Abstract
To reach sites of inflammation, neutrophils execute a series of adhesion and migration events that include transmigration through the vascular endothelium and chemotaxis through the vicinal extracellular matrix until contact is made with the point of injury or infection. These in vivo microenvironments differ in their mechanical properties. Using polyacrylamide gels of physiologically relevant elasticity in the range of 5 to 100 kPa and coated with fibronectin, we tested how neutrophil adhesion, spreading, and migration were affected by substrate stiffness. Neutrophils on the softest gels showed only small changes in spread area, whereas on the stiffest gels they showed a 3-fold increase. During adhesion and migration, the magnitudes of the distortions induced in the gel substrate were independent of substrate stiffness, corresponding to the generation of significantly larger traction stresses on the stiffer gels. Cells migrated more slowly but more persistently on stiffer substrates, which resulted in neutrophils moving greater distances over time despite their slower speeds. The largest tractions were localized to the posterior of migrating neutrophils and were independent of substrate stiffness. Finally, the phosphatidylinositol 3-kinase inhibitor LY294002 obviated the ability to sense substrate stiffness, suggesting that phosphatidylinositol 3-kinase plays a mechanistic role in neutrophil mechanosensing.
Introduction
Cells interact with their environment both biochemically and physically. The biochemical interaction has been widely studied in several different systems. Only more recently has the study of how the physical environment affects cell function gained momentum.1–3 This process, termed mechanosensing, has been shown to be relevant in several different cell types. Endothelial cells are known to become activated in response to shear flow,4 fibroblasts have been observed to migrate toward stiffer substrates,5 and cells undergoing mitosis have been shown to redistribute proteins in response to both intrinsic and externally induced shape asymmetries.6 These findings have shown that cells are able to respond to mechanical cues in their local environment. Changes in cell shape and speed based on substrate stiffness have been shown for fibroblasts,1,7 neurites,8 myocytes,9 and smooth muscle cells.10 Substrate elasticity has also been shown to direct differentiation of bone marrow–derived mesenchymal stem cells.11 These findings illustrate the importance of determining cellular functions in mechanical environments that more accurately mimic in vivo conditions. As these environments can have a Young modulus ranging from a few pascals to hundreds of kilopascals,11 substrates of tunable stiffness, such as those made of polyacrylamide, have been used.1,5,7,9–14 Specifically, this study made use of gels with a Young's modulus in the range of 5 to 100 kPa, consistent with reported values in endothelial cells,15–17 breast tissue,18,19 and skeletal muscle16,20 (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Furthermore, this range is reflective of pathophysiologic changes in tissue, such as atherosclerosis21 and fibrosis.22
Of the studies investigating the interaction between cells and flexible substrates, relatively few12,14 have focused on leukocytes. Neutrophils, also known as polymorphonuclear leukocytes, are the most abundant of the circulating white blood cells and are early responders to sites of infection and injury.23 They are highly motile cells, moving from the blood vessels, through tissue and the extracellular matrix, to the point of injury or infection. Contact with components of the extracellular matrix, including fibronectin, has been shown to augment intracellular signaling and lead to cellular responses to soluble mediators that do not occur in the absence of adhesion.24 Through polarization and repetitive cycles of adhesion, extension, contraction, and de-adhesion, neutrophils are able to migrate through various extravascular environments in a short period of time.25 During this process, neutrophils interpret several extracellular signals from a variety of sources to direct their migration.26 In addition to these biochemical cues, the local physical environment offers mechanical cues,27 whose role in directing neutrophil activity has yet to be explained. Because the mechanical properties of cells and tissues can be affected by disease and inflammation,3,21,22,28 we investigate whether a neutrophil's response to inflammatory signals is mechanosensitive.
In this report, we explore the role of substrate rigidity on the adhesion, migration, and force generation of human neutrophils. Using fibronectin-coated polyacrylamide substrates across a physiologically relevant range of stiffness, we show that the elastic properties of the substrate dictate the ability and efficiency of the neutrophil to adhere and migrate. Moreover, blockade of phosphatidylinositol 3-kinase (PI3K) activity is sufficient to obviate stiffness-dependent differences in neutrophil chemotaxis, suggesting a regulatory role in neutrophil mechanosensing.
Methods
Reagents
Histopaque 1077, dextran (average molecular weight, 400-500 kDa), formyl-methionyl-leucyl-phenylalanine (fMLP), and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich; Liebovitz L-15 medium and Hanks Balanced Salt Solutions were purchased from Invitrogen; polymyxin B and Sulfo-Sanpah were purchased from Pierce Biotechnology. Fibronectin was purchased from Fisher Scientific; LY294002 was purchased from Calbiochem.
Cell preparation
Neutrophils were isolated from healthy human volunteers by collection into ethylenediaminetetraacetic acid–containing Vacutainer tubes (BD Biosciences). Histopaque was used for the initial cell separation followed by gravity sedimentation through 3% dextran. Contaminating erythrocytes were removed by hypotonic lysis, yielding a neutrophil purity of more than 95%. Neutrophils were suspended in Hanks Balanced Salt Solution (without Ca2+/Mg2+) on ice until use in the experiments. All reagents contained less than 0.1 pg/mL endotoxin as determined by Limulus amoebocyte lysate screening (BioWhittaker Inc). When necessary, endotoxin removal was achieved using immobilized polymyxin B (Affinity Pak Detoxi-Gel; Pierce Biotechnology) followed by repeat Limulus testing. Institutional Review Board approval was obtained from the Rhode Island Hospital's Committee on Protection of Human Subjects to allow donation of venous blood from normal, healthy donors for isolation of peripheral blood neutrophils. In addition, informed consent was obtained in accordance with the Declaration of Helsinki.
For spreading, traction, and chemokinetic migration assays, approximately 106 neutrophils were resuspended at 37°C in 2 mL Liebovitz L-15 medium with 2 mg/mL glucose added. Neutrophils were allowed to settle for 5 minutes before introducing fMLP (100 nM final concentration) to activate and induce migration. For chemotaxis, a femptotip (Eppendorf North America) was filled with a 1-mM concentration of fMLP and placed with the tip at the gel surface. When indicated, neutrophils were pretreated on ice for 30 minutes with either 20 μM LY294002 or 0.02% DMSO as a vehicle control, in L-15 with 2 mg/mL glucose.
Substrate preparation
Substrates were prepared at 4°C following a method described by Pelham and Wang.1 Briefly, a solution of acrylamide and bis-acrylamide was mixed with 0.2 μm fluorescent beads (Invitrogen) and polymerized using tetramethylethylenediamene and ammonium persulfate. Gels were made in heatable glass-bottom DeltaT dishes (Bioptechs Inc) using AbGene frames (AbGene Ltd) as molds. The gel mixture was centrifuged at 300g for 20 minutes at 4°C before polymerization, ensuring that the fluorescent beads remained in a subapical layer of the surface. After centrifugation, the gels were allowed to polymerize at room temperature. Once polymerized, gels were soaked overnight in phosphate-buffered saline, allowing unpolymerized acrylamide to diffuse out. The final size of the gels was approximately 1 cm × 1 cm × 300 μm.
Gel stiffness was regulated by varying the percentage of bis-acrylamide in relation to the percentage of acrylamide in the initial mixture,12 and elasticity was confirmed with a magnetically driven plate rheometer.29,30 The elasticity of the gels has been shown to be unaffected by the protein-coating procedure,1 and the density of protein on the surface of the gel was unaffected by the elasticity of the gels (supplemental data; supplemental Figure 1).5,8 The gels were coated with fibronectin using the chemical crosslinker Sulfo-Sanpah. Sulfo-Sanpah was allowed to covalently bond to the acrylamide gel for approximately 1 hour at room temperature. Fibronectin, 10 μg/mL, was then added to the gel and exposed to ultraviolet light for 10 minutes, cross-linking the protein.
Microscopy
A Nikon TE-2000U inverted microscope, outfitted with a Bioptechs Inc stage heater and a 20× Nikon Plan APO objective, was used for all experiments. Both differential interference contrast (DIC) and fluorescence images were captured over 30 minutes on a 10-second interval using the Elements program (Nikon Inc). All data were analyzed using ImageJ (National Institutes of Health) and MATLAB (MathWorks) computational software.
Migration analysis
Cells were tracked using a program written in MATLAB. The cell boundary was determined through thresholding of the DIC images, and the cell center determined through a center of mass calculation based on the cell border. Using these trajectories, the mean squared displacement (MSD) was calculated as
 |
where r⃗ is the position of the cell, Δt is the time interval, and (〈⋯〉) brackets represent a time average. The MSD is discussed in terms of the slope, as this relates directly to a classification of the behavior of the motion. The MSD can be described as
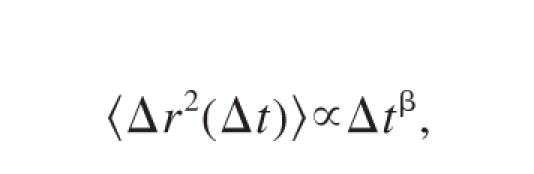 |
where the value of β characterizes the behavior. A value of β = 1 describes random motion. A value of β = 2 represents linear (ballistic) motion at a constant speed and is a theoretical upper limit. As the persistence of the motion increases, β increases from 1 to 2. When plotted as a log-log plot, β is measured as the slope of the data. For further discussion of the MSD, refer to our previous work.29 The root mean square speed was calculated by taking the square root of the calculated MSD at the smallest time interval, (Δt = 10 s), and dividing by (Δt).
The distribution of turning angles during cell migration was determined by the angle of displacement with respect to the previous step for the time interval indicated. Positive angles indicate a direction counterclockwise to the previous step. Negative angles indicate a direction clockwise with respect to the previous step. Persistence, as measured in the inset of Figure 4B, was defined as the percentage of steps for which −π/2 < θ < π/2.
Figure 4.
Analysis of mobility and directionality of neutrophils migrating on substrates of different stiffness. (A) The MSD is plotted as a function of time between steps. Neutrophils on the softer gels (5 and 10 kPa) had a slope of ∼ 1, indicative of diffusive behavior. As the stiffness increased, the slope of the MSD increased, indicating more persistent behavior. The MSD was also larger for neutrophils on softer gels. (B) The angular distribution of steps is shown for each stiffness based on a 10-second interval between frames. The stiffer gels showed a more peaked distribution ∼ 0°, confirming that corresponding motion was more directed. Inset: The persistence, defined in the inset as the percentage of steps for which −π/2 < θ < π/2, is shown for time steps of 10, 20, and 30 seconds. The cells were more persistent on the stiffer gels. Error bars represent SE. *P < .05 vs 10 kPa.
Displacement measurements and Fourier transform traction cytometry
The displacement field of the substrate was estimated by comparing images of the diffuse fluorescence intensity of the beads embedded in the gel over time. Each gel was checked to ensure that the beads were contained in a layer at the surface within the focal depth of the objective. Before calculating displacements, stacks of images were corrected for drift using ImageJ. Images of the unstrained gel surface, before any neutrophil attachment, were used as reference images for bead displacement. The bead density at the surface of the gel was approximately 50 000 beads per field of view (1392 × 1040 pixels). The intensity distribution of the unstrained image u(x,y) was transformed by a displacement function f(x,y) to fit the intensity distribution of the strained image g(x,y) using the Gauss-Newton-Algorithm, minimizing the sum of squares of the errors between the transformed image v(x,y) = f(x,y) · u(x,y) and the strained image g(x,y).
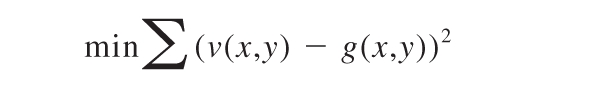 |
The transformation of the reference fluorescence image by the displacement function was performed for each individual pixel, and the squares of the errors (differences) between the transformed and the strained image were summed over all pixels. To achieve an accurate displacement field, the computation of f(x,y) was done iteratively. The images were first strongly smoothed with a Gaussian filter and rescaled to a small image size. This filtering process ensured that large-scale deformations could be followed and that the fit did not end up in a local minimum. The first computation of f(x,y) gave a rough estimate, v1(x,y), for the displacement field. With every iteration, the smoothing filter size was reduced and the image size increased, using the previous iteration as the source image, ie,
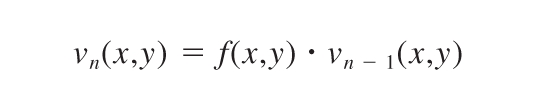 |
The final f(x,y), which represented the displacement field, was computed as the last iteration without smoothing the source image vn(x,y). To avoid fitting to noise in the image, the intensity of a pixel was only taken into account if the gradient between neighboring pixels was sufficiently large. Consequently, to avoid discontinuities in the displacement function f(x,y), it was smoothed with a Gaussian filter with a sigma of 2, 3, or 5 pixels, depending on the iteration step. The resultant displacement matrix f(x,y), which gave a displacement value for each pixel representing the function that best fit the unstrained image to the strained image, was used in the traction calculations.
Tractions were computed following the approach described by Butler et al.31 This method solves the inverse Bousinessq equations32 in Fourier space to simplify calculations. This technique has been shown to be equivalent33 to other computational methods.34 After transforming the displacement field into Fourier space, Fourier components at wave vectors k = 0 are set to zero to eliminate translational artifacts (ie, drift). The displacements are then multiplied by K(k)−1, which is the Fourier transform of the Bousinessq solution to the Green function that maps tractions to displacements on an infinite elastic half-space. By applying the inverse Fourier transform to the resultant product, the tractions exerted on the gel are found.
Root mean square displacement and shear stress magnitudes were determined using only the region directly underneath the neutrophil of interest. The cell boundary was determined using the corresponding DIC image taken during the experiment.
Statistics
Data were pooled from a minimum of at least 3 different donors, with the total number of cells for each condition indicated in the figures. Analysis of variance with Newman-Keuls post-hoc analysis as appropriate was performed using MATLAB. The null hypothesis was rejected if P was less than .01.
Results
Neutrophil spreading increases with substrate stiffness
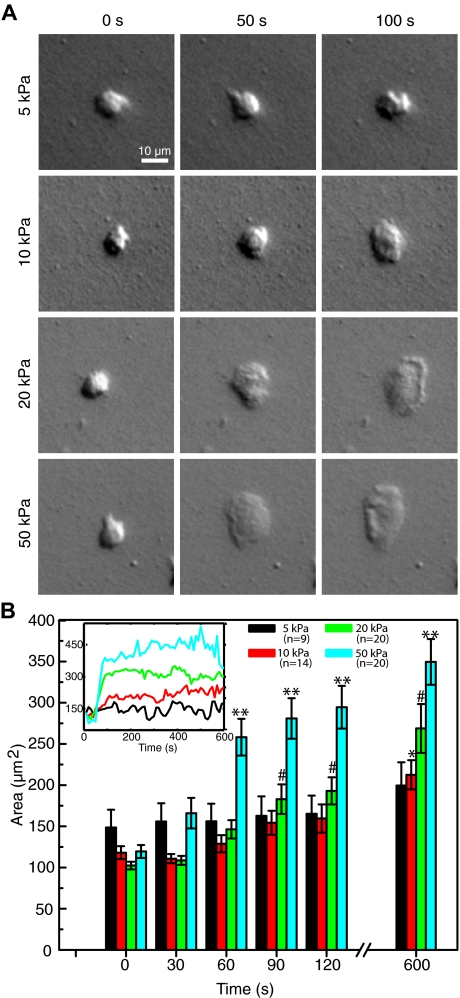
Neutrophils were allowed to settle on fibronectin-conjugated polyacrylamide substrates ranging in stiffness from 5 to 50 kPa. Example DIC images of neutrophils on each stiffness are shown in Figure 1A. On settling, the cells were approximately 100 μm2 in area, or the same size as cells suspended in the fluid phase, as expected.35 Cell area is shown as a function of time in Figure 1B. Cells on the 5-kPa gel showed no statistically significant change in average size over a period of 10 minutes. During the same time period, cells on the 10-kPa gel showed a significant increase in size (80% ± 15%, P < .001). The largest differences over this time span, however, were seen in the cells on the 20-kPa (162% ± 29% increase, P < .001) and 50-kPa (186% ± 22% increase, P < .001) gels. P values reflect significance with respect to t = 0 values.
Figure 1.
Comparison of neutrophil spreading on substrates of different stiffness. (A) DIC images show neutrophils spreading over a period of 100 seconds on 4 gels of different stiffness (5, 10, 20, and 50 kPa), each at 3 time points. (B) The average spread area for each stiffness condition is plotted over time. Neutrophils on the softer gels maintained a smaller area and spread significantly less than the neutrophils on the stiffer gels. The majority of spreading was completed within the first 120 seconds. Neutrophils on the softest substrate showed no statistical change in size, even after 10 minutes, whereas those on the stiffest substrate more than tripled in area in the same amount of time. The number of cells measured for each condition is indicated in the legend. Error bars represent SE measurements. *Significant increase for the 10-kPa gels (with respect to the t = 0 point). #Significant increase for the 20-kPa gels (with respect to the t = 0 point). **Significant increase for the 50-kPa gels (with respect to the t = 0 point). Inset: The areas of the 4 neutrophils shown in the DIC images are plotted for the full 10 minutes.
When spreading, the cell area increased rapidly, reaching approximately 75% or more of its maximum area within the first 120 seconds. This is seen in the inset of Figure 1B, which shows the area as a function of time for the 4 individual cells that are shown in Figure 1A. This sharp increase was followed by a steady state where cell area showed only small fluctuations but on average remained constant for the duration of the experiment (30 minutes). In addition, cells on the stiffer substrates (20 and 50 kPa) often showed distinctive morphologic features not seen on the softer substrates. These included ridges forming along what would become the leading edge and a distinctive edge ruffling pattern during migration (Figure 1A; supplemental Video 4).
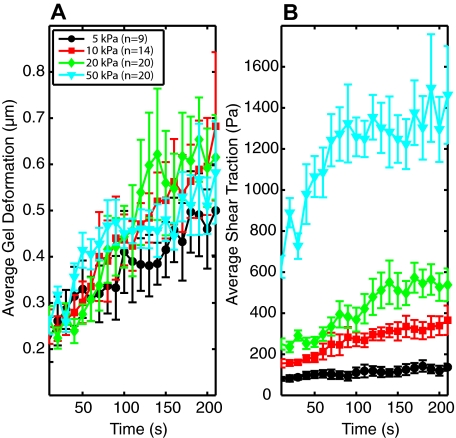
Neutrophils exert greater forces on stiffer substrates
The average magnitude of the displacements in the gel, as determined by the fluorescent beads, was found to be independent of gel stiffness (Figure 2A). This resulted, however, in an increase in the average stress generated by the neutrophils on the stiffer gels. In other words, neutrophils were able to exert significantly stronger forces on the stiffer substrates (Figure 2B). We report the root mean squared magnitudes of stress to avoid any complications of a cell having a larger area and, thus, more adhesion sites, on a stiffer gel. Because traction and displacement values were calculated for each pixel, root mean square values were calculated by summing the squared magnitudes for each pixel covered by the cell and dividing by the number of pixels summed before taking the square root. Thus, the values presented in Figure 2 represent the root mean squared magnitude of a pixel under the neutrophil and are unaffected by the spread or size of the cell. All stress values were at least an order of magnitude greater than the background noise measured in the system. Root mean squared traction stress values showed little fluctuation after the first 3 minutes.
Figure 2.
Comparison of root mean square gel deformation and traction stress exerted by neutrophils on substrates of different stiffness. (A) The root mean square value of the displacements of the fiducial markers embedded in the gel under the area of the cell is plotted as a function of time. There is no statistical difference between gels of different stiffness. (B) The root mean square stress applied by the neutrophil on the gel in the area occupied by the cell is plotted as a function of time for each stiffness. Cell outlines for calculations were determined from DIC images. Error bars represent SEM.
Chemokinetic migration efficiency is a function of substrate stiffness
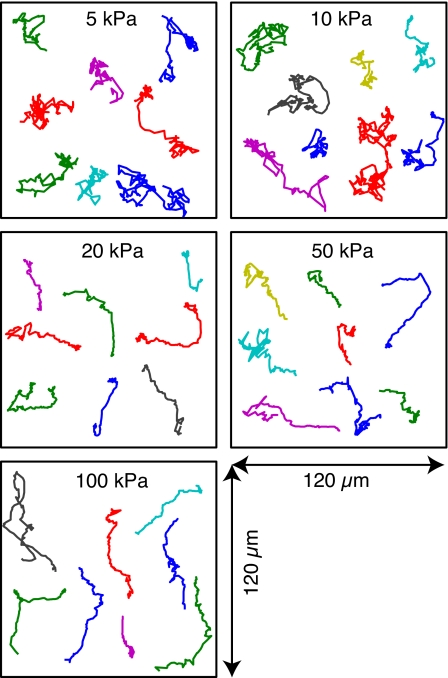
An inverse correlation between cell speed and substrate stiffness has been reported for fibroblasts5,7 and smooth muscle cells.36 For short time scales, the neutrophils on the stiffer gels moved with significantly smaller average speeds, indicating that this inverse correlation holds for neutrophils as well. Random migration paths of neutrophils on gels of different stiffness are shown in Figure 3. On the softer gels (5 and 10 kPa), the neutrophils exhibited paths characteristic of random walks. This result was confirmed by evaluating the MSD, which showed a slope of approximately 1 (Figure 4A), where a value of 1.0 defines random diffusive behavior. As the stiffness of the substrate was increased, the migration paths of the neutrophils appeared to become more directed (Figure 3). Furthermore, the cells moved more slowly (Figure 4A) and with increased persistence (Figure 4B). In this case, a value of 2.0 defines purely linear motility at a constant speed.
Figure 3.
Migration plots showing 8 random neutrophil migration paths for gels of 5 different stiffnesses. On the softer gels (5 and 10 kPa), the neutrophils moved quickly and changed direction often. On the stiffer gels, the neutrophils were more efficient, moving slower but showing a much greater persistence and changing direction less often.
The increase in the slope of the MSD suggests that directionality develops with substrate stiffness. Angular distributions showed that, while the neutrophils moved faster on the softer gels, they changed direction more often. Figure 4B shows the angular distribution of turning angles for each gel stiffness. The distribution peak increased with increasing stiffness. The inset in Figure 4B shows the persistence value, a measure of how often a cell continues to travel in its current direction of motion. The persistence is defined as the probability that a step deviates in a direction within 90° of the previous step. This value also increased with substrate stiffness, consistent with the results of the MSD and the migration paths shown in Figure 3. On the softer gels (5 and 10 kPa), the neutrophils moved a greater total distance, as determined by path length, but changed direction more often, resulting in less displacement. The cells were seen to traverse the same areas repeatedly, moving quickly but not getting very far. On the stiffer gels, the cells showed more directed migration, changing direction less often, but moving less with each step. As a result, the neutrophils actually traveled a greater distance after several minutes while moving more slowly.
Stiffness-dependent migration behaviors remain during chemotaxis
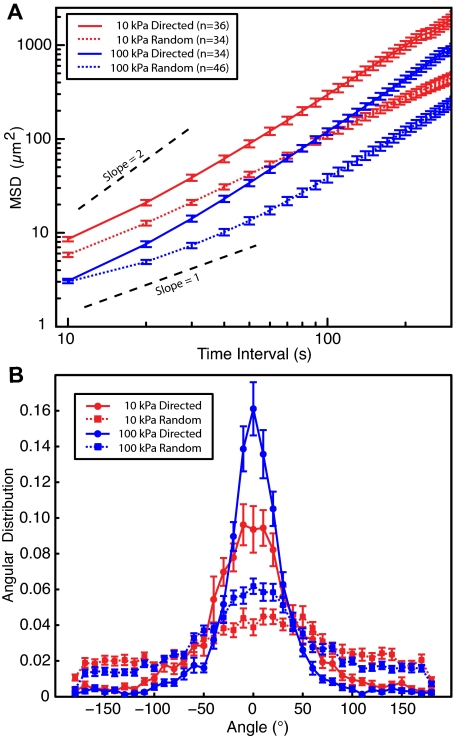
Neutrophils migrating toward a point source of fMLP exhibited the same trends as neutrophils undergoing random migration. Gels with a stiffness of 10 and 100 kPa were chosen as representative of the 2 ends of the stiffness spectrum studied. It was necessary to make a small modification to the tracking algorithm for the case of chemotactic migration. This modification was more sensitive to small variations in cell shape, especially along the leading edge, and resulted in greater noise in the persistence measurements with the 10-second interval used for the chemokinetic experiments. As such, a 60-second interval was used for measuring the angular distribution and persistence for the directed migration. The chemokinetic data plotted in Figure 5 represent the equivalent data from Figure 4 analyzed by the new method. Both methods revealed equivalent trends in the data.
Figure 5.
Comparison of chemokinetic and chemotactic behavior. (A) The MSDs for neutrophils on 10- and 100-kPa substrates are shown for both random and directed migration. The cells move more efficiently on the stiffer substrate. (B) The angular distribution of steps is shown for neutrophils undergoing both chemokinesis and chemotaxis on a 60-second interval. Neutrophils chemotaxing toward a pipette tip showed more peaked distributions than neutrophils randomly migrating. For both chemokinetic and chemotactic behavior, however, the angular distribution of steps showed a greater peak on the 100-kPa substrates compared with the 10-kPa substrate for each condition. Migration is thus more persistent on stiffer substrates.
Neutrophils on the softer gels showed a greater MSD during chemotaxis as shown in Figure 5A. For both the soft and the stiff gels, the slope of the directed MSD was close to 2, consistent with the prediction for directed behavior. Furthermore, the values of the MSD on the smallest interval were consistent with the results of the chemokinesis. Directionality on the stiffer gels also remained greater during chemotaxis (Figure 5B). The angular distributions of steps on both gels were more peaked during directed migration than random migration, as expected. There was still, however, a significant difference between the 10- and the 100-kPa gels, with the neutrophils on the stiffer substrate having a more peaked distribution. This narrower distribution resulted in straighter migration trajectories and thus greater directionality. These results indicate that the behaviors displayed during chemokinesis remain while the cells undergo chemotaxis.
Neutrophil tractions are concentrated in the posterior of the cell
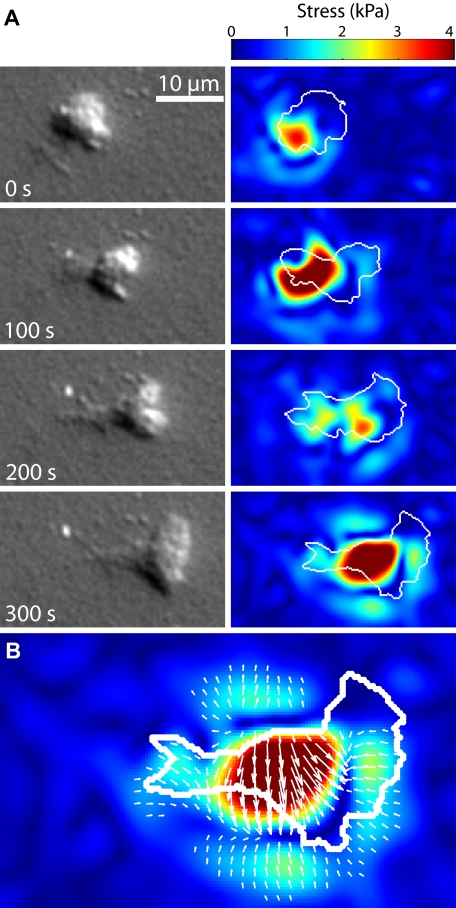
Smith et al14 reported that a migrating neutrophil exerts a concentration of tractions in the rear of the polarized cell, driving the cell body forward. Our results confirmed that the greatest tractions were localized in the rear of the cell relative the cell front, and that this distribution was independent of substrate stiffness. An example of a cell migrating on a 10-kPa gel is shown in Figure 6A and in supplemental Video 2. The white outlines of the cell were determined from DIC images. The greatest tractions exerted by the migrating cell remained in its posterior throughout the time lapse. Figure 6B shows that the large traction in the posterior was balanced by more numerous small tractions located along the periphery and the leading edge. In addition, the evolution of tractions concentrated in the rear of the cell could be seen to evolve cyclically, resulting in an intermediate step with a reduction of traction as seen in the third panel of Figure 6A. This result is consistent with typical migration models involving cyclical steps of protrusion, adhesion, and retraction,37 and provides explicit evidence that such stepwise models are still applicable, even for the fast-moving neutrophil.
Figure 6.
Distribution of tractional stresses in a migrating neutrophil. (A) A time series of images showing a neutrophil migrating chemokinetically to the right on a 10-kPa gel. The DIC images show that the cell has moved over a full body length in 300 seconds (supplemental Video 2). The images to the right of the DIC images are the corresponding traction images for those time points. The white outline over the traction images represents the approximate boundary as determined from the DIC images. The largest stresses occurred in the posterior of the neutrophil, a feature independent of gel stiffness. The full series of images for this particular example can be seen in supplemental Video 3. (B) A larger version of the last traction image in the series from panel A is shown. The white arrows represent the overall direction and magnitude of the applied tractional stresses. The few large tractions in the center of the cell were balanced by more numerous small tractions in the surrounding areas.
Inhibition of PI3K eliminates a neutrophil's ability to sense the differences in substrate stiffness during chemotaxis
The role of PI3K in leukocyte chemotaxis to fMLP has been shown to include fine regulation rather than absolute dependence.38,39 For example, PI3K inhibition delayed but did not abrogate polarization and chemotaxis of murine neutrophils.38 Inhibition of PI3K in neutrophilic differentiated HL60 cells impaired the ability of migrating cells to respond rapidly to changes in direction of a CXCL8 gradient while otherwise allowing normal chemotaxis.39 Here we determined whether PI3K is a mechanism underlying the sensitivity of PMNs to substrate stiffness.
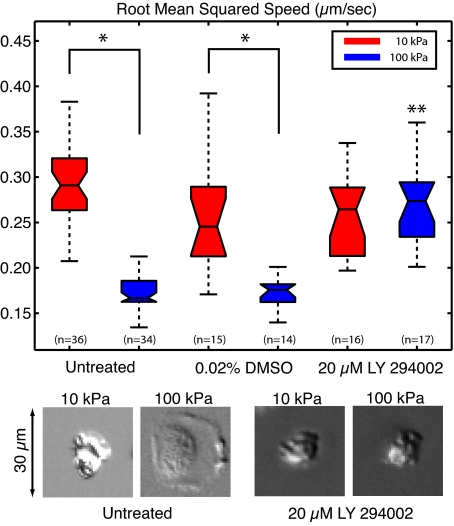
Neutrophils treated with the PI3K inhibitor LY 294002 behaved identically whether they were placed on 10- or 100-kPa gel substrates. Cells were pretreated for 30 minutes on ice with 20 μM LY294002, before being placed on a substrate with a micropipette filled with the chemoattractant fMLP. Drug activity was confirmed by inhibition of phospho-AKT by immunoblotting (data not shown). Cells treated with 0.02% DMSO, a vehicle control, showed no difference from cells that were untreated. In both cases, the root mean squared cell speed was approximately 2 times greater on the 10-kPa substrates, compared with the 100-kPa substrate (Figure 7). The difference between 10- and 100-kPa substrates was significant in both the untreated and DMSO-treated neutrophils, as indicated in Figure 7. When the neutrophils were pretreated with the PI3K inhibitor, the cells were indistinguishable on the 10- and 100-kPa substrates. The PI3K inhibited cells showed a root mean square speed that was significantly different from both the untreated and DMSO-treated cells on the 100-kPa substrates (Figure 7). Furthermore, cells pretreated with the PI3K inhibitor showed a reduced ability to spread during migration on the stiffer substrates, as shown in the DIC images at the bottom of Figure 7 and supplemental Videos 3 through 6. These findings suggest a mechanistic role for PI3K in neutrophil mechanosensing during chemotaxis toward fMLP.
Figure 7.
LY294002 inhibits the ability of the neutrophil to sense stiff substrates. The root mean squared speed was calculated for neutrophils chemotaxing toward a pipette tip loaded with fMLP. Untreated cells and cells treated with 0.02% DMSO (vehicle control) and 20 μM LY294002 were recorded for a period of 30 minutes. No statistical difference was seen between the untreated cells and vehicle (DMSO)–treated cells. *Both the untreated and vehicle (DMSO)–treated cells showed a statistical difference between 10- and 100-kPa gels. Cells treated with LY294002 were indistinguishable on the 10- and 100-kPa gels. **The cells treated with LY294002 on the 100-kPa gels were significantly different from the untreated and vehicle cells on 100-kPa gels. In addition, cells treated with LY294002 showed limited ability to spread as shown in the representative DIC images below the plot. The DIC images are snapshots of single cells taken from supplemental Videos 3 through 6, respectively.
Discussion
Neutrophils represent an interesting cell type for study of movement because of their quick migration dynamics and their important role in the body's immune response. Within only a few minutes, the neutrophil experiences drastically different physical environments, ranging from the viscous fluid in the blood vessel, to the elastic extracellular matrix, to the highly variable points of stiffness at sites of inflammation. Each of these physical environments offers its own unique mechanical cues, which can affect a neutrophil's function and guide its behavior. Furthermore, the range of elasticity studied in this report correlate well with the reported values of a variety of relevant normal16 and inflamed21,22 tissue types.
We have shown that the substrate elasticity has an immediate impact on cell morphology, with cells spreading more quickly and to larger areas on the stiffer substrates (Figure 1). Other cell types, such as myotubes, fibroblasts, and endothelial cells,1,9–11,40 have also shown changes in area as a function of matrix stiffness, but none has responded as rapidly as neutrophils. Neutrophils on the 20- and 50-kPa gels showed more than a 2-fold increase within the first 2 minutes, illustrating both the speed and the magnitude of this response. It is puzzling that our findings contrast with those reported by Yeung et al12 who used a similar fibronectin-coupled polyacrylamide gel system but found no change in neutrophil size on gels of different stiffness. It may be relevant to note that their reported circumference ranged from 12 to 15 μm, which is significantly smaller than the expected range of 30 to 40 μm for unstimulated human neutrophils.35 Future work aimed at elucidating the effect of matrix stiffness on neutrophil phenotype and function should provide further clarification.
We find that neutrophils were able to pull significantly harder on the stiffer gels, consistent with reports of other cell types.5,7,41 Our results also show, however, that, although neutrophils exert stronger forces on the stiffer substrates, the average displacements in the gel are statistically equivalent to the displacements seen on the softer substrates. This indicates that, within a physiologically relevant range of substrate stiffness, neutrophils produce relatively constant displacements, which appear to be limited by the force generation apparatus within the cell. As such, the forces reported here can be considered as a lower bound for the maximum possible force a neutrophil can generate. This finding is consistent with recent strain-dependent findings in fibroblasts.42 Additional experiments were performed with neutrophils on a 100-kPa gel. At this stiffness, displacement measurements could not be distinguished from background noise in the algorithm, suggesting that the gel was too stiff for the neutrophil to deform.
Neutrophil motility showed a dependence on substrate stiffness, with the cell speed decreasing with increasing substrate stiffness during both chemokinesis and chemotaxis. A decrease in instantaneous speed on stiffer substrates has been previously reported in fibroblasts1,5,7 and vascular smooth muscle cells.43 These reports, however, do not take into account directionality, an advantage of measuring the MSD. Because the persistence in direction increases with substrate stiffness, neutrophils are able to compensate for the reduction in instantaneous speed. Both of these effects, the decrease in speed and the increase in persistence, are probably related to the dynamic adhesion and de-adhesion of the neutrophil to the substrate. Given that neutrophil spread area increases with substrate stiffness, it is reasonable to postulate that there is a proportional increase in the number of adhesion receptors via integrins in contact with the substrate. These receptors are continuously binding and unbinding as the cell migrates. With a large number of adhesion sites, ie, on stiffer substrates, cell movement could decrease because the coordinated detachment of a larger area requires greater amounts of time. In addition, as new adhesion sites are formed, they probably form near previous adhesion sites because those areas of the membrane are already in close proximity to the substrate, leading to an increase in persistence. During chemotaxis, the smaller step size requires smaller angular corrections to maintain a straight path toward the point source and thus results in a more peaked angular distribution of steps. This effect would be similar to previous reports that the density of adhesion receptors in the extracellular matrix impacts cell migration,40,44 only in this case it is the number of adhesion receptors in the membrane that is changing based on the substrate stiffness.
Substrate stiffness was seen to have no effect on the spatial distribution of tractions exerted by neutrophils. On gels of all stiffness, neutrophils exhibited large contractile stresses in the posterior of the polarized cell. These posterior contractions often appeared to push the front of the cell forward, as first suggested by Smith et al14 and more recently as shown in fibroblasts.45 At times, however, these strong concentrated tractions in the rear seemed to impede the migration progress of the neutrophil. Indeed, it is possible to explain the larger posterior tractions as a result of a higher density of adhesions needed to balance the more disperse tractions along the leading edge. Unfortunately, because each traction map represents a brief equilibrium state (ie, all tractions must sum to zero), the exact mechanism responsible for the forward motion cannot be inferred from the traction maps. Further experiments are necessary to determine whether protrusions along the leading edge typical of slow-moving cells,37,46 or pressure gradient driven amoeboid motion45 dominates in neutrophil migration. Regardless of mechanism, our results still clearly show the migration of the large contractile stress from the posterior of the cell forward. This observation suggests that an inchworm, or stepwise, model of cell migration is valid, although in the case of neutrophils it is more evident by the concentrated region of stresses in the posterior of the cell. It is important to note that many of the small balancing tractions spread beyond the apparent boundary of the cell as determined by the DIC image. This suggests that there are several small thin protrusions, similar to those seen in Cukierman et al,47 that are unresolved using DIC. This practical limitation, however, highlights one technical advantage of using the unconstrained Fourier transform traction cytometry method because it does not impose an artificial boundary around the cell to determine the tractions, as required by other methods.34
Efforts directed at elucidating the mechanistic basis of neutrophil mechanosensing found precedence for the role of PI3K in the response of endothelial cells to shear stress. Tzima et al48 showed a tripartate complex mediating the response of cells to shear with PI3K activation downstream of VEGFR activation. In those studies, PI3K inhibition with LY294002 was sufficient to obviate the stress response of endothelial cells. PI3K inhibition was also shown to maintain endothelial αvβ3 integrin in an inactive state after application of shear forces, suggesting that PI3K activates integrins in an inside-out pathway as has been shown previously in other cell types.49 An effect on neutrophil integrin function may play a role in neutrophil mechanosensing and is consistent with the selective effect of LY294002 on the morphology of neutrophils migrating on stiff but not soft matrix. These cells (Figure 7) acquired a more rounded, less spread, appearance, as may result from diminished integrin activitation. Chemotaxis of neutrophils toward fMLP has been shown to be minimally affected by PI3K inhibition.38,39 Therefore, findings reported here are probably not a consequence of direct PI3K activation by an fMLP gradient. Whether PI3K plays a direct or indirect role in neutrophil mechanosensing, or perhaps regulates other cell functions, such as cell spreading, awaits further clarification.
In conclusion, we find that the mechanical properties of the substrate strongly affect the principal functions of spreading, force generation, and migration in polymorphonuclear leukocytes. Additional research will explore how substrate elasticity may affect other biologic tests of neutrophil function, such as respiratory burst, degranulation, and phagocytosis. Finally, future work will also be needed to reveal whether neutrophil mechanosensing is regulated differentially among other extracellular matrix components compared with fibronectin as studied here, and in that regard, whether PI3K is a global or specific regulator of mechanosensing. The soft substrates used in this study may ultimately prove to be a more accurate mimic of the in vivo conditions neutrophils encounter than the traditional experimental conditions of working with cells on glass and plastic, thus providing a more appropriate approach to many existing studies on cell adhesion and migration.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation's Materials, Research, Science and Engineering Center at Brown University (DMR 0079964; J.X.T.), the National Institutes of Health (GM066194 and AI079582; J.S.R.), and a Brown University Seed Fund (J.X.T., J.S.R.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: P.W.O. designed research, analyzed results, made the figures, and wrote the paper; D.C.P., and N.A.M. performed experiments; D.P.Z. performed experiments and contributed to the development of analysis software; B.F. contributed to the development of analysis software; and J.S.R. and J.X.T. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jonathan S. Reichner, Department of Surgery, Rhode Island Hospital, 593 Eddy St, Providence, RI 02903; e-mail: Reichner@brown.edu.
References
- 1.Pelham RJ, Jr, Wang YL. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janmey PA, Weitz DA. Dealing with mechanics: mechanisms of force transduction in cells. Trends Biochem Sci. 2004;29:364–370. doi: 10.1016/j.tibs.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 4.Ali MH, Schumacker PT. Endothelial responses to mechanical stress: where is the mechanosensor? Crit Care Med. 2002;30(suppl):S198–S206. doi: 10.1097/00003246-200205001-00005. [DOI] [PubMed] [Google Scholar]
- 5.Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Effler JC, Kee YS, Berk JM, Tran MN, Iglesias PA, Robinson DN. Mitosis-specific mechanosensing and contractile-protein redistribution control cell shape. Curr Biol. 2006;16:1962–1967. doi: 10.1016/j.cub.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghosh K, Pan Z, Guan E, et al. Cell adaptation to a physiologically relevant ECM mimic with different viscoelastic properties. Biomaterials. 2007;28:671–679. doi: 10.1016/j.biomaterials.2006.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flanagan LA, Ju YE, Marg B, Osterfield M, Janmey PA. Neurite branching on deformable substrates. Neuroreport. 2002;13:2411–2415. doi: 10.1097/01.wnr.0000048003.96487.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engler AJ, Griffin MA, Sen S, Bonnemann CG, Sweeney HL, Discher DE. Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J Cell Biol. 2004;166:877–887. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D. Substrate compliance versus ligand density in cell on gel responses. Biophys J. 2004;86:617–628. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 12.Yeung T, Georges PC, Flanagan LA, et al. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 13.Wang HB, Dembo M, Hanks SK, Wang Y. Focal adhesion kinase is involved in mechanosensing during fibroblast migration. Proc Natl Acad Sci U S A. 2001;98:11295–11300. doi: 10.1073/pnas.201201198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith LA, Aranda-Espinoza H, Haun JB, Dembo M, Hammer DA. Neutrophil traction stresses are concentrated in the uropod during migration. Biophys J. 2007;92:L58–L60. doi: 10.1529/biophysj.106.102822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato M, Suzuki K, Ueki Y, Ohashi T. Microelastic mapping of living endothelial cells exposed to shear stress in relation to three-dimensional distribution of actin filaments. Acta Biomater. 2007;3:311–319. doi: 10.1016/j.actbio.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 16.Mathur AB, Collinsworth AM, Reichert WM, Kraus WE, Truskey GA. Endothelial, cardiac muscle and skeletal muscle exhibit different viscous and elastic properties as determined by atomic force microscopy. J Biomech. 2001;34:1545–1553. doi: 10.1016/s0021-9290(01)00149-x. [DOI] [PubMed] [Google Scholar]
- 17.Kataoka N, Iwaki K, Hashimoto K, et al. Measurements of endothelial cell-to-cell and cell-to-substrate gaps and micromechanical properties of endothelial cells during monocyte adhesion. Proc Natl Acad Sci U S A. 2002;99:15638–15643. doi: 10.1073/pnas.242590799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Houten EE, Doyley MM, Kennedy FE, Weaver JB, Paulsen KD. Initial in vivo experience with steady-state subzone-based MR elastography of the human breast. J Magn Reson Imaging. 2003;17:72–85. doi: 10.1002/jmri.10232. [DOI] [PubMed] [Google Scholar]
- 19.Samani A, Bishop J, Luginbuhl C, Plewes DB. Measuring the elastic modulus of ex vivo small tissue samples. Phys Med Biol. 2003;48:2183–2198. doi: 10.1088/0031-9155/48/14/310. [DOI] [PubMed] [Google Scholar]
- 20.Uffmann K, Maderwald S, Ajaj W, et al. In vivo elasticity measurements of extremity skeletal muscle with MR elastography. NMR Biomed. 2004;17:181–190. doi: 10.1002/nbm.887. [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto T, Abe H, Ohashi T, Kato Y, Sato M. Local elastic modulus of atherosclerotic lesions of rabbit thoracic aortas measured by pipette aspiration method. Physiol Meas. 2002;23:635–648. doi: 10.1088/0967-3334/23/4/304. [DOI] [PubMed] [Google Scholar]
- 22.Georges PC, Hui J-J, Gombos Z, et al. Increased stiffness of the rat liver precedes matrix deposition: implications for fibrosis. Am J Physiol Gastrointest Liver Physiol. 2007;293:G1147–G1154. doi: 10.1152/ajpgi.00032.2007. [DOI] [PubMed] [Google Scholar]
- 23.Nathan C. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol. 2006;6:173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 24.Choi M, Salanova B, Rolle S, et al. Short-term heat exposure inhibits inflammation by abrogating recruitment of and nuclear factor-κB activation in neutrophils exposed to chemotactic cytokines. Am J Pathol. 2008;172:367–377. doi: 10.2353/ajpath.2008.070532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheetz MP, Felsenfeld DP, Galbraith CG. Cell migration: regulation of force on extracellular-matrix-integrin complexes. Trends Cell Biol. 1998;8:51–54. doi: 10.1016/s0962-8924(98)80005-6. [DOI] [PubMed] [Google Scholar]
- 26.Van Haastert PJ, Devreotes PN. Chemotaxis: signalling the way forward. Nat Rev Mol Cell Biol. 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- 27.Bershadsky AD, Balaban NQ, Geiger B. Adhesion-dependent cell mechanosensitivity. Annu Rev Cell Dev Biol. 2003;19:677–695. doi: 10.1146/annurev.cellbio.19.111301.153011. [DOI] [PubMed] [Google Scholar]
- 28.Martin P. Wound healing-aiming for perfect skin regeneration. Science. 1997;276:75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- 29.Raupach C, Zitterbart DP, Mierke CT, Metzner C, Muller FA, Fabry B. Stress fluctuations and motion of cytoskeletal-bound markers. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;76 doi: 10.1103/PhysRevE.76.011918. 011918. [DOI] [PubMed] [Google Scholar]
- 30.Müller O, Gaub H, Bärmann M, Sackmann E. Viscoelastic moduli of sterically and chemically cross-linked actin networks in the dilute to semidilute regime: measurements by an oscillating disk rheometer. Macromolecules. 1991;24:3111–3120. [Google Scholar]
- 31.Butler JP, Tolic-Norrelykke IM, Fabry B, Fredberg JJ. Traction fields, moments, and strain energy that cells exert on their surroundings. Am J Physiol Cell Physiol. 2002;282:C595–C605. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- 32.Landau L, Lifshitz E. Theory of Elasticity. Oxford, MA: Butterworth-Heinemann; 1986. [Google Scholar]
- 33.Sabass B, Gardel ML, Waterman CM, Schwarz US. High resolution traction force microscopy based on experimental and computational advances. Biophys J. 2008;94:207–220. doi: 10.1529/biophysj.107.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dembo M, Wang YL. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J. 1999;76:2307–2316. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young B, Lowe S, Stevens A, Heath J. Wheater's Functional Histology: A Text and Colour Atlas. 5th Ed. New York, NY: Churchill-Livingstone; 2006. pp. 49–64. [Google Scholar]
- 36.Zaman MH, Trapani LM, Sieminski AL, et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci U S A. 2006;103:10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ridley AJ, Schwartz MA, Burridge K, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 38.Heit B, Liu L, Colarusso P, Puri KD, Kubes P. PI3K accelerates, but is not required for, neutrophil chemotaxis to fMLP. J Cell Sci. 2008;121:205–214. doi: 10.1242/jcs.020412. [DOI] [PubMed] [Google Scholar]
- 39.Sai J, Raman D, Liu Y, Wikswo J, Richmond A. Parallel phosphatidylinositol 3-kinase (PI3K)-dependent and Src-dependent pathways lead to CXCL8-mediated Rac2 activation and chemotaxis. J Biol Chem. 2008;283:26538–26547. doi: 10.1074/jbc.M805611200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peyton SR, Putnam AJ. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J Cell Physiol. 2005;204:198–209. doi: 10.1002/jcp.20274. [DOI] [PubMed] [Google Scholar]
- 41.Kong HJ, Polte TR, Alsberg E, Mooney DJ. FRET measurements of cell-traction forces and nano-scale clustering of adhesion ligands varied by substrate stiffness. Proc Natl Acad Sci U S A. 2005;102:4300–4305. doi: 10.1073/pnas.0405873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghibaudo M, Saez A, Trichet L, et al. Traction forces and rigidity sensing regulate cell functions. Soft Matter. 2008;4:1836–1843. [Google Scholar]
- 43.Wong J, Velasco A, Rajagopalan P, Pham Q. Directed movement of vascular smooth muscle cells on gradient-compliant hydrogels. Langmuir. 2003;19:1908–1913. [Google Scholar]
- 44.Gupton SL, Waterman-Storer CM. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 2006;125:1361–1374. doi: 10.1016/j.cell.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 45.Iwasaki T, Wang YL. Cytoplasmic force gradient in migrating adhesive cells. Biophys J. 2008;94:L35–L37. doi: 10.1529/biophysj.107.124479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 47.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 48.Tzima E, Irani-Tehrani M, Kiosses WB, et al. A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature. 2005;437:426–431. doi: 10.1038/nature03952. [DOI] [PubMed] [Google Scholar]
- 49.Hughes PE, Pfaff M. Integrin affinity modulation. Trends Cell Biol. 1998;8:359–364. doi: 10.1016/s0962-8924(98)01339-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.