Abstract
The modification of proteins by the small ubiquitin-like modifier (SUMO) is known to regulate an increasing array of cellular processes. SUMOylation of the mitochondrial fission GTPase dynamin-related protein 1 (DRP1) stimulates mitochondrial fission, suggesting that SUMOylation has an important function in mitochondrial dynamics. The conjugation of SUMO to its substrates requires a regulatory SUMO E3 ligase; however, so far, none has been functionally associated with the mitochondria. By using biochemical assays, overexpression and RNA interference experiments, we characterized the mitochondrial-anchored protein ligase (MAPL) as the first mitochondrial-anchored SUMO E3 ligase. Furthermore, we show that DRP1 is a substrate for MAPL, providing a direct link between MAPL and the fission machinery. Importantly, the large number of unidentified mitochondrial SUMO targets suggests a global role for SUMOylation in mitochondrial function, placing MAPL as a crucial component in the regulation of multiple conjugation events.
Keywords: mitochondria, MAPL, SUMO1, DRP1, fission
Introduction
Post-translational modifications, including the conjugation of small ubiquitin-like modifier (SUMO) to its substrates, allow the rapid response to dynamic changes in cellular signals. The SUMOylation of protein targets leads to diverse consequences, including the formation or disassembly of protein complexes, the regulation of protein localization and the modulation of other post-translational modifications such as phosphorylation and ubiquitination (Geiss-Friedlander & Melchior, 2007; Martin et al, 2007). SUMOylation is a multi-step process involving an E1 heterodimer, an E2 ligase Ubc9 (ubiquitin-like protein SUMO-1 conjugating enzyme 9) and a SUMO E3 ligase, which is thought to provide specificity and enhance the reaction rate of the final step (Geiss-Friedlander & Melchior, 2007). All SUMO E3 ligases characterized so far are localized primarily within the nucleus. These include the Protein Inhibitor of Activated STATS (PIAS) family (Kahyo et al, 2001; Bischof et al, 2006), polycomb 2 (Kagey et al, 2003) and Ran binding protein 2 (RanBP2; Pichler et al, 2002). Initially, SUMO E3 ligases were identified by the presence of a conserved RING-finger domain; however, at least two family members do not require this domain for their SUMOylation activity (Pichler et al, 2002; Ivanov et al, 2007). Consequently, SUMO E3 ligases are defined by their ability to (i) bind to Ubc9, (ii) bind to SUMO1 and (iii) enhance the transfer of SUMO from Ubc9 onto a substrate. As with the SUMO proteases, each of the SUMO E3 ligases has been shown to function on multiple substrates, so the pairing of the substrate to the E3 ligase is restricted by their subcellular localizations.
Although most SUMO substrates have been studied in the nucleus, there are several essential targets localized in the cytosol and associated with other intracellular organelles (Johnson & Blobel, 1999; Rajan et al, 2005; Dadke et al, 2007; Makhnevych et al, 2007; Martin et al, 2007; Kang et al, 2008). We have previously identified SUMOylation as a post-translational modification for several mitochondrial proteins (Harder et al, 2004; Wasiak et al, 2007; Zunino et al, 2007). In particular, we have shown that the SUMOylation of the fission GTPase dynamin-related protein 1 (DRP1) regulates mitochondrial dynamics both in steady state and during cell death (Harder et al, 2004; Wasiak et al, 2007). Specifically, the increase in DRP1 SUMOylation leads to an increase in DRP1 protein levels and a more stable association with the membrane (Wasiak et al, 2007). Importantly, the abundance of mitochondrial SUMOylated substrates suggests that SUMOylation has a global role in mitochondrial processes. We were therefore interested in identifying a SUMO E3 ligase involved in the regulation of mitochondrial SUMOylation. Here, we show that the mitochondrial-anchored protein ligase (MAPL; Neuspiel et al, 2008) functions as a SUMO E3 ligase involved in the conjugation of specific mitochondrial targets, including DRP1.
Results
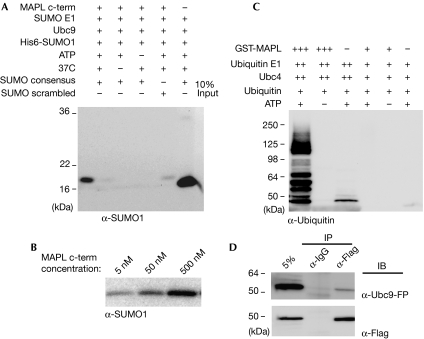
MAPL was first identified as a novel mitochondrial outer membrane protein, the expression of which led to an increase in mitochondrial fragmentation in a RING-finger-dependent manner (Neuspiel et al, 2008). As the SUMOylation of DRP1 is known to enhance fission, we tested whether the RING domain of MAPL shows SUMO E3 ligase activity. We first examined whether MAPL has SUMO1 E3 ligase activity in vitro. Incubation of a biotinylated peptide containing the SUMO1 consensus sequence ΨKxE with 500 nM of the recombinant RING-finger domain of MAPL (MAPL(257−352)), the E1 heterodimer and Ubc9 led to the conjugation of His-SUMO (Fig 1). This conjugation was significantly increased over the minimal reaction observed with only the E1 heterodimer and Ubc9. In addition, the reaction was ATP-, temperature- and dose-dependent (Fig 1A,B).
Figure 1.
MAPL has SUMO E3 ligase activity in vitro. (A) Biotinylated peptide was incubated in the indicated conditions. After the reaction, the peptide was isolated and the isolates probed for SUMO1. (B) The same reaction conditions were used as in lane 1 (A) but with increasing amounts of MAPL(257−352). (C) GST-MAPL(257−352) was used in an in vitro auto-ubiquitination assay. In the presence of 30 μM GST-MAPL(257−352), 0.5 μM of ubiquitin E1, 5 μM of Ubc4 and 10 μM ubiquitin, the RING domain of MAPL facilitates the conjugation of ubiquitin. In the presence of 500 nM GST-MAPL(257−352), 50 nM of ubiquitin E1, 250 nM of Ubc4 and 10 μM ubiquitin, the RING domain of MAPL does not facilitate the conjugation of ubiquitin. (D) After the transfection of HeLa cells with MAPL-Flag and Ubc9-YFP, cell lysates were immunoprecipitated (IP) with either anti-Flag coupled resin or mouse IgG, as indicated. GST, glutathione S-transferase; IB, immunoblotting; MAPL, mitochondrial-anchored protein ligase; SUMO, small ubiquitin-like modifier; Ubc9, ubiquitin-like protein SUMO-1 conjugating enzyme 9; YFP, yellow fluorescent protein.
The RING-finger domain of MAPL was previously reported to function as an E3 ubiquitin ligase in vitro (Li et al, 2008). We also observed auto-ubiquitination activity with glutathione S-transferase (GST)-MAPL(257−352); however, the reaction required the presence of a 30 μM RING domain, and increased levels of both the E1 and E2 (Fig 1C). Under lower concentrations of E1 and E2, 100 nM of MAPL(257−352) is sufficient to sustain a robust SUMOylation reaction, but even 500 nM of GST-MAPL(257−352) is not enough to support the ubiquitination reaction (Fig 1C, right three lanes). Therefore, although it is possible for MAPL to facilitate ubiquitin conjugation in vitro, under physiological concentrations MAPL preferentially acts as a SUMO E3 ligase. In addition to the conjugation assays described so far, all known SUMO E3 ligases interact directly with the E2 enzyme Ubc9 (Tang et al, 2008. To confirm whether MAPL could also bind to Ubc9, HeLa cells were transfected with both Ubc9-yellow fluorescent protein (YFP) and MAPL-Flag, solubilized and immunoprecipitates were examined by Western blot (Fig 1D). The data show that a fraction of Ubc9-YFP is precipitated efficiently with MAPL-Flag, which is consistent with the evidence that they function within the same enzymatic cascade.
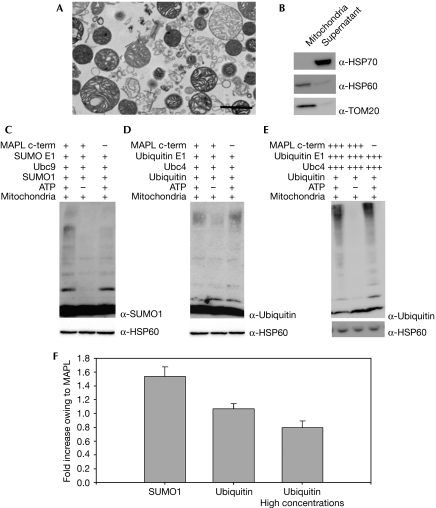
As MAPL is a mitochondrial-anchored protein, we tested whether its RING domain could mediate the SUMOylation of mitochondrial targets on intact organelles. As revealed by electron microscopy and Western blot analysis, highly purified mitochondria were isolated from bovine heart (Fig 2A,B) and used as substrates for in vitro SUMOylation reactions. The addition of excess E1 heterodimer and Ubc9 enzymes as well as His-SUMO1 to mitochondria isolated from bovine heart resulted in the efficient conjugation of several mitochondrial targets in an ATP-dependent manner (Fig 2C, compare lanes 1 and 2). This result is consistent with our previous report that there are multiple mitochondrial SUMO1 conjugates (Harder et al, 2004), and highlights the presence of an active endogenous SUMO E3 ligase on the mitochondria. The basal reaction was enhanced 1.5-fold on the addition of 500 nM MAPL(257−352) (sn=0.14, n=10, P<0.05; Fig 2C, compare lanes 1 and 3, quantified in Fig 2F). Isolated mitochondria were also able to support the ubiquitination of substrates in an ATP-dependent manner (Fig 2D). This is most likely to be driven, at least in part, by a previously identified mitochondrial ubiquitin E3 ligase, MARCH-V/MITOL (membrane associated RING-CH-V/mitochondrial ubiquitin protein ligase; Nakamura et al, 2006; Yonashiro et al, 2006; Karbowski et al, 2007). However, the addition of 500 nM MAPL(257−352) to the ubiquitination reaction on isolated mitochondria did not result in an increase in the basal ubiquitination reaction observed during the 90-min incubation (sn=0.07, n=6, P>0.05; Fig 2D, compare lanes 1 and 3). Importantly, increasing the concentrations of E1, E2 and MAPL(257−352) to the levels that allowed the self-ubiquitination reaction in Fig 1C did not result in an increase in the basal ubiquitination reaction (sn=0.09, n=5, P>0.05; Fig 2E,F). This indicates that MAPL functions preferentially as a SUMO1 E3 ligase not only on synthetic peptides, but also on native mitochondrial targets.
Figure 2.
MAPL SUMOylates native mitochondrial substrates. The purity of isolated bovine heart mitochondria was visualized by (A) electron microscopy (scale bar, 10 μM) and (B) Western blot analysis. (C) Isolated mitochondria from bovine heart were incubated with 50 nM SUMO E1, 250 nM Ubc9 and 10 μM his-SUMO1 under the conditions indicated; the addition of 500 nM MAPL(257−352) is indicated. (D) Isolated mitochondria from bovine heart were incubated in the presence of 50 nM ubiquitin E1, 250 nM Ubc4 and 10 μM ubiquitin under the conditions indicated; the addition of 500 nM MAPL(257−352) is indicated. (E) Isolated mitochondria from bovine heart were incubated in the presence of 0.5 μM ubiquitin E1, 5 μM Ubc4 and 10 μM ubiquitin under the conditions indicated; the addition of 30 μM MAPL(257−352) is indicated. (F) Quantification of the increase in the SUMOylation/ubiquitination reactions owing to the addition of exogenous MAPL (lanes 1 versus 3 in C, D and E); standard errors are represented. HSP, heat-shock protein; MAPL, mitochondrial-anchored protein ligase; SUMO, small ubiquitin-like modifier; TOM, translocase of the mitochondrial outer membrane; Ubc, ubiquitin-conjugating enzyme.
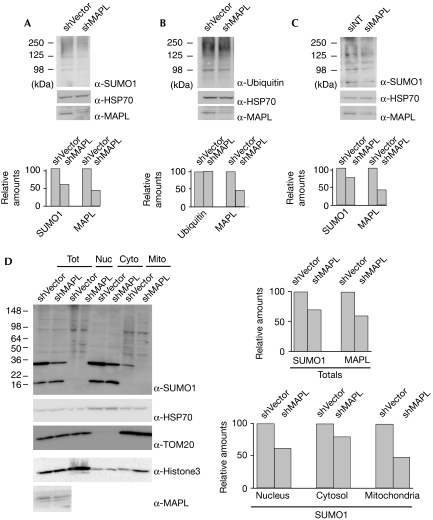
We then turned our attention to the activity of MAPL in intact cells by downregulating the expression of MAPL. Stable cell lines transfected with short hairpin RNA (shRNA) directed against MAPL allowed us to reduce the levels of MAPL by 58% compared with a scrambled control (Fig 3A,B). This decrease in MAPL levels led to a concomitant 46% (sn=5, n=3, P<0.05) decrease in the levels of endogenous SUMO1 conjugates (Fig 3A), but the levels of ubiquitinated proteins remained unchanged (Fig 3B). To ensure that this effect was not due to off-target silencing, we used a pooled short interfering RNA (siRNA) mixture to silence MAPL in HeLa cells (Fig 3C). The data show that a 58% silencing of MAPL led to a 24% reduction in SUMO conjugates (sn=8.6, n=3, P<0.05). This further supports the evidence that MAPL predominantly functions as a SUMO E3 ligase under physiological conditions. Finally, to determine whether the silencing of MAPL primarily affected mitochondrial SUMO1 substrates, we isolated the nuclear, cytosolic and mitochondrial fractions of HeLa cells stably transfected with shRNA directed against MAPL or a control shRNA (Fig 3D). The data show that although the reduction of MAPL expression leads to a decrease in SUMO1 conjugates in all three fractions, the greatest loss of SUMO1 conjugates occurs on the mitochondria.
Figure 3.
Silencing of MAPL reduces total SUMO1 conjugates. (A) HeLa cells were stably transfected with either shRNA vector alone (shVector) or with MAPL-specific shRNA (shMAPL). Total lysates were probed for SUMO1. (B) As in (A), but the extracts were probed with anti-ubiquitin antibodies. (C) HeLa cells were transfected with either a pooled siRNA mixture directed against MAPL or a control siRNA. Total lysates were probed for SUMO1. (A–C) The graphs represent the average of three independent experiments. (D) Isolation of the nuclear (Nuc), cytosolic (Cyto) and mitochondrial (Mito) fractions of HeLa cells stably transfected with an shRNA directed against MAPL or a control shRNA. A 40% reduction in the expression of MAPL leads to a 30% reduction in total SUMO1 conjugates, 38% reduction in nuclear SUMO1 conjugates, 20% reduction in cytosolic SUMO1 conjugates and a 53% reduction in mitochondrial SUMO1 conjugates. HSP, heat-shock protein; MAPL, mitochondrial-anchored protein ligase; NT, non-targeted; shRNA, short hairpin RNA; siRNA, short interfering RNA; SUMO, small ubiquitin-like modifier; TOM, translocase of the mitochondrial outer membrane; Tot, total.
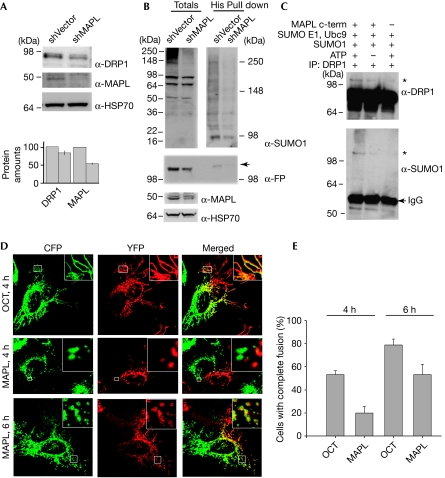
As the SUMOylation of DRP1 has been shown to stabilize the protein and stimulate mitochondrial fission (Harder et al, 2004; Wasiak et al, 2007; Zunino et al, 2007), we tested whether silencing of MAPL leads to the destabilization of DRP1. The downregulation of MAPL by 46% (sn=2.3, n=4) led to a 17% decrease in the DRP1 levels (sn=6, n=4; Fig 4A). This decrease in the DRP1 protein levels, although statistically significant (P<0.05), did not lead to significant changes in mitochondrial morphology (Neuspiel et al, 2008). To test whether this loss of DRP1 is associated with a direct reduction in its SUMO1 conjugation, cells expressing His6-SUMO1 and DRP1-YFP were solubilized and all SUMO1 conjugates were isolated using nickel nitrilotriacetic acid (Ni-NTA)-agarose beads (Fig 4B). An examination of the total extracts shows that the transfection levels of His6-SUMO were similar, as several conjugate bands are of equal intensity (Fig 4B). However, the high molecular weight conjugation pattern for SUMOylation is reduced on the silencing of MAPL. Isolation of these conjugates further confirms the reduction in SUMOylation. The ratio of DRP1 in the total extracts to that found in the His6-SUMO1 isolates is reduced from 1 to 0.3 (Fig 4B). The molecular weight of DRP1-YFP is shifted in the His6-SUMO column, which is consistent with a sodium dodecyl sulphate (SDS)-resistant His6-SUMO1 conjugate (Fig 4B, arrow).
Figure 4.
MAPL SUMOylates dynamin-related protein 1 and modulates mitochondrial dynamics. (A) HeLa cells were stably transfected with either shRNA vector alone (shVector) or MAPL-specific shRNA (shMAPL). Total lysates were analysed by Western blots with the indicated antibodies. The graph represents the quantification from four independent experiments; standard errors are represented. (B) Vector or shMAPL-expressing HeLa cells were transfected with DRP1-YFP and His6-SUMO1, harvested and SUMO1 conjugates were isolated using nickel nitrilotriacetic acid (Ni-NTA)-agarose. Western blots of SUMO1, DRP1-FP and controls are indicated. SUMO conjugates in the totals were separated using a 4–20% SDS–PAGE gradient (left), and the His6-SUMO1 isolates were separated on a 6% SDS–PAGE gel (right). (C) Endogenous DRP1 was immunoprecipitated (IP) from HeLa cells and used as the substrate in an in vitro SUMO assay. (D) HeLa cells transfected with the indicated constructs were seeded. Images of both fluorophores were taken from cells fixed after 4 and 6 h of PEG addition. (E) Quantification of the percentage of heterokaryons with complete fusion. Ten heterokaryons were analysed for each condition in three independent experiments. CFP, cyan fluorescent protein; DRP1, dynamin-related protein 1; FP, fluorescent protein; HSP, heat-shock protein; MAPL, mitochondrial-anchored protein ligase; OCT, ornithine carbamyl transferase (amino acids 1–32); PEG, polyethylene glycol; SDS, sodium dodecyl sulphate; shRNA, short hairpin RNA; SUMO, small ubiquitin-like modifier; YFP, yellow fluorescent protein.
To show that recombinant MAPL can SUMOylate DRP1 directly, we isolated endogenous DRP1 from HeLa cell extracts by immunoprecipitation and used this as a substrate in the in vitro SUMOylation assay (Fig 4C). Quantification reveals that the presence of ATP and MAPL (Fig 4C, lane 1) led to an increase in the signal (asterisk) relative to the background (Fig 4C, lane 3) by 37% in the SUMO1 blot and 47% in the DRP1 blot. In the absence of ATP (Fig 4C, lane 2), there is no detectible increase in the SUMO1 signal, with the DRP1 antibody detecting a 19% increase. In addition to the in vitro reaction, immunoprecipitation of endogenous MAPL also revealed a fraction of DRP1 present in the precipitates (supplementary Fig 1 online), which is consistent with the evidence that MAPL SUMOylates DRP1. Taken together, our data show that the overexpression of MAPL stimulates fission (Neuspiel et al, 2008), that the silencing of MAPL leads to a reduction in DRP1 levels and SUMO conjugates, and that MAPL SUMOylates DRP1 in vitro.
The fragmented phenotype observed on the overexpression of MAPL can be explained by the stabilization and activation of DRP1 fission activity. However, it is also clear that there are several unidentified SUMO1 targets on the mitochondria. Therefore, we decided to test whether the rates of fusion might also be affected by the increased mitochondrial SUMOylation. To assess directly the fusion capacity of mitochondria in cells overexpressing MAPL, we used a polyethylene glycol (PEG) fusion assay (Neuspiel et al, 2005). Mitochondria from heterokaryons transfected with matrix marker proteins alone were 53.3% fused within 4 h (sn=3.3, n=3; Fig 4D,E). By contrast, cells expressing MAPL-cyan fluorescent protein (CFP) and MAPL-YFP showed significantly less mitochondrial fusion at 4 h, with only 20% of cells having completely mixed mitochondrial contents (sn=5.77, n=3, P<0.05). However, by 6 h after cell fusion, there was no longer a significant difference between the control cells and MAPL-overexpressing cells (79% for cells overexpressing the markers (sn=5, n=3) and 53.3% for MAPL-overexpressing cells (sn=8.8, n=3, P>0.05); Fig 4D,E). This indicates that in addition to promoting fission, MAPL leads to a reduction in fusion rates.
Discussion
The data presented here identify MAPL as the first mitochondrial SUMO E3 ligase. MAPL supports SUMOylation on a synthetic peptide containing the SUMO consensus sequence, on isolated mitochondrial proteins and on native DRP1. Our previous data have shown that MAPL stimulates mitochondrial fragmentation (Neuspiel et al, 2008), and this study provides new mechanistic insights into the role of MAPL in regulating the SUMOylation of DRP1. In SUMOylating DRP1, MAPL functions as a positive regulator of mitochondrial fission; however, the silencing data indicate that MAPL is not an essential component of the division machinery (Neuspiel et al, 2008). The complex pattern of SUMO conjugates found on the mitochondria that are formed upon incubation with excess MAPL indicates that MAPL probably also functions in other mitochondrial processes through targets distinct from DRP1, including potential regulators of the fusion machinery.
Our data confirm that MAPL is also able to facilitate ubiquitination. However, the MAPL-catalysed ubiquitination was supported only when the concentrations of the enzymes were in the micromolar range. As the cellular concentrations of ubiquitin and SUMO do not enter the micromolar range, it is unlikely that MAPL provides significant ubiquitination activity in vivo. Furthermore, MAPL knockdown did not affect the mitochondrial ubiquitination pattern, and the addition of MAPL-RING to isolated mitochondria did not stimulate the ubiquitination reaction on native mitochondrial targets. Therefore, although MAPL might show ubiquitination activity under some conditions, the data show that MAPL functions in vivo as an efficient SUMO E3 ligase.
Finally, the identification of a mitochondrial SUMO E3 ligase significantly expands the repertoire of SUMOylation enzymes, confirms the importance of non-nuclear SUMOylation and provides a new platform from which to study the function of SUMOylation outside of the nucleus. Although some of the changes in the global SUMOylation on silencing of MAPL might be indirect, it is clear that MAPL has an important role in cellular SUMOylation. The localization of the RING domain on the cytosolic side of the mitochondrial outer membrane might also result in the SUMOylation of targets on closely apposed organelles such as the endoplasmic reticulum or peroxisomes through transfer of mitochondrial-derived vesicles (Neuspiel et al, 2008), as well as of other cytosolic factors. Characterization of the MAPL-specific SUMO proteome will clarify these possibilities and help to define the role of MAPL in both mitochondrial dynamics and a broader cellular biological context.
Methods
SUMOylation and ubiquitination assay. The SUMO conjugation assay was performed using 50 nM SUMO E1, 250 nM Ubc9, 10 μM His6-SUMO1, 20 μM of a consensus peptide in buffer A (50 mM Tris pH 8.0, 100 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol) and either an ATP-regenerating system (2.5 U creatine kinase, 125 nM creatine phosphate, 5 mM ATP) or 0.1 U apyrase. Streptavidin-coupled magnetic beads (Invitrogen, Mississauga, CA, USA) were washed in buffer A supplemented with 2 mg/ml BSA and 1% Triton X-100, then incubated with the conjugation reaction for 60 min at 25°C, and finally washed in buffer A supplemented with 500 mM NaCl and 1% Triton X-100. Alternatively, bovine heart mitochondria were centrifuged at 800 g for 15 min and resuspended in Tris mitochondria buffer (50 mM Tris pH 7.4, 220 mM mannitol, 68 mM sucrose, 80 mM KCl, 0.5 mM EGTA, 2 mM Mg(Ac)2). A measure of 100 μg of these mitochondria was then used as substrate. The in vitro ubiquitination reaction was performed using buffer A with the conditions described by Li et al (2008; 0.5 μM E1, 5 μM Ubc4, 30 μM GST-RING) or with 50 nM ubiquitin E1, 250 nM Ubc4, 10 μM ubiquitin and 500 nM MAPL-GST. All reactions were incubated for 90 min at 30°C.
PEG fusion assay. The PEG assay was performed as described previously (Neuspiel et al, 2005). HeLa cells were transfected for 6 h with each of four markers: OCT:CFP, OCT:YFP, CFP:MAPL and MAPL:YFP, and seeded together. After 16 h, cells were incubated for 1 h with 20 μg/ml cycloheximide, 50% PEG solution (Sigma, Oakville, CA, USA) was added for 60 s and then replaced with media containing 20 μg/ml cycloheximide.
Immunoprecipitations. HeLa cells transfected with MAPL-Flag and Ubc9-YFP were solubilized in 50 mM Tris pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1% Triton, 20 mM N-ethyl maleimide and protease inhibitor cocktail (Roche, Mississauga, CA, USA). After centrifugation at 9,300g for 30 min, 1 mg of the supernatant was incubated at 4°C with Flag resin (Sigma) or 2.5 μg G-protein agarose-coupled mouse IgG (Sigma). For the endogenous MAPL immunoprecipitations, 20 μl of anti-MAPL serum or 2.5 μg rabbit IgG (Sigma) were incubated with Protein A agarose. NTA-agarose pull down experiments to isolate His6-SUMO conjugates have been described previously (Harder et al, 2004).
siRNA. The siGENOME SMARTpool reagent directed against MAPL and the siCONTROL non-targeting were obtained from Dharmacon (Lafayette, CO, USA). Transfection was performed according to the manufacturer's protocol. HeLa cells were seeded in six-well dishes at approximately 50% confluency and transfected every 72 h three times.
Supplementary information is available at EMBO reports online (http://www.emboreports.org)
Supplementary Material
Supplementary Information
Acknowledgments
This work was funded by a Canadian Institutes of Health Research (CIHR) operating grant MOP68833 awarded to H.M.M. E.B. is a recipient of a CIHR Frederick Banting and Charles Best Canada Graduate Scholarship. The authors thank Vincent Soubannier and Peter Rippstein (University of Ottawa Heart Institute, Canada) for isolating and preparing the bovine mitochondrial samples for electron microscopy. E.B and R.Z. performed all of the experiments. E.B. and H.M.M. designed the experiments and co-wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bischof O, Schwamborn K, Martin N, Werner A, Sustmann C, Grosschedl R, Dejean A (2006) The E3 SUMO ligase PIASy is a regulator of cellular senescence and apoptosis. Mol Cell 22: 783–794 [DOI] [PubMed] [Google Scholar]
- Dadke S et al. (2007) Regulation of protein tyrosine phosphatase 1B by sumoylation. Nat Cell Biol 9: 80–85 [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F (2007) Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8: 947–956 [DOI] [PubMed] [Google Scholar]
- Harder Z, Zunino R, McBride H (2004) Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr Biol 14: 340–345 [DOI] [PubMed] [Google Scholar]
- Ivanov AV et al. (2007) PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell 28: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Blobel G (1999) Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol 147: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Melhuish TA, Wotton D (2003) The polycomb protein Pc2 is a SUMO E3. Cell 113: 127–137 [DOI] [PubMed] [Google Scholar]
- Kahyo T, Nishida T, Yasuda H (2001) Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol Cell 8: 713–718 [DOI] [PubMed] [Google Scholar]
- Kang JS, Saunier EF, Akhurst RJ, Derynck R (2008) The type I TGF-beta receptor is covalently modified and regulated by sumoylation. Nat Cell Biol 10: 654–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Neutzner A, Youle RJ (2007) The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol 178: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, Joazeiro CA (2008) Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the Organelle's dynamics and signaling. PLoS ONE 3: e1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhnevych T, Ptak C, Lusk CP, Aitchison JD, Wozniak RW (2007) The role of karyopherins in the regulated sumoylation of septins. J Cell Biol 177: 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Wilkinson KA, Nishimune A, Henley JM (2007) Emerging extranuclear roles of protein SUMOylation in neuronal function and dysfunction. Nat Rev Neurosci 8: 948–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S (2006) MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep 7: 1019–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuspiel M, Zunino R, Gangaraju S, Rippstein P, McBride HM (2005) Activated Mfn2 signals mitochondrial fusion, interferes with Bax activation and reduces susceptibility to radical induced depolarization. J Biol Chem 280: 25060–25070 [DOI] [PubMed] [Google Scholar]
- Neuspiel M, Schauss AC, Braschi E, Zunino R, Rippstein P, Rachubinski RA, Andrade-Navarro MA, McBride HM (2008) Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol 18: 102–108 [DOI] [PubMed] [Google Scholar]
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108: 109–120 [DOI] [PubMed] [Google Scholar]
- Rajan S, Plant LD, Rabin ML, Butler MH, Goldstein SA (2005) Sumoylation silences the plasma membrane leak K+ channel K2P1. Cell 121: 37–47 [DOI] [PubMed] [Google Scholar]
- Tang Z, Hecker CM, Scheschonka A, Betz H (2008) Protein interactions in the sumoylation cascade: lessons from X-ray structures. FEBS J 275: 3003–3015 [DOI] [PubMed] [Google Scholar]
- Wasiak S, Zunino R, McBride HM (2007) Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol 177: 439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonashiro R et al. (2006) A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J 25: 3618–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunino R, Schauss A, Rippstein P, Andrade-Navarro M, McBride HM (2007) The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J Cell Sci 120: 1178–1188 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information