Abstract
Local rates of recombination positively correlate with DNA sequence diversity in many species. To test whether this relationship stems from mutagenicity of meiotic recombination, studies often look for a similar association between local rates of recombination and sequence “divergence” between species. Because recombination is mutagenic in yeast, I evaluate this assay by testing whether noncoding DNA sequence divergence between Saccharomyces species is related to measures of meiotic double-strand DNA breaks or crossover rates derived from Saccharomyces cerevisiae. Contrary to expectation, I find that sequence divergence is either uncorrelated or negatively correlated with rates of both double-strand break and crossover. Several caveats are mentioned, but these results suggest that mutagenesis from meiotic recombination is not the primary driver of sequence divergence between Saccharomyces species. This study demonstrates that the association between interspecies nucleotide divergence and local recombination rates is not always a reliable indicator of recombination's mutagenicity.
Keywords: recombination, divergence, diversity, Saccharomyces
Introduction
Over the past two decades, molecular evolutionary studies have identified a strong positive relationship between meiotic recombination rate in regions of the genome and DNA sequence diversity in those regions (Aguade et al. 1989; Aquadro 1997; Nachman 2002). A simple explanation for such a pattern could be that meiotic recombination is mutagenic. In a seminal work, Begun and Aquadro (1992) identified an association between recombination rate and nucleotide diversity within Drosophila melanogaster, but they did not observe an association between recombination and nucleotide “divergence” between D. melanogaster and Drosophila simulans. The lack of an association with divergence suggested that recombination itself was not mutagenic—the association with nucleotide diversity was better explained by large stretches of the genome hitchhiking with positively selected variants (MaynardSmith and Haigh 1974) or with deleterious mutations being eliminated (Charlesworth et al. 1993; Nordborg et al. 1996) in low recombination regions. Both processes increase stochastic variance in the inheritance of gene copies and, consequently, reduce nucleotide diversity. As in Drosophila, studies of Solanum wild tomatoes also did not observe a significant relationship of recombination rate and divergence between species (Stephan and Langley 1998; Roselius et al. 2005), but recombination and divergence are correlated in human–chimpanzee (Hellmann et al. 2003) and Zea mays–Tripsacum dactyloides (Tenaillon et al. 2004). Overall, recombination rate is inconsistently related to nucleotide divergence between species, suggesting either variability in the operation of evolutionary forces among taxa or that this association is an inconsistent test of meiotic recombination's mutagenicity.
Most of the evidence for recombination's mutagenicity is correlative (Lercher and Hurst 2002; Filatov and Gerrard 2003), and direct empirical evidence comes primarily from studies of yeast. Esposito and Bruschi (1993) found that lesions in the Saccharomyces cerevisiae LEU1 locus were both recombinogenic and mutagenic, and Strathern et al. (1995) showed that recombinational repair of double-strand breaks (DSBs) elevated the local mutation rate by 100-fold relative to S-phase replication. Although mitotic recombination was investigated in the studies above, similar results have been obtained for meiotic recombination (Strathern J, personal communication), and mutation rates are known to be higher in yeast meiosis relative to mitosis (Magni and Von Borstel 1962).
Given the mutagenicity of recombination in yeast, one may expect to find that local meiotic recombination rates, or DSBs in general, are associated with nucleotide divergence between Saccharomyces species. This system is appropriate for such a test because assembled, aligned, and annotated full-genome sequences are available for this species and several congenerics (Goffeau et al. 1996; Kellis et al. 2003), and extensive fine-scale meiotic DSB maps (Gerton et al. 2000; Blitzblau et al. 2007; Buhler et al. 2007) and crossover/noncrossover maps (Mancera et al. 2008) have been constructed. The approach taken here is simple: I test whether there is an association between local rates of DSB (or crossover/noncrossover rate) and divergence between species, factoring out several potential confounding factors.
Materials
Multiple sequence alignments of Saccharomyces species were obtained from the University of California Santa Cruz (UCSC) genome browser (http://hgdownload.cse.ucsc.edu/downloads.html#yeast). These alignments were prepared using version 7 of the MULTIZ program, which builds a multiple alignment from local pairwise alignments of a designated reference genome with each other genome of interest (Blanchette et al. 2004). The S. cerevisiae SK1 sequence alignment was produced by the Saccharomyces Genome Resequencing Group at the Sanger Institute and can be obtained from ftp://ftp.sanger.ac.uk/pub/dmc/yeast/latest. All alignments were annotated with sequence features listed in the Saccharomyces Genome Database (Fisk et al. 2006), and focal regions were unannotated intergenic regions and introns (both 5′ and within coding sequence [CDS]). Analyses of intergenic regions excluded the first and last ∼150 bp outside of CDS or other RNA-encoding sequences (e.g., rRNAs) to reduce the impact of conserved promoters and other regulatory elements on the results (also minimizing the effects of a few small [<5 bp] indels found in the UCSC aligned sequences). Analyses of introns excluded the first and last ∼15 bp within introns to reduce potentially conserved splicing-related regions.
Estimates of meiotic DSBs were taken from the recent study of Buhler et al. (2007), supplementary table S1 (Supplementary Material online). As in that study, I focused on denoised ratios of background-normalized fluorescence obtained from a dmc1Δ strain of S. cerevisiae. Their method to localize DSB hot spots detected the single-stranded DNA (ssDNA) intermediates that surround DSB sites via microarray hybridization (see also Blitzblau et al. 2007), estimating the frequency of DSB-linked ssDNA at 40,766 sites across the yeast genome. For simplicity, I refer to these estimates simply as DSBs or DSB rate throughout this paper. I repeated my analyses using their denoised ratios of background-normalized fluorescence obtained from a rad50S-derived mutant strain. Crossover (4,163) and noncrossover (2,126) distributions were taken from Mancera et al. (2008), and these positions were binned into 20-kb windows along the sequence alignment for analysis.
Data sets were formatted using custom Perl scripts (available upon request) and statistical analyses used StatView software. Except where noted, any windows bearing less than 100 bp of aligned sequence were excluded from analysis because of the imprecise divergence estimate that would necessarily result.
Results
Association between Divergence and Meiotic DSBs
I tested for correlations between nucleotide divergence among Saccharomyces species and rates of meiotic DSB. Preliminarily, I examined the full-genome sequence alignment between S. cerevisiae and Saccharomyces paradoxus without exclusion. A total of 38,905 windows had at least 100 bp of aligned sequence. DSB rate (or more precisely, frequency of DSB-linked ssDNA) and percent sequence difference were significantly (albeit weakly) correlated, but the relationship was “negative” rather than positive (N = 38,905, r = 0.045, P < 0.0001): regions of high DSB rate (including known hot spots) tended to have slightly less sequence differentiation between species.
To avoid regions of the genome under selective constraint as much as possible, I focused analyses on unannotated intergenic regions of the genome: excluding CDS, repetitive regions, binding sites, and anything else with a Saccharomyces Genome Database (Fisk et al. 2006) annotation. Patterns observed were confirmed in intron sequences, but because there are only approximately 400 annotated in the yeast genome and many are small, the power was necessarily limited. Because strong codon bias is known in yeast (e.g., Bennetzen and Hall 1982) and is associated with recombination rate in its intensity (Kliman et al. 2003), I did not analyze synonymous sites.
For examining sequence differences between S. cerevisiae and S. paradoxus, 4,712 intergenic windows fit the criteria described above. Again, DSBs and sequence differentiation in intergenic windows were significantly negatively correlated (fig. 1; N = 4,712, r = 0.252, P < 0.0001). Because this relationship seemed to be driven heavily by the variance in divergence among regions of lowest DSB-linked ssDNA (fig. 1), I excluded all regions associated with denoised ratios below 3.0 (32% of the overall data set), and the association was still significant (N = 3,182, r = 0.156, P < 0.0001). A similar negative relationship between DSBs and divergence was also observed in intron sequences (N = 196, r = 0.196, P = 0.0058; fig. 1). These relationships further held for sequence differentiation between S. cerevisiae and more distant relative Saccharomyces mikatae (intergenic: N = 4,164, r = 0.193, P < 0.0001; intron: N = 193, r = 0.213, P = 0.0030).
FIG. 1.—
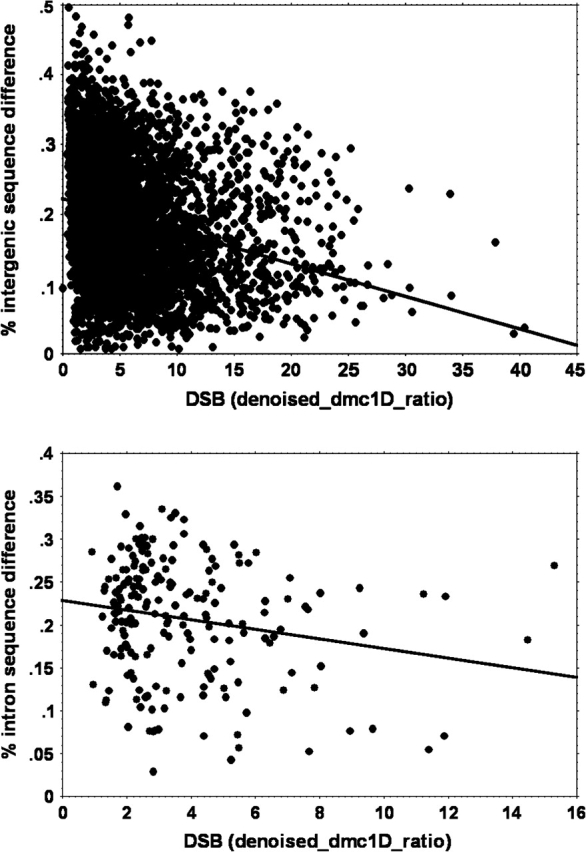
Relationship between local rates of DSB (dmc1D-ratio) and nucleotide difference between Saccharomyces cerevisiae and Saccharomyces paradoxus in intergenic regions (upper panel) or introns (lower panel).
Because DSB rates were measured in S. cerevisiae alone, and because recombinational or DSB hot spots may differ between species, I performed additional analyses wherein only bases that were conserved between S. mikatae and S. paradoxus were examined. This limitation forces a greater fraction of the divergence studied to reflect changes that occurred in the S. cerevisiae lineage (though sites with multiple changes are also included) and thus to potentially better reflect processes that may match the DSB distribution in S. cerevisiae. However, even with this polarization, the negative relationship between DSBs and divergence remained for both intergenic sequences (N = 2,926, r = 0.235, P < 0.0001) and introns (N = 126, r = 0.249, P = 0.0049). This result did not change if an additional outgroup species, Saccharomyces kudriavzevii, was used in addition to S. mikatae to limit the sequence differences between S. cerevisiae and S. paradoxus.
GC content is also known to influence DSBs in Saccharomyces (e.g., Gerton et al. 2000; Petes and Merker 2002), so I further performed a multiple regression between GC content and DSBs on divergence. The highly significant negative relationship between DSBs and divergence remained, and I also observed a significantly negative relationship with GC content in intergenic but not intron divergence (intergenic: overall r = 0.266, overall P < 0.0001, P (DSB) < 0.0001, P (GC content) < 0.0001; intron: overall r = 0.277, overall P = 0.0074, P (DSB) = 0.0115, P (GC content) = 0.1647). These results did not change qualitatively if specific types of mutations (e.g., AT→CG) were considered separately among base differences.
I further tested whether sequences on individual chromosomes may be driving the overall negative relationship. Hence, I repeated the analysis above but analyzed intergenic regions of each chromosome separately. Fourteen of the sixteen chromosomes (all but chromosomes 1 and 6) displayed the significant negative association of DSBs to polarized divergence between S. cerevisiae and S. paradoxus.
Intergenic sequences contain constrained regulatory motifs (or unannotated transcripts), and such sequences may affect the rate of accumulation of interspecies sequence differences (Doniger et al. 2005). This concern is particularly acute in a streamlined genome such as yeast's. Several studies (Haddrill et al. 2005; Halligan and Keightley 2006) have suggested that the “most neutral” sequences are those within short introns (excluding the splice-related sites at the ends). Hence, I repeated the analysis of polarized divergence between S. cerevisiae and S. paradoxus on just introns less than 100 bp in size, but for which there was at least 30 bp of sequence within a particular DSB window. The resultant regression of the few introns that fit these criteria was not statistically significant but with a trend toward negative (N = 62, r = 0.066, P = 0.61).
As all the above analyses used a dmc1 repair-defective mutation to examine DSB distribution, I repeated the analyses using estimates derived from a rad50S mutation, as used in other studies (e.g., Gerton et al. 2000). The results were qualitatively unchanged (though generally weaker or nonsignificant), with or without adding GC content as a covariate.
Association between Divergence and Crossovers or Noncrossovers
To determine whether there was a strong difference between crossover and noncrossover gene conversion outcomes of DSB in the association with divergence, I repeated the analysis using data from Mancera et al. (2008), which used 52,000 markers to infer these positions in all four viable spores derived from 51 meioses. I observed a strong negative association of intergenic DNA sequence divergence with crossovers (N = 587, r = 0.170, P < 0.0001) and noncrossovers (N = 587, r = 0.122, P = 0.0030, see fig. 2) but weaker or no significant relationship of intron divergence with these recombination measures (crossovers: N = 147, r = 0.004, P = 0.9630; noncrossovers: N = 147, r = 0.147, P = 0.0958). As above, I limited the data set to bases conserved between S. mikatae and S. paradoxus and also added GC content to the multiple regression. The patterns described above were unchanged (intergenic/crossover: N = 583, r = 0.161, P < 0.0001; intergenic/noncrossover: N = 583, r = 0.131, P = 0.0015; intron/crossover: N = 105, r = 0.003, P = 0.9749; intron/noncrossover: N = 105, r = 0.138, P = 0.1613).
FIG. 2.—

Relationship between nucleotide difference between Saccharomyces species in intergenic regions and local rates of crossover (upper panel) or noncrossover gene conversion (lower panel).
Association between Strain Divergence within Species and Recombination Measures
Because of the nonsignificant or negative relationships observed between recombination measures and divergence between species, I also examined the relationship between DSBs and sequence diversity within species, as measured by sequence differences between the partially incompatible (Heck et al. 2006; Demogines et al. 2008) S. cerevisiae S288c and SK1 strains. I found that, again, DSB was significantly negatively related to sequence differentiation between strains for intergenic regions (N = 5,090, r = 0.081, P < 0.0001, see fig. 3) but not significantly related to intron differentiation (N = 201, r = 0.016, P = 0.8193). Adding GC content as a covariate did not change these results. The association of DSB with intergenic differentiation became only borderline significant if regions with denoised ratios below 3.0 (36% of the data) were excluded (N = 3,249, r = 0.035, P = 0.0461).
FIG. 3.—
Relationship between local rates of DSB (dmc1D-ratio) and nucleotide difference between Saccharomyces cerevisiae strains in intergenic regions.
Discussion
I tested the hypothesis that mutagenicity from meiotic recombination is a primary driver of sequence divergence between Saccharomyces species. Such a relationship may be predicted given empirical evidence that meiotic recombination is mutagenic in yeast. Contrary to this prediction, intergenic and intronic divergence between species were either uncorrelated or negatively correlated with local estimates of meiotic DSB or crossovers and noncrossovers (figs. 1 and 2). A complicating factor to these analyses is that some of the noncoding regions surveyed surely had selective constraints affecting their rates of divergence (Fay and Benavides 2005), and these constraints may differ even between closely related species (Doniger and Fay 2007). Further, if recombination hot spots move very rapidly on an evolutionary timescale, as studied in humans and chimpanzees (e.g., Coop and Myers 2007), local DSB estimates in S. cerevisiae may not be representative of ancestral DSB estimates. Nonetheless, with the data available, I conclude that the signature of any mutagenic effect of meiotic recombination appears to have been wiped out by other, unknown forces and is not apparent in sequence differences between Saccharomyces species. This finding demonstrates that one cannot always rely upon an association between interspecies nucleotide divergence and local recombination rates to determine whether recombination is mutagenic.
Drosophila studies have typically rejected mechanistic and favored selective explanations for the association of recombination rate to nucleotide diversity within species because of the absence of association between local recombination rate and divergence between species (e.g., Begun and Aquadro 1992; Begun et al. 2007; but see Kulathinal et al. 2008). The results here support the idea that most divergence between species does not result from mutagenicity of meiotic recombination, at least in yeast. Both Begun et al. (2007) and Kulathinal et al. (2008) observed associations between very fine-scale crossover rates and nucleotide diversity within Drosophila species—if these associations result from the action of natural selection, then selection's effect on nucleotide diversity across the genome is essentially ubiquitous.
Kulathinal et al. (2008) recently analyzed whole-genome sequences and recombination rates in the Drosophila pseudoobscura species group and found a statistically significant and positive correlation between nucleotide differences between species and local rates of crossing-over. They proposed a hypothesis that DSB precursors to crossing-over (and gene conversion) may be mutagenic, as perhaps through mutagenicity from strand invasion in the single-stranded phase (akin to results observed in mitochondria, see Faith and Pollock 2003). However, Kulathinal et al. (2008) could not rule out that their measures of divergence may have been confounded with segregating ancestral variation (lineage sorting). My results fail to support their hypothesis of meiotic DSB mutagenicity driving nucleotide divergence in yeast, suggesting falsification of their hypothesis, confounding factors, and/or a possible taxon-specific difference between yeast and Drosophila.
These results suggest that mutagenicity of meiotic DSBs is of minor importance in generating nucleotide divergence between yeast species. Spontaneous mutation rates appear higher in meiotic than mitotic replication (Magni and Von Borstel 1962), and it could be that mismatch repair is less efficient in meiosis than mitosis independent of rate of DSBs. As such, a high basal mutation rate may “swamp” the mutagenic signature of DSBs in particular. Alternatively, factors causing mutations during vegetative reproduction may be more important to generating the base changes that result in nucleotide divergence between species. However, in contrast to this hypothesis, I failed to find that spontaneous base substitution mutations arising in vegetatively cultured yeast (Lynch et al. 2008) are more likely to appear in regions of high interspecies nucleotide divergence or regions of high meiotic DSB (supplementary fig. 1, Supplementary Material online).
One curiosity with the analysis presented here is the apparent negative association between local estimates of meiotic DSB and nucleotide divergence between species in some of my analyses, particularly in intergenic regions. Although GC content was related to levels of meiotic DSB in this data set, it was not consistently related to nucleotide divergence in a multiple regression with DSB (see also Hawk et al. 2005). The observed negative association could suggest that regions of high meiotic DSB are less likely to produce or accumulate new mutations or rates of meiotic DSB and recombination are reduced in regions of high spontaneous mutation potentially resulting from weaker pairing at heterozygous sites (e.g., Hunter et al. 1996). Biased gene conversion (BGC) could be involved in the former possibility and could reduce sequence diversity within species. DSB sites are usually recipients of genetic information, so a distally regulated recombination hot spot (e.g., Neumann and Jeffreys 2006) could persist but lose diversity because of BGC. Such a BGC-driven process could also potentially reduce sequence differences between two closely related species if they share ancestral polymorphisms or exchange genetic material. I cannot rule out this possibility, but these species are fairly highly diverged (average 18% divergence in both intergenic and intron regions between S. cerevisiae and S. paradoxus, compared with average <1% between S288c and SK1), suggesting that few ancestral polymorphisms persist. Further, I failed to observe that only particular types of changes between species were associated with DSBs (e.g., AT→GC) as might be predicted by BGC (e.g., Galtier and Duret 2007). Alternatively, the negative relationship could simply result from regions of high meiotic DSB being more constrained by natural selection. As meiotic DSBs are especially common in promoter-containing regions (Wu and Lichten 1994; Baudat and Nicolas 1997; Mancera et al. 2008), this possibility cannot be excluded, but this hypothesis fails to explain why a negative association between DSBs and divergence is also observed in some intron analyses. With the data presented, we can only speculate as to the cause of this apparent negative association, but mutagenesis from meiotic recombination is clearly not the primary driver of sequence divergence between Saccharomyces species.
Supplementary Material
Supplementary table S1 and figure 1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
I thank J. Fay, C. Jones, R. Kliman, R. Kulathinal, T. Petes, members of the Noor laboratory, and two anonymous reviewers for helpful comments and discussions. I also thank J. Strathern for sharing his unpublished results. This research was funded by National Science Foundation grants 0509780 and 0715484 and National Institutes of Health grant GM076051.
References
- Aguade M, Miyashita N, Langley CH. Reduced variation in the yellow-achaete-scute region in natural populations of Drosophila melanogaster. Genetics. 1989;122:607–615. doi: 10.1093/genetics/122.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquadro CF. Insights into the evolutionary process from patterns of DNA sequence variability. Curr Opin Genet Dev. 1997;7:835–840. doi: 10.1016/s0959-437x(97)80048-2. [DOI] [PubMed] [Google Scholar]
- Baudat F, Nicolas A. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc Natl Acad Sci USA. 1997;94:5213–5218. doi: 10.1073/pnas.94.10.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature. 1992;356:519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, et al. (13 co-authors) Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Hall BD. Codon selection in yeast. J Biol Chem. 1982;257:3026–3031. [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, et al. (12 co-authors) Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitzblau HG, Bell GW, Rodriguez J, Bell SP, Hochwagen A. Mapping of meiotic single-stranded DNA reveals double-strand-break hotspots near centromeres and telomeres. Curr Biol. 2007;17:2003–2012. doi: 10.1016/j.cub.2007.10.066. [DOI] [PubMed] [Google Scholar]
- Buhler C, Borde V, Lichten M. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e324. doi: 10.1371/journal.pbio.0050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Myers SR. Live hot, die young: transmission distortion in recombination hotspots. PLoS Genet. 2007;3:e35. doi: 10.1371/journal.pgen.0030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demogines A, Wong A, Aquadro C, Alani E. Incompatibilities involving yeast mismatch repair genes: a role for genetic modifiers and implications for disease penetrance and variation in genomic mutation rates. PLoS Genet. 2008;4:e1000103. doi: 10.1371/journal.pgen.1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger SW, Fay JC. Frequent gain and loss of functional transcription factor binding sites. PLoS Comput Biol. 2007;3:e99. doi: 10.1371/journal.pcbi.0030099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger SW, Huh J, Fay JC. Identification of functional transcription factor binding sites using closely related Saccharomyces species. Genome Res. 2005;15:701–709. doi: 10.1101/gr.3578205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito MS, Bruschi CV. Diploid yeast cells yield homozygous spontaneous mutations. Curr Genet. 1993;23:430–434. doi: 10.1007/BF00312630. [DOI] [PubMed] [Google Scholar]
- Faith JJ, Pollock DD. Likelihood analysis of asymmetrical mutation bias gradients in vertebrate mitochondrial genomes. Genetics. 2003;165:735–745. doi: 10.1093/genetics/165.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Benavides JA. Hypervariable noncoding sequences in Saccharomyces cerevisiae. Genetics. 2005;170:1575–1587. doi: 10.1534/genetics.105.042283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov DA, Gerrard DT. High mutation rates in human and ape pseudoautosomal genes. Gene. 2003;317:67–77. doi: 10.1016/s0378-1119(03)00697-8. [DOI] [PubMed] [Google Scholar]
- Fisk DG, Ball CA, Dolinski K, Engel SR, Hong EL, Issel-Tarver L, Schwartz K, Sethuraman A, Botstein D, Cherry JM. Saccharomyces cerevisiae S288C genome annotation: a working hypothesis. Yeast. 2006;23:857–865. doi: 10.1002/yea.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Duret L. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 2007;23:273–277. doi: 10.1016/j.tig.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Gerton JL, DeRisi J, Shroff R, Lichten M, Brown PO, Petes TD. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, et al. (16 co-authors) Life with 6000 genes. Science. 1996;274(546):563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Haddrill PR, Charlesworth B, Halligan DL, Andolfatto P. Patterns of intron sequence evolution in Drosophila are dependent upon length and GC content. Genome Biol. 2005;6:R67. doi: 10.1186/gb-2005-6-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan DL, Keightley PD. Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Res. 2006;16:875–884. doi: 10.1101/gr.5022906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawk JD, Stefanovic L, Boyer JC, Petes TD, Farber RA. Variation in efficiency of DNA mismatch repair at different sites in the yeast genome. Proc Natl Acad Sci USA. 2005;102:8639–8643. doi: 10.1073/pnas.0503415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck JA, Argueso JL, Gemici Z, Reeves RG, Bernard A, Aquadro CF, Alani E. Negative epistasis between natural variants of the Saccharomyces cerevisiae MLH1 and PMS1 genes results in a defect in mismatch repair. Proc Natl Acad Sci USA. 2006;103:3256–3261. doi: 10.1073/pnas.0510998103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann I, Ebersberger I, Ptak SE, Paabo S, Przeworski M. A neutral explanation for the correlation of diversity with recombination rates in humans. Am J Hum Genet. 2003;72:1527–1535. doi: 10.1086/375657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N, Chambers SR, Louis EJ, Borts RH. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 1996;15:1726–1733. [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kliman RM, Irving N, Santiago M. Selection conflicts, gene expression, and codon usage trends in yeast. J Mol Evol. 2003;57:98–109. doi: 10.1007/s00239-003-2459-9. [DOI] [PubMed] [Google Scholar]
- Kulathinal RJ, Bennett SM, Fitzpatrick CL, Noor MA. Fine-scale mapping of recombination rate in Drosophila refines its correlation to diversity and divergence. Proc Natl Acad Sci USA. 2008;105:10051–10056. doi: 10.1073/pnas.0801848105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Hurst LD. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 2002;18:337–340. doi: 10.1016/s0168-9525(02)02669-0. [DOI] [PubMed] [Google Scholar]
- Lynch M, Sung W, Morris K, et al. (11 co-authors) A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci USA. 2008;105:9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magni GE, Von Borstel RC. Different rates of spontaneous mutation during mitosis and meiosis in yeast. Genetics. 1962;47:1097–1108. doi: 10.1093/genetics/47.8.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MaynardSmith J, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- Nachman MW. Variation in recombination rate across the genome: evidence and implications. Curr Opin Genet Dev. 2002;12:657–663. doi: 10.1016/s0959-437x(02)00358-1. [DOI] [PubMed] [Google Scholar]
- Neumann R, Jeffreys AJ. Polymorphism in the activity of human crossover hotspots independent of local DNA sequence variation. Hum Mol Genet. 2006;15:1401–1411. doi: 10.1093/hmg/ddl063. [DOI] [PubMed] [Google Scholar]
- Nordborg M, Charlesworth B, Charlesworth D. The effect of recombination on background selection. Genet Res. 1996;67:159–174. doi: 10.1017/s0016672300033619. [DOI] [PubMed] [Google Scholar]
- Petes TD, Merker JD. Context dependence of meiotic recombination hotspots in yeast: the relationship between recombination activity of a reporter construct and base composition. Genetics. 2002;162:2049–2052. doi: 10.1093/genetics/162.4.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roselius K, Stephan W, Stadler T. The relationship of nucleotide polymorphism, recombination rate and selection in wild tomato species. Genetics. 2005;171:753–763. doi: 10.1534/genetics.105.043877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W, Langley CH. DNA polymorphism in lycopersicon and crossing-over per physical length. Genetics. 1998;150:1585–1603. doi: 10.1093/genetics/150.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern JN, Shafer BK, McGill CB. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon MI, U'Ren J, Tenaillon O, Gaut BS. Selection versus demography: a multilocus investigation of the domestication process in maize. Mol Biol Evol. 2004;21:1214–1225. doi: 10.1093/molbev/msh102. [DOI] [PubMed] [Google Scholar]
- Wu TC, Lichten M. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science. 1994;263:515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



