-expressed genes with predicted secretion signals are a good class in which to screen for adaptive evolution because their products are more likely to be present in seminal fluid. Similarly, the secreted set had a significantly higher rate of amino acid substitution (dN) compared with the nonsecreted set (P < 0.0001), and this elevated rate cannot be attributed to a difference in nucleotide mutation rate because values of synonymous substitution were similar (secreted mean dS = 0.178; nonsecreted mean dS = 0.173; P = 0.4667). Of the secreted set, 7 proteins (1.6%) had dN/dS values exceeding 1, and 31 proteins (7.1%) exceeded 0.5. We chose to study proteins with dN/dS values in excess of 0.5 because the averaging effect of dN/dS computed over all amino acid sites may result in a value less than 1 for a protein with a portion of sites under selection. A screening threshold of 0.5 is supported by experimental evidence in which strong evidence of positive selection is revealed by a site-by-site test in proteins with overall dN/dS values that are elevated but less than 1 (Swanson et al. 2004; Clark and Swanson 2005). We wished to identify candidate proteins for studies of adaptive evolution, that is, proteins in the secreted set that were both highly expressed in the prostate and had elevated dN/dS values. The number of ESTs found for each transcript served as a rough measure of expression level; values for individual genes ranged from 3 to 4,177 ESTs. Some prominent examples of highly expressed and rapidly evolving genes are “androgen-binding protein delta” (Abpd), and two genes from a group of proteins that are named for their appearance in Svs: Svs5 and Svs7.
FIG. 1.—
Plots of dN/dS for rodent and primate seminal fluid protein genes. (A) Seminal fluid protein ESTs identified in the MPED for mouse versus rat. (B) Genes encoding seminal fluid proteins identified by mass spectrometry in human versus chimpanzee (from Clark and Swanson [2005]). The diagonal represents neutral evolution, a dN/dS ratio of one. Most genes are subject to purifying selection and fall below the diagonal, whereas several genes fall above or near the line suggesting positive selection. Comparison of the two plots shows elevated dN/dS ratios in seminal fluid genes of both taxonomic groups.
Svs Genes
We selected the rapidly evolving secreted proteins (dN/dS > 0.5) for more detailed evolutionary analysis. The prostate proteins with dN/dS > 0.5 were sorted on the representation of their ESTs (highest–lowest) in the mPEDB. Those proteins lacking signal peptides and those with transmembrane domains were removed. Some prominent examples of highly expressed and rapidly evolving genes are the Abpd, Svs7, Svs5, probasin (Pbsn), lipocalin 5 (Lcn5), and the gene encoding hypothetical protein LOC77397. One of these, ABPd (encoded by Abpd), is a member of the androgen-binding protein family, encoded by a highly duplicated, rapidly evolving family of rodent genes (Emes et al. 2004; Laukaitis et al. 2008). Among the others were Svs3 (aka Svs3a, see below), Svs5, Svs6, and Svs7, four of the seven mouse Svs proteins identified as Svs1–Svs7. In addition to being highly expressed in the prostate (this report), Svs proteins are found in the most prevalent secretion of mouse seminal fluid, that of the seminal vesicles (Luo et al. 2001). This makes them of special interest for studying evolution of the soluble proteins in semen, the environment in which sperm are passed from the male to the female during insemination.
Table 1 shows the group of mouse proteins that have been identified as Svs proteins. This is a heterogeneous group of proteins as evidenced by their putative functions and their chromosomal locations. At first glance, it would appear that Svs proteins have little in common beyond their appearance in seminal vesicle fluid; however, there is evidence that Svs2–6 are encoded by paralogous genes (Svs2–6) produced by duplication from a WFDC-type proteinase gene ancestor (Clauss et al. 2005). WFDC gene products are characterized by a whey acidic protein (WAP) four-disulfide core, called the WFDC domain. The genes Svs2–6 appear to be restricted to rodents in that they lack orthologs in the genomes of human and dog, although it has been suggested that human Semenogelins I (SgI) and II (SgII) are related to the Svs2 and Svs3 genes in the mouse (Clauss et al. 2005). The other two proteins characterized as mouse Svs are Svs1 and Svs7. The gene for Svs1 is found on mouse chromosome 6 and the gene Svs7 is found on mouse chromosome 9. Svs1 has been characterized as a copper diamine hydrolase, a flavoprotein that catalyzes the aerobic oxidation of amines to the corresponding aldehyde and ammonia (Lundwall et al. 2003), and appears to be quite different from the other Svs proteins in size and primary structure. Svs7 has been described as mouse caltrin, a protein that inhibits calcium uptake by epididymal spermatozoa by binding to calcium transporters on the membrane (Coronel et al. 1992).
Table 1.
Mouse Svs Genes
| Protein | Accession Number | Putative Function (and Reference) | Core Nucleotide Accession Number | Mus musculus Chromosomal Location | Strand | Mouse/Rat dN/dS (#N, #S) |
| Svs 1 | NP_766476 | Copper amine oxidase (Lundwall et al. 2003) | NM_172888 | chr6: 48,937,059–48,941,066 | + | 0.39 (99, 91) |
| Svs 2 | NP_059086 | Semen-coagulating protein (Kawano and Yoshida 2007) | NM_017390 | chr2: 164,062,597–164,063,979 | + | 0.28 (24, 28) |
| Svs3a | NP_067338 | Copulatory plug formation (Lin et al. 2002) | NM_021363 | chr2: 164,115,025–164,116,041 | + | 0.55 (77, 53) |
| Svs 3b | NP_775553 | Copulatory plug formation (Lin et al. 2002) | NM_173377 | chr2: 164,081,341–164,082,357 | + | 0.53 (74, 54) |
| Svs 4 | NP_033326 | Anti-inflammatory and immune-modulating agent (Ialenti et al. 2001) | NM_009300 | chr2: 164,102,812–164,104,021 | + | Not found in the evolutionary screen reported here |
| Svs 5 | NP_033327 | Serine proteinase inhibitor (Clauss et al. 2005) | NM_009301 | chr2: 164,158,522–164,159,331 | + | 1.47 (14, 4) |
| Svs 6 | NP_038707 | Serine proteinase inhibitor (Clauss et al. 2005) | NM_013679 | chr2: 164,142,504–164,143,351 | + | 0.83 (29, 13) |
| Svs 7 | NP_064660 | Caltrin/sperm capacitation (Luo et al. 2001) | NM_020264 | chr9: 35,415,358–35,419,431 | − | 1.74 (106, 21) |
Table 1 shows two mouse Svs3 genes, designated Svs3a and Svs3b. Using the BLAT tool and either Svs3 sequence in the mouse genome as a query string, we obtained two, closely linked Svs3-like sequences in the rat genome (NM_001007605 and NM_001102417). Because we do not have genome sequences for the other rodent taxa (e.g., M. m. domesticus, M. m. musculus, M. m. castaneus, and M. caroli), we cannot at this time determine whether there is more than a single Svs3-like gene in these taxa. Phylogenetic analysis of the Svs3 sequences suggests that the Svs3 duplications in the mouse and rat genomes occurred independently (shown in supplementary fig. 1, Supplementary Material online), although it is possible that continuous gene conversion between the duplicates makes them appear to be more recently duplicated.
Positive Selection in Svs Genes
We tested the four Svs genes that met our criteria (Svs3, Svs5, Svs6, and Svs7) for signs of positive selection. We used PCR to obtain the coding sequences for these proteins from seven murid rodents and confirmed orthology by reciprocal Blast searches against the mouse and rat genomes. Our primer set amplified an Svs3 in each of the other rodent taxa we studied, but we cannot determine at this time which paralog, a or b, each represents. We assessed variation in dN/dS at codon sites by comparing neutral models to selection models of codon evolution. Model parameters were estimated using a maximum likelihood method employed in the CODEML program of the PAML package (Yang 1997; Nielsen and Yang 1998; Yang et al. 2000). Svs3 (Svs3a and Svs3b in mouse and rat), Svs5, and Svs6 did not show signs of positive selection in rodents under the sites model described above. By contrast, Svs7 showed significant signs of positive selection within rodents (P < 0.0001), with an estimated 33% of codons showing a dN/dS ratio of 12.38 (the amino acid sequence alignment is shown in supplementary fig. 2A, Supplementary Material online). We also looked for signs of lineage-specific variation in selective pressure by estimating dN/dS along phylogenetic lineages. Among the four Svs genes, we found signs of such selection only in Svs5 where there was an indication of lineage-specific selection in the rat. In that analysis, the branch model of the CODEML program yielded a dN/dS estimate of 1.92 over the entire sequence for the rat branch. When we compared that model to a branch model with the rat branch fixed to 1, the difference was not significant. For a more sensitive test, we used the branch-sites model to allow variation between codon sites and branches with the rat branch as the foreground branch (Zhang et al. 2005). It estimated that about half of the codon sites had a dN/dS of 4.85 on the rat branch. Comparison to the null model shows the inference of positive selection to be statistically significant. (χ2 = 5.747, degree of freedom = 1, P = 0.0165).
Given the strong evidence for positive selection on Svs7 in rodents, we sought to test this in primates as well. We obtained human Svs7 from the Homo sapiens genome and confirmed its conserved gene order with rodent Svs7 by comparison of those regions in the mouse and rat genomes. Completed primate genomes were interrogated for putative Svs7 sequences, using the human Svs7 as the query term. Complete sequences were obtained from the Macaca mulatta, Pongo pygmaeus, Callithrix jacchus, and Tarsius syrichta genomes. Interrogating the Pan troglodytes genome did not yield a complete Svs7 ortholog for the chimpanzee due to a sequencing gap. Interestingly, although the 76 amino acid sequence we obtained from the chimpanzee genome was an incomplete Svs7, it was identical to that portion (80%) of human Svs7. We confirmed the sequences we obtained as Svs7 orthologs by reciprocal best-hit Blast searches of their sequences on the human genome. CODEML analysis of primate Svs7 also showed significant signs of positive selection (P = 0.0047; Model 8a vs. 8) with an estimated 19% of codons showing a dN/dS of 10.74 (table 2; the amino acid sequence alignment is shown in supplementary fig. 2B, Supplementary Material online).
Table 2.
Selection Test on Candidate Svs Genes from the Screen
| Gene | Mouse–Rat dN/dSa | Species in Divergent Analysisb | Ratio of dN/dS (%Codons)c | P Value: All Speciesd | Codon Sites under Selectione |
| Svs3 | 0.55 | 2, 3, 4, 6, 7, 9 | 1.0 (41) | 0.99 | None |
| Svs5 | 1.47 | 1, 3, 5, 8, 9 | 4.85 (1.3) | 0.87 | None |
| Svs6 | 0.83 | 2, 3, 4, 6, 7, 8, 9 | 1.0 (0) | 1.0 | None |
| Svs7 | 1.74 | 1, 2, 3, 5, 6, 7, 8, 9 | 12.38 (33) | <0.0001 | 11S, 14K, 16R, 23S, 24Q, 25S, 28V, 31P, 40H, 42V, 44T, 50K, 52M, 54S, 64T, 66K, 67K, 75E |
| Primate Svs7 | 10, 11, 12, 13, 14 | 10.74 (19) | 0.0047 | 8Y, 18R, 38D, 52R, 59R, 60G, 75V |
Mouse–Rat dN/dS was determined using the evolutionary screen.
The genomes/species used in this analysis are 1) mouse genome (C57BL/6), 2) Mus musculus domesticus, 3) Mus musculus musculus, 4) Mus musculus castaneus, 5) Mus spicilegus, 6) Mus spretus, 7) Mus caroli, 8) Mus pahari, 9) rat genome (Rattus norvegicus), 10) human genome (Homo sapiens), 11) rhesus macaque genome (Macaca mulatta), 12) orangutan genome (Pongo pygmaeus), 13) marmoset genome (Callithrix jacchus), and 14: tarsier genome (Tarsius syrichta).
The dN/dS ratio of the class of codons under positive selection is given with the percentage of codon sites predicted to be in that class.
The P value rejecting the model of neutral evolution (M8A) over that of selection (M8) is given.
Sites with posterior probabilities greater than 0.9 are indicated in regular typeface; P > 0.95 indicated in bold typeface; and P > 0.99 indicated in bold, underlined typeface.
Spatial Distributions of Selected Svs7 Sites on Three-Dimensional Structures
Because of the evidence for positive selection on rodent and primate Svs7, we predicted positively selected codon sites using a Bayes empirical Bayes (BEB) method (Yang et al. 2005). The sites under selection in rodent Svs7 and in primate Svs7 are listed in table 2. Eighteen codon sites in rodent Svs7 and seven sites in primate Svs7 were identified as positively selected at a BEB posterior probability threshold of 90%. The difference in the number of predicted sites between rodents and primate could be due to the difference in the number of sequences in each group because power improves with addition of taxa (Anisimova et al. 2002). These sites are plotted on three-dimensional models in figure 2, along with other sites identified below the 90% posterior probability level. We tested whether the spatial distribution of these selected sites was nonrandom. Positively selected amino acid sites in rodent Svs7 tended to form clusters in the protein structure. The degree of clustering for selected sites with greater than 90% BEB posterior probability is statistically significant when compared with random permutations of surface sites (P = 0.01728; 100,000 permutations). Primate Svs7 selected amino acid sites predicted with 90% BEB probability showed a tendency to be more clustered than random surface sites, but this result was not significant (P = 0.11632); however, the test may lack power when considering so few sites. Evidence suggests that the Svs7 protein is under positive selection in both rodents and primates, but it is not clear if the driving force behind selection is the same in both taxonomic groups. One way to assess the similarity of their adaptive pressures is to ask whether selection acted on similar structural regions of the protein. When compared on the same structural model, rodent and primate selected sites were either identical or at neighboring residues significantly more often than expected by chance (90% BEB sites, P = 0.0044). This test provides evidence that positive selection acted at similar regions of the Svs7 protein in rodents and primates.
FIG. 2.—
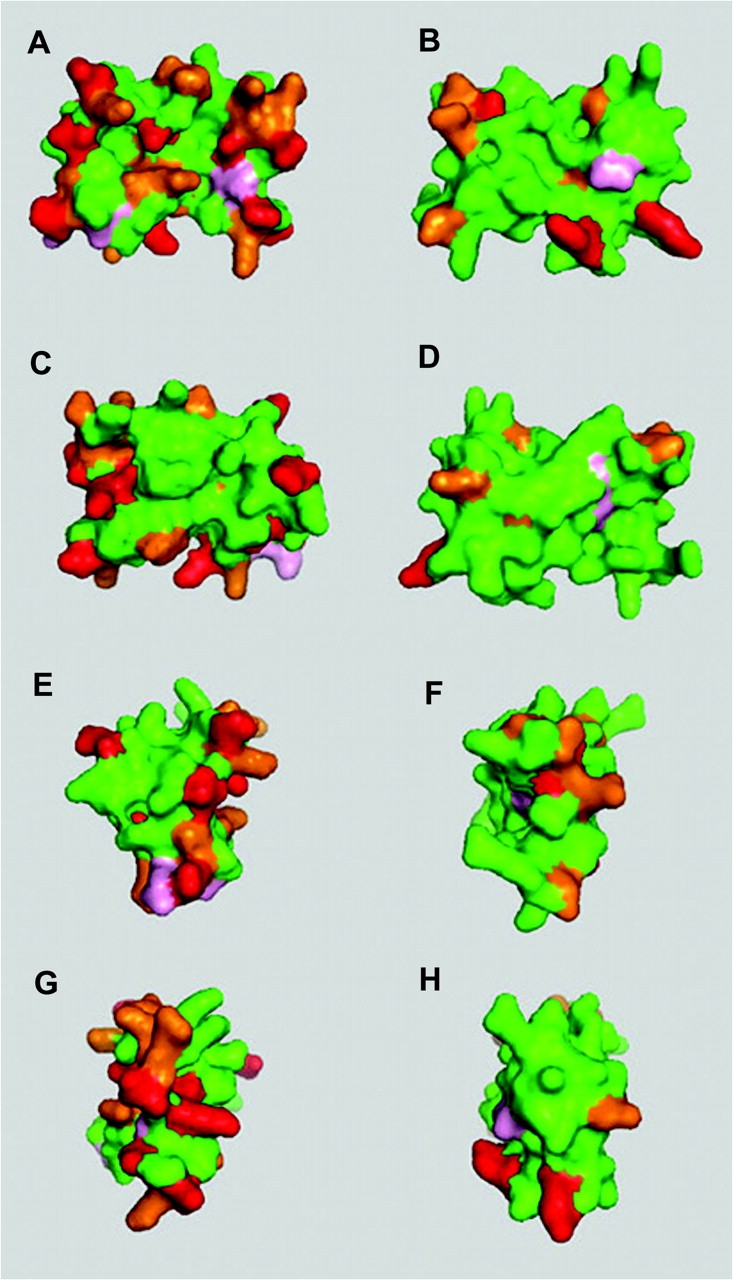
Positive selection at sites on molecular models of rodent (A, C, E, and G) and primate (B, D, F, and H) Svs7. Because all the Svs7 proteins with confirmed orthologous genes mapped on the d1rewc model, that was used by the structure threading program for these sequences. Residues in red are selected with a BEB posterior probability >99%; those in pink with a BEB posterior probability >95%; and those in gold with a BEB posterior probability >50%. (A) Rodent Svs7—top; (B) primate Svs7—top; (C) rodent Svs7—bottom; (D) primate Svs7—bottom; (E) rodent Svs7—left; (F) primate Svs7—left; (G) rodent Svs7—right; and (H) primate Svs7—right.
Evolution of Svs7
In addition to the seven rodent taxa and the four primate taxa described above, we searched other vertebrate genomes for Svs7-like proteins in order to determine their evolutionary history. Using PSI-Blast, we found Svs7-like proteins in the genome data of the cat (Felis catus), dog (Canis familiaris), cow (Bos taurus), elephant (Loxodonta africana), hedgehog (Erinaceus europaeus), guinea pig (Cavia porcellus), platypus (Ornithorhynchus anatinus), chicken (Gallus gallus), several snakes (e.g., Dispholidus typus, the boomslang), and several fish (e.g., Danio rerio, the zebrafish). We did not find an Svs7-like protein in the genome of opossum (Monodelphis domestica). Table 3 summarizes the accession numbers (where known), the chromosomal location/strand, and the threading models of these Svs7-like proteins (the alignment of the protein sequences corresponding to these genes is shown in supplementary fig. 3, Supplementary Material online).
Table 3.
Svs7 Genes in Various Organisms
| Organism | Accession Number | Chromosomal Location (strand) | Orthology | Threaded Structurea (resultsb) |
| Human | XP_001132703 | chr11: 125,208,474–125,213,529 (+) | Orthologous with other Svs7s | d1rewc (90%; 86; 22%) |
| Orangutan | Nonec | Unknown | Unknown | d1rewc (95%; 86; 24%) |
| Rh. macaque | XP_001108343 | chr14: 123,994,746–123,999,824 (+) | Orthologous with other Svs7s | d1rewc (5%; 46; 24%) |
| Marmoset | Nonec | Unknown | Unknown | d1rewc (20%; 86; 21%) |
| Tarsier | Nonec | Unknown | Unknown | d1rewc (45%; 86; 20%) |
| Mouse | NP_064660 | chr9: 35,415,358–35,419,431 (−) | Orthologous with other Svs7s | d1rewc (95%; 86; 21%) |
| Rat | NP_001102227 | chr8: 35,408,660–35,410,707 (−) | Orthologous with other Svs7s | d1rewc (95%; 86; 22%) |
| Catd | Nonec | Unknown | Unknown | d1mr6a (90%; 68; 19%) |
| Elephant | Nonec | Unknown | Unknown | d1rewc (95%; 86; 22%) |
| Hedgehog | Nonec | Unknown | Unknown | d1rewc (95%; 86; 19%) |
| Guinea pig | Nonec | Unknown | Unknown | d1rewc (95%; 86; 23%) |
| Cow | NP_001098948 | chr29: 24,532,076–24,537,414 (+) | Orthologous with other Svs7s | d1rewc (70%; 86; 23%) |
| Dogd | XP_849126e | chr5: 11,809,092–11,814,548 (−) | Orthologous with other Svs7s | d1rewc (90%; 86; 20%) |
| Platypus | XP_001519355 | Unknown | Orthologous with other Svs7s | d1rewc (90%; 86; 22%) |
| Chicken | NP_990106 | chr 2: 154,011,241–154,012,614 (+) | Not orthologous with Svs7s | d1cdq (95%; 77; 25%) |
| Snake | ABU68483 | Unknown | Unknown | d1txb (100%; 73; 25%)f |
All structures are versions of elapid snake-like toxins; three-fingered proteins.
Data consist of structural model, % confidence, length, and % identity.
Submitted to GenBank.
May be part of Acrv1.
Ensembl ENSF00000006303.
This may indeed be an elapid snake toxin as the snake in question is a boomslang (Dispholidus typus).
We determined orthology with mouse and rat Svs7 genes based on the criterion of conserved gene order (table 3). This criterion only held for the Svs7 genes of human, rhesus macaque, dog, cow, and platypus (where each maps near the PUS3 locus, the Cdon locus, the Acrv1 locus, and other loci in a conserved gene region). The opossum genome contains a large sequencing gap in this region, and the Svs7 gene may well be hidden within it. The incomplete nature of the genomes of cat, elephant, hedgehog, and guinea pig do not allow a determination of conserved gene order with the other mammal Svs7 genes we report here.
The genes encoding the Svs7-like proteins of chicken and zebra fish do not share a conserved gene order with mammal Svs7 genes. The 10 half-Cys residue pattern found in Svs7 is shared by a number of other proteins whose genes do not share a conserved gene order with Svs7. Examples of these are mammal Ly-6 genes (Fleming et al. 1993), elapid snake toxin genes (Altschul et al. 1990; Fry et al. 2003), and the C-terminal regions of acrosomal-1 protein genes (Luo et al. 2001). We attribute the identification of these proteins by the PSI-Blast program to the 10-Cys residue core structure shared by proteins that are not products of genes orthologous to mammal Svs7 genes (Fleming et al. 1993; Fry et al. 2003).
Discussion
Evolutionary Screen
As a class, rodent prostate secretory genes show patterns of evolution similar to seminal fluid protein genes in other taxonomic groups. Several primate seminal fluid protein genes also have elevated dN/dS ratios (Clark and Swanson 2005), and male accessory gland genes are also rapidly evolving in Drosophila flies (Swanson et al. 2001) and both Allonemobius and Gryllus crickets (Braswell et al. 2006). Furthermore, our observation that secreted rodent prostate proteins evolve approximately twice as fast as those of nonsecreted proteins is paralleled in the primate prostate and in the Drosophila male accessory gland. Although rodent and primate prostate genes evolve similarly, their complement of highly expressed genes are largely different. Of the seven Svs genes only Svs1 and Svs7 have probable human orthologs. Rodent Svs2 and Svs3 are highly repetitive and have N-terminal regions that are homologous to human Semenogelin I and Semenogelin II, which encode the primate semen coagulum proteins. Also, Svs2 and Svs3 are located in the chromosomal position corresponding to the human Semenogelin locus (Clauss et al. 2005). Several of the most abundant rodent prostate genes have no clear ortholog in the human genome, such as the genes encoding spermine-binding protein (SBP; highest expression level), Pbsn (fourth highest), and Svs5 (sixth highest). The lack of conservation of the prostate expression profile between rodents and primates could be due to changes in expression level, rapid sequence divergence that makes identification of orthologs difficult, or a high rate of gene turn over. Within primates, seminal fluid protein genes have become pseudogenes in multiple species (Lundwall and Olsson 2001; Olsson et al. 2004; Clark and Swanson 2005), and a high rate of gene birth and death was observed in Drosophila accessory gland genes (Begun et al. 2006).
The mouse epididymal transcriptome has also been analyzed in an evolutionary genomics context. Recently, Dean et al. (2007) reanalyzed the data of Johnston et al. (2005) and found evidence for recurrent positive selection on a small proportion of epididymis-expressed genes compared with the whole genome, although the difference was not statistically significant. They also showed that a subset of epididymis-specialized genes showed an overrepresentation on the X chromosome, consistent with the preferential X-linkage of genes involved in adaptive evolution (Vicoso and Charlesworth 2006). They concluded that, overall, the patterns suggest that at least some epididymis-specialized, secreted genes are important in male fertility. Our evolutionary screen of prostate ESTs identified four Svs genes with elevated pairwise dN/dS ratios. Using CODEML analysis, we subsequently verified branch-specific selection (rat branch) in Svs5 (encoded on mouse chromosome 2) and selection on numerous Svs7 (mouse chromosome 9) sites in both rodents and primates. Another group (Ramm et al. 2007) studied seven rodent genes expressed in the testes, prostate, or seminal vesicles, with the goal of ascertaining high rates of adaptive gene evolution linked to postcopulatory sexual selection. Of these, they found high dN/dS in Prm1 (mouse chromosome 16) and Sva (mouse chromosome 6), Acrv1 (mouse chromosome 9) and Svs2 (mouse chromosome 2), but not for Svs4 (aka Svp2; mouse chromosome 2), Msmb (mouse chromosome 14), or Spink3 (mouse chromosome 18).
Selection on Svs Genes
Of the many candidates for rapidly evolving rodent prostate genes revealed by our evolutionary screen, we focused this study on the Svs genes because all except for Svs4 appeared in the screen results. Furthermore, our evolutionary screen identified four Svs protein genes, Svs3 (Svs3a and Svs3b in mouse and rat), Svs6, Svs5, and Svs7, which could be evolving more rapidly than the others (i.e., dN/dS > 0.5). We also found that Svs3 has been duplicated in mouse and rat. Our primer set amplified an Svs3 in each of the other rodent taxa we studied, but we cannot determine at this time which paralog, a or b, each represents. Others have shown that rodent Svs2–6 genes are an expansion from an ancestral protein containing a WAP four-disulfide core, called the WFDC domain (Clauss et al. 2005). Duplication of Svs3 appears to be a continuation of that process and, strikingly, it may have occurred twice in rodents (supplementary fig. 1, Supplementary Material online). When the completed genomes of the other taxa become available, it should be possible to determine how widespread this particular duplication is as well as when it originated and in what lineages in the history of murid Svs genes.
Our CODEML sites analysis did not find evidence for adaptive evolution of Svs3, Svs6, and Svs5, though we did find evidence for selection on the rat branch for Svs5. By contrast, we found that rodent Svs7 is evolving very rapidly and we extended our analysis to five primates for comparison. Our analysis identified 18 residues under positive selection in rodent Svs7 and seven under positive selection in primate Svs7 (table 2 and fig. 2). We suspect that, when more primate Svs7 sequences become available, it will be possible to identify additional sites under selection, particularly sites shared with rodents. Nonetheless, our statistical assessment of the similarity of adaptive pressures on Svs7 in primates and rodents suggests that positive selection acted at similar regions of the Svs7 protein. Thus, although it seems tempting to speculate that the selective pressure is similar in the two mammalian lineages, more information about the function of Svs7 is necessary to support that idea.
Evolutionary History of Svs7
Svs7 appears to be a relatively new gene in the history of vertebrate evolution insofar as we have been able to identify orthologs only in mammalian lineages. The platypus is the basal taxon in which we identified an Svs7 ortholog. The opossum genome likely also contains an Svs7 ortholog but we could not identify the gene due to the presence of a large gap in the colinear region of its genome. Otherwise, the gene appears to be widespread in mammals, including dog, cow, primates, and rodents. We have found putative Svs7 genes also in the cat, elephant, and hedgehog genomes; however, the incomplete coverage of those genomes at present does not allow us to identify them unequivocally as Svs7 orthologs.
The Svs7-like genes we found in chicken and other nonmammalian vertebrates are unlikely to be orthologs because they do not share a conserved gene order with mammal Svs7 genes. Instead, they share a similar overall structure that is detected by PSI-Blast analysis. The core ten half-Cys residue pattern and the CCXXDLCN consensus sequence that are shared by Svs7 and these nonorthologous genes hint at a common ancestral protein structure that predates the divergence of diapsid and synapsid reptiles. These two features of the primary structure appear to dictate the three-finger snake toxin-like three-dimensional structure shared by all these proteins (Fleming et al. 1993; Fry et al. 2003). We suggest that this basal structure has been co-opted by a number of gene families, including mammal Ly-6 genes (Fleming et al. 1993), elapid snake venom protein genes (Fry et al. 2003), Svs7 genes (this report), and the region of the Acrv1 gene family that encodes the C-terminal protein sequence.
Although it is tempting to envision the Svs7 gene as a mammalian novelty, there is not enough information available to support that conclusion. What seems to be clear from the results we report here, however, is that Svs7 genes in rodents and primates are evolving rapidly, under the strong influence of positive selection. To understand why this is occurring, it is necessary to identify a putative function for the Svs7 protein.
What Is the Biological Role of Svs7?
Svs7 has been described as mouse caltrin, although the sequences reported differed in their C-terminal ends (Lardy 1985; Luo et al. 2001). The concept of the caltrin function began with the observation that ejaculated sperm do not take up calcium ions, whereas epididymal sperm do (reviewed in Lardy 1985, 2003). Experiments in which epididymal sperm were exposed to seminal fluid before the addition of calcium showed that a component of seminal fluid was necessary to inhibit calcium uptake in ejaculated sperm. Lardy and his colleagues characterized the responsible component and showed that it was a protein bound to the plasma membranes of ejaculated sperm but not to membranes of epididymal sperm. They named it “caltrin,” calcium transport inhibitor.
Caltrin has an important role in sperm capacitation and thus fertilization (summarized by Lardy [2003]). Calcium uptake disrupts the acrosome and also facilitates acquisition of hyperactivated sperm motility, which is characterized by rapid lashing and wider excursion of the sperm tail. Subsequently, the sperm swim in tight arcs that allow them to drive through the zona and penetrate the egg. Upon ejaculation, caltrin binds to the acrosome plasma membrane and thus prevents premature expression of these calcium uptake–mediated events that are critical to fertilization. This direct interaction between caltrin and sperm in the process of being ejaculated appears to play an important role in male fertility. Given the identification of Svs7 as mouse caltrin, we suggest that its rapid evolution that we report here for rodents and primates reflects the importance of its role in mammalian reproduction and thus potentially in speciation. We speculate that an explanation of the adaptive evolution of Svs7 that we have observed in rodents and primates stems from an important function in sperm competition; however, other possibilities include polyspermy avoidance or cryptic female choice. If the efficiency of caltrin function can be enhanced by amino acid changes, the possessing male might enjoy a competitive edge over others in an environment of multiple inseminations of females by different males.
Unfortunately, assigning such a role to Svs7 in mammals generally is confounded by the variety of molecules identified as caltrins in diverse mammalian groups. Indeed, caltrin has been identified as Svs7 only in mouse (Luo et al. 2001; Lardy 2003). In rat, cow, and guinea pig, other molecules quite different from Svs7 have been identified as having that function (Lardy 1985, 2003). This dilemma appears to be the result of our inability at this time to distinguish true convergent evolution, in which multiple molecular forms have come to fulfill the caltrin function in diverse mammal groups, from a lack of appreciation of subtle differences in the functions of these different protein forms, possibly because our physiological definition of the ‘caltrin’ function is too loose. Indeed, it might be better to characterize caltrin as a function, rather than assigning the name to a specific protein. Looking at the problem differently, we suggest that there may be a conflict between the limitations of a physiologically defined function versus the power of genomics to identify orthologous genes whose products may or may not share such a function. For us better to understand why this pattern of selection is observed on Svs7 genes in primates and rodents, we need a better understanding of the functions played by the Svs7 proteins in diverse mammals. Thus, assignment of a broader role for Svs7 as a caltrin must await a comparative biochemical, physiological, and genomic analysis of the protein across the whole mammalian phylogeny.
Supplemental Material
Supplementary figures 1–3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
The authors wish to thank Janet M. Young for the use of her computer script to search primate genomes and Christina M. Laukaitis for help with the searches. R.C.K. was supported by a Senior Postdoctoral Fellowship, grant number 5F33HD055016-02, from the National Institute of Child Health and Human Development (NICHHD). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the NICHHD. N.L.C. was supported by a National Institutes of Health (NIH) Postdoctoral Fellowship F32GM084592 from the National Institute Of General Medical Sciences and E.D.N. was supported by the 2007 Amgen Scholars program. W.J.S. was supported by NIH grants HD42563, HD054631, HD057974, and National Science Foundation (NSF) grant DEB-0743539.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Bielawski J, Yang Z. Accuracy and power of Bayes prediction of amino acid sites under positive selection. Mol Biol Evol. 2002;19:950–958. doi: 10.1093/oxfordjournals.molbev.a004152. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Lindfors HA, Thompson ME, Holloway AK. Recently evolved genes identified from Drosophila yakuba and D. erecta accessory gland expressed sequence tags. Genetics. 2006;172:1675–1681. doi: 10.1534/genetics.105.050336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen J, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: signalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bielawski J, Yang Z. Maximum likelihood methods for detecting adaptive evolution after gene duplication. J Struct Funct Genomics. 2003;3:201–212. [PubMed] [Google Scholar]
- Braswell W, Andres J, Maroja L, Harrison R, Howard D, Swanson W. Identification and comparative analysis of accessory gland proteins in orthoptera. Genome. 2006;49:1069–1080. doi: 10.1139/g06-061. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Fledel-Alon A, Williamson S (11 co-authors) Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- Chevret P, Veyrunes F, Britton-Davidian J. Molecular phylogeny of the genus Mus (Rodentia: Murinae) based on mitochondrial and nuclear data. Biol J Linn Soc Lond. 2005;84:417–427. [Google Scholar]
- Clark N, Aagard J, Swanson W. Evolution of reproductive proteins from animals and plants. Reproduction. 2006;131:11–22. doi: 10.1530/rep.1.00357. [DOI] [PubMed] [Google Scholar]
- Clark N, Swanson W. Pervasive adaptive evolution in primate seminal proteins. PLos Genet. 2005;1:335–342. doi: 10.1371/journal.pgen.0010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauss A, Lilja H, Lundwall A. The evolution of a genetic locus encoding small serine proteinase inhibitors. Biochem Biophys Res Commun. 2005;333:383–389. doi: 10.1016/j.bbrc.2005.05.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronel C, Winnica D, Novella M, Lardy H. Purification, structure, and characterization of caltrin proteins from seminal vesicle of the rat and mouse. J Biol Chem. 1992;267:20909–20915. [PubMed] [Google Scholar]
- Dean M, Good J, Nachman M. Adaptive evolution of proteins secreted during sperm maturation: an analysis of the mouse epididymal transcriptome. Mol Biol Evol. 2007;25(2):383–392. doi: 10.1093/molbev/msm265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emes R, Goodstadt L, Winter EE, Ponting C. Comparison of the genomes of human and mouse lays the foundation of genome zoology. Hum Mol Genet. 2003;12:701–709. doi: 10.1093/hmg/ddg078. [DOI] [PubMed] [Google Scholar]
- Emes R, Riley M, Laukaitis C, Goodstadt L, Karn R, Ponting C. Comparative evolutionary genomics of androgen-binding protein genes. Genome Res. 2004;14:1516–1529. doi: 10.1101/gr.2540304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming T, O'hUigin C, Malek T. Characterization of two novel Ly-6 genes. J Immunol. 1993;150:5379–5390. [PubMed] [Google Scholar]
- Franzkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comput Chem. 1998;19:319–333. [Google Scholar]
- Fry B, Wuster W, Kini R, Brusic V, Khan A, Venkataraman D, Rooney A. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J Mol Evol. 2003;57:110–129. doi: 10.1007/s00239-003-2461-2. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, Weinstock GM, Metzker ML, et al. (230 co-authors) Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Hughes A, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- Ialenti A, Santagada V, Caliendo G, et al. (10 co-authors) Synthesis of novel anti-inflammatory peptides derived from the amino acid sequence of the bioactive protein SV-IV. Eur J Biochem. 2001;268:3399–3406. doi: 10.1046/j.1432-1327.2001.02236.x. [DOI] [PubMed] [Google Scholar]
- Jensen J, Wong A, Aquadro C. Approaches for identifying targets of positive selection. Trends Genet. 2007;23:568–577. doi: 10.1016/j.tig.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Johnston D, Jelinsky S, Bang H, Candeloro P, Wilson E, Kopf G, Turner T. The mouse epididymal transcriptome: transcriptional profiling of segmental gene expression in the epididymis. Biol Reprod. 2005;73:404–413. doi: 10.1095/biolreprod.105.039719. [DOI] [PubMed] [Google Scholar]
- Kawano N, Yoshida M. Semen-coagulating protein, Svs2, in mouse seminal plasma controls sperm fertility. Biol Reprod. 2007;76:353–361. doi: 10.1095/biolreprod.106.056887. [DOI] [PubMed] [Google Scholar]
- Lardy H. A half century of biochemistry. In: Semenza G, editor. A history of biochemistry. New York: Elserier; 1985. pp. 297–325. [Google Scholar]
- Lardy H. Happily at work. J Biol Chem. 2003;278:3499–3509. doi: 10.1074/jbc.X200003200. [DOI] [PubMed] [Google Scholar]
- Laukaitis C, Heger A, Blakley T, Munclinger P, Ponting C, Karn R Forthcoming. Rapid bursts of androgen-binding protein (Abp) gene duplication occurred independently in diverse mammals. BMC Evol Biol. 2008;8:46–55. doi: 10.1186/1471-2148-8-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H-J, Luo C-W, Chen Y-H. Localization of the transglutaminase cross-linking site in Svs III, a novel glycoprotein secreted from mouse seminal vesicle. J Biol Chem. 2002;277:3632–3639. doi: 10.1074/jbc.M107578200. [DOI] [PubMed] [Google Scholar]
- Lundwall A, Malm J, Clauss A, Valtonen-Andre C, Olsson AY. Molecular cloning of complementary DNA encoding mouse seminal vesicle-secreted protein SVS I and demonstration of homology with copper amine oxidases. Biol Reprod. 2003;69:1923–1930. doi: 10.1095/biolreprod.103.019984. [DOI] [PubMed] [Google Scholar]
- Lundwall A, Olsson A. Semenogelin II gene is replaced by a truncated line 1 repeat in the cotton-top tamarin. Biol Reprod. 2001;65:420–425. doi: 10.1095/biolreprod65.2.420. [DOI] [PubMed] [Google Scholar]
- Luo C-W, Lin H-J, Chen Y-H. A novel heat-labile phospholipid-binding protein, SVS VII, in mouse seminal vesicle as a sperm motility enhancer. J Biol Chem. 2001;276:6913–6921. doi: 10.1074/jbc.M006954200. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nelson PS, Pritchard C, Abbott D, Clegg N. The human (PEDB) and mouse (mPEDB) prostate expression databases. Nucleic Acids Res. 2002;30:218–220. doi: 10.1093/nar/30.1.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A, Valtonen-Andre C, Lilja H, Lundwall A. The evolution of the glandular kallikrein locus: identification of orthologs and pseudogenes in the cotton-top tamarin. Gene. 2004;343:347–355. doi: 10.1016/j.gene.2004.09.020. [DOI] [PubMed] [Google Scholar]
- Pilch B, Mann M. Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biol. 2006;7:R40. doi: 10.1186/gb-2006-7-5-r40. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramm S, Oliver P, Ponting C, Stockley P, Emes R Forthcoming. Sexual selection and the adaptive evolution of mammalian ejaculate proteins. Mol Biol Evol. 2008;25:207–219. doi: 10.1093/molbev/msm242. [DOI] [PubMed] [Google Scholar]
- Swanson W, Clark A, Waldrip-Dail H, Wolfner M, Aquadro C. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc Natl Acad Sci USA. 2001;98:7375–7379. doi: 10.1073/pnas.131568198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson W, Nielsen R, Yang Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol. 2003;20:18–20. doi: 10.1093/oxfordjournals.molbev.a004233. [DOI] [PubMed] [Google Scholar]
- Swanson W, Vacquier V. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Swanson W, Wong A, Wolfner M, Aquadro C. Evolutionary expressed sequence tag analysis of Drosophila female reproductive tracts identifies genes subjected to positive selection. Genetics. 2004;168:1457–1465. doi: 10.1534/genetics.104.030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, et al. (221 co-authors) Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Bielawski J. Statistical methods for detecting molecular adaptation. Trends Ecol Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Hasegawa M. Models of amino acid substitution and applications to mitochondrial protein evolution. Mol Biol Evol. 1998;15:1600–1611. doi: 10.1093/oxfordjournals.molbev.a025888. [DOI] [PubMed] [Google Scholar]
- Yang Z, Swanson W, Vacquier V. Maximum-likelihood analysis of molecular adaptation in abalone sperm lysin reveals variable selective pressures among lineages and sites. Mol Biol Evol. 2000;17:1446–1455. doi: 10.1093/oxfordjournals.molbev.a026245. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong W, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Young J, Waters H, Dong C, Fülle H-J, Liman E. Degeneration of the olfactory guanylyl cyclase D gene during primate evolution. PLoS ONE. 2007;2:e884. doi: 10.1371/journal.pone.0000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.