Abstract
Ephrin (Eph) receptor tyrosine kinases fall into two subclasses (A and B) according to preferences for their ephrin ligands. All published structural studies of Eph receptor/ephrin complexes involve B-class receptors. Here, we present the crystal structures of an A-class complex between EphA2 and ephrin-A1 and of unbound EphA2. Although these structures are similar overall to their B-class counterparts, they reveal important differences that define subclass specificity. The structures suggest that the A-class Eph receptor/ephrin interactions involve smaller rearrangements in the interacting partners, better described by a ‘lock-and-key'-type binding mechanism, in contrast to the ‘induced fit' mechanism defining the B-class molecules. This model is supported by structure-based mutagenesis and by differential requirements for ligand oligomerization by the two subclasses in cell-based Eph receptor activation assays. Finally, the structure of the unligated receptor reveals a homodimer assembly that might represent EphA2-specific homotypic cell adhesion interactions.
Keywords: Eph, ephrin, receptor tyrosine kinase, crystallography
Introduction
The Eph receptors and their ephrin ligands control a diverse array of cell–cell interactions in the nervous and vascular systems (Himanen & Nikolov, 2003; Pasquale, 2005). On ephrin binding, the Eph kinase domain is activated, initiating ‘forward' signalling in the receptor-expressing cells. Simultaneously, signals are also induced in the ligand-expressing cells, a phenomenon referred to as ‘reverse' signalling. Both the Eph receptors and the ephrins are divided into two subclasses (A and B) based on their affinities for each other and sequence conservation. In general, the A-subclass Eph receptors (EphA1–A10) bind to the A-class ephrins (ephrin-A1–A6), whereas the EphB-subclass receptors (EphB1–B6) interact with the B-subclass ephrins (ephrin-B1–B3). As both Eph receptors and ephrins are membrane bound, their interaction occurs only at the sites of cell–cell contact. It is thought that in the absence of cell–cell interactions, the molecules exist in loosely associated clusters (microdomains), which become more compact on Eph receptor/ephrin complex formation, generating clearly defined signalling centres (Pasquale, 2005).
The extracellular Eph receptor region contains a conserved amino-terminal ligand-binding domain (LBD). An adjacent cysteine-rich region might be involved in receptor–receptor oligomerization often observed on ligand binding (Lackmann et al, 1998), whereas the next two fibronectin repeats have yet to be assigned a clear biological function. The cytoplasmic Eph receptor region contains a kinase domain, a sterile α-motif domain, and a PDZ-binding motif. All ephrins have a 20 kDa extracellular receptor-binding domain, with B-type ephrins also containing a short cytoplasmic region.
Several crystal structures of Eph receptor/ephrin complexes involving B-class receptors have been reported (Himanen et al, 2001, 2004; Chrencik et al, 2006b). As understanding of the molecular mechanisms that bring about the unique partner preferences of the two molecular subclasses requires the comparison of A- and B-class structures, we determined the structure of an A-class receptor, EphA2, both alone and in complex with its cognate ligand, ephrin-A1.
Results And Discussion
Structure of the unbound EphA2-LBD
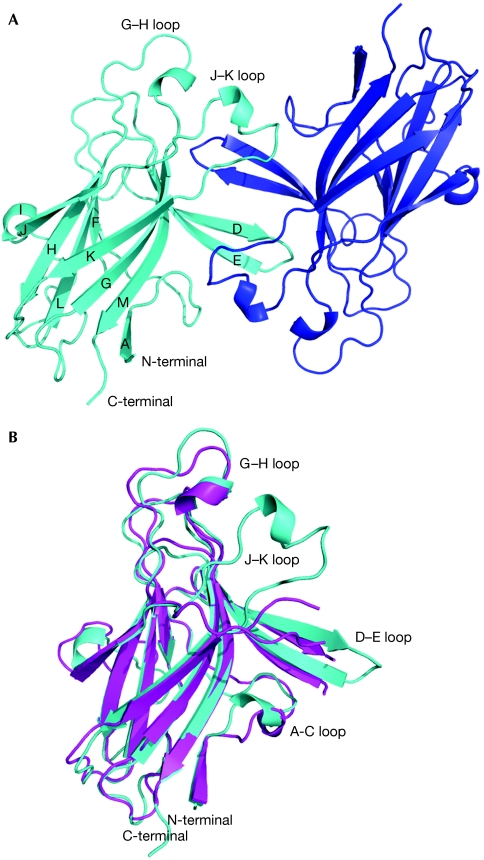
The structure of the human EphA2-LBD was determined by using molecular replacement at 2.5 Å resolution (supplementary Table S1 online). As in all Eph receptors, the EphA2-LBD forms a compact globular structure (Fig 1) with a β-sandwich jellyroll folding topology. The β-strands are connected by loops of varying length, including a long, well-ordered loop (H–I) that packs against one of the β-sheets, and two long loops (D–E and J–K), which protrude from the middle of the opposing β-sheet forming the Eph ligand-binding cleft. EphA2 shares 25–35% sequence identity with the other Eph receptors and, accordingly, its overall structure is similar to the other known Eph receptor structures, including the location of the two disulphide bridges (Cys 105–Cys 115 and Cys 70–Cys 188). The structures of the unbound EphA2 and EphB2, for example, can be superimposed with root-mean-square deviations (r.m.s.d.) between corresponding Cα positions of 1.7 Å (Fig 1B).
Figure 1.
Structure of the EphA2 ligand-binding domain. Secondary structure elements are labelled. (A) Structure of EphA2-ligand-binding domain dimer in the asymmetric unit. One monomer is shown in cyan, the other one in blue. (B) Comparison of the structures of unbound EphA2 (cyan) and EphB2 (magenta).
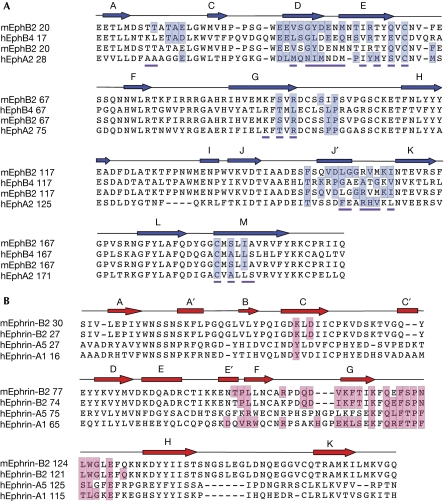
The main structural differences between EphA2 and the B-class Eph receptors are the conformations of the D–E, J–K and H–I loops. The length of the Eph receptor H–I loop is the only clear feature that distinguishes the sequences of the A- and B-class molecules, with EphA receptors having H–I loops that are four residues shorter than the B-class molecules (Fig 2A; Himanen & Nikolov, 2003). This loop is not part of the high-affinity Eph receptor/ephrin interface, but in the EphB2/ephrin-B2 structure is involved in low-affinity packing interactions generating Eph/ephrin heterotetramers (Himanen et al, 2001). In the EphB2/ephrin-A5 and EphB4/ephrin-B2 structures, conversely, only the high-affinity ligand–receptor heterodimers have been observed, and the precise role of the Eph receptor H–I loop in the formation of Eph receptor/ephrin signalling clusters is not yet fully understood. The structures of Eph receptor D–E, J–K loops, which form the ligand-binding cleft, also differ between the EphA2 and EphB2/B4 receptors. Although in the unligated B-class receptors they adopt more open conformations (Goldgur et al, 2009), in EphA2 they form a compact ligand-binding channel even in the absence of the ligand (see later).
Figure 2.
Structure-based sequence alignment of Eph receptors and ephrins. Secondary structure elements are shown above the sequences. The residues forming the high-affinity ligand–receptor interfaces are highlighted in blue and red. (A) Sequence alignment of Eph receptors the structure of which, bound to their ligands, has been reported. The residues forming the EphA2 homodimer interface observed in the unligated structure have a magenta underscore. (B) Sequence alignment of ephrins the structure of which, bound to their receptors, has been reported.
EphA2 dimers in the crystal
Analytical ultracentrifugation and gel-filtration experiments (supplementary Fig S1 online) reveal that the EphA2-LBD is a monomer in a solution at a low concentration (∼10 μM). Nevertheless, it forms a dimer at high concentrations in the asymmetric unit of the crystals (Fig 1A). The two EphA2 monomers are similar with r.m.s.d. between equivalent Cα positions of ∼0.1 Å. The dimer interface is formed mostly by ligand-binding D–E and J–K loop residues (see Figs 2A and 3A). Indeed, as Fig 2A shows, most of the EphA2 ligand-interacting residues are part of the Eph homodimer interface in the unligated receptor. Therefore, this interface is quite expansive and predominantly hydrophobic, burying a total of 2,200 Å2. This observation is consistent with the theory that, at high Eph receptor concentrations at the cell surface, these receptors might exist as pre-formed inactive (non-signalling) dimers, which rearrange on ligand binding. Alternatively, as the EphA2 dimer is anti-parallel with the carboxyl terminal ends facing opposite directions, the structure suggests that EphA2 could mediate cell–cell adhesion through homophilic interactions in trans between adjacent cells. Although dimerization of EphA receptors in cis, on the same cells, is well documented and involves many regions of the ectodomain (Lackmann et al, 1998), homophilic interactions in trans have not been shown directly before. Such interactions might be specific for EphA2. In the well-polarized Madin–Darby canine kidney and MCF10A human breast epithelial cells, there is low basal activation of EphA2, which is localized primarily to adherens junctions (Miao et al, 2003), indicating that most of it is not actively engaged with ephrin-As but might, instead, mediate homotypic cell adhesions. On the basis of the structure of the unligated EphA2 dimer, we propose that in the absence of ephrin-As, EphA2 participates in specific head-to-head (in trans) interactions at adherens junctions. The binding affinities are not high, but the large number of interacting EphA2 receptors could still significantly contribute to cell–cell adhesion and might have signalling mechanisms distinct from those induced by the heterophilic EphA2/ephrin-A interactions.
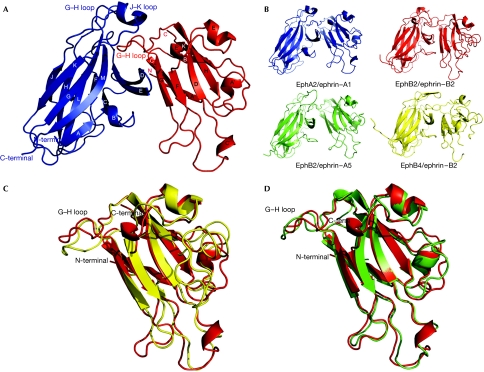
Figure 3.
Structure of the EphA2/ephrin-A1 complex. (A) Structure of the complex of EphA2 (residues Glu 28–Cys 201; blue) and ephrin-A1 (residues Ala 18–Ile 151; red). Secondary structure elements are labelled. (B) Comparison of the structures of the known A- and B-class Eph receptor/ephrin complexes. EphB2: Val 27–Arg 207; ephrin-B2: Ile 31–Gly 168; ephrin-A5: Val 28–Met 165; EphB4: Glu 17–Lys 196. (C) Comparison of the structures of Eph receptor-bound ephrin-A1 (red) and ephrin-B2 (yellow). (D) Comparison of the structures of Eph receptor-bound ephrin-A1 (red) and ephrin-A5 (green).
Structure of the EphA2/ephrin-A1 complex
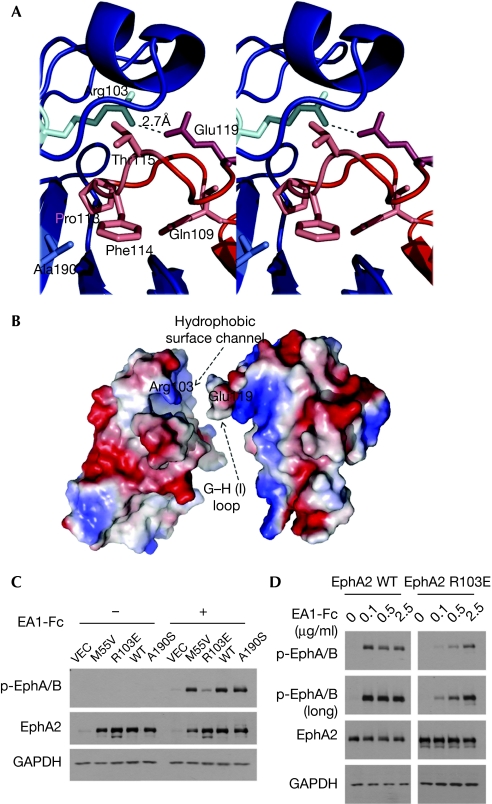
The EphA2/ephrin-A1 complex is a heterodimer in solution (supplementary Fig S1 online), and the crystal structure reveals a 1:1 ligand/receptor complex. There are eight EphA2/ephrin-A1 heterodimers in the asymmetric unit that are similar, with the r.m.s.d. between equivalent Cα positions in the range of 0.1–0.2 Å. The EphA2/ephrin-A1 heterodimer is architecturally similar to the B-class complexes (Fig 3). The ligand/receptor interface centres around the G–H loop of ephrin-A1, which is inserted in a channel on the surface of EphA2 (Figs 3 and 4A). Eph receptor strands D, E and J, define the two sides of the channel, whereas strands G and M line its back. The ligand binds by approximating the side of its β-sandwich to the outside surface of the channel and then inserting its long G–H loop into the channel, which finally becomes buttressed by the G–H loop of the receptor closing in from the top. The binding is dominated by the Van der Waals contacts between two predominantly hydrophobic surfaces, as the ligand buries Gln 109, Phe 111, Thr 112, Pro 113, Phe 114, Thr 115, Leu 116 and Gly 117 (Fig 4A). Gln 109 interacts not only with the sides of the channel, but also with EphA2 residues, Phe 100 and Pro 101, from the long G–H loop at the top of the interface. At the tip of the inserted ephrin loop are Pro 113, Phe 114 and Thr 115, with the side chain of Phe 114 reaching to the very end of the channel. Pro 113 is in direct contact with the Cys 70--Cys 188 disulphide bridge in EphA2. Adjacent to the channel/G–H-loop interactions, a second, structurally separate, contact area encompasses the ephrin-A1 docking site along the upper surface of the receptor. Here, the ephrin β-sandwich (strands C, G and F) interacts through a network of hydrogen bonds and salt bridges (Eph receptor/ephrin: Arg 103–Glu 119; Arg 159–Asp 86; Asp 53–Lys 107) with EphA2 strands D, E and the B–C loop region (Fig 3A).
Figure 4.
Steric and electrostatic complementarities at the EphA2/ephrin-A1 interface. (A) Stereoview of the ephrin-A1 G–H loop (red) buried in the EphA2 channel (blue). (B) The complementary molecular surfaces of EphA2 and ephrin-A1 colour coded according to electrostatic potential, illustrating the ‘lock, key and latch' binding mechanism (see text for further details). (C) R103E but not M55V or A190S substitutions reduced ligand-induced activation of EphA2. HEK293 cells were infected with retrovirus-expressing vector control (VEC), wild-type or mutant EphA2, and stimulated with ephrin-A1 (1 μg/ml) for 10 min. EphA2 activation was monitored by blotting with a p-EphA/B antibody, which detects active Eph kinases (Guo et al, 2006). (D) HEK293 cells were stimulated for 10 min with indicated concentrations of ephrin-A1-Fc, and checked for EphA2 activation by immunoblot.
The overall structure of the EphA2-LBD in the complex is similar to that of the unbound protein (Fig 5A,B). The most significant conformational changes involve loops at the ephrin-binding interface, including D–E, J–K, G–H and the A–C region. It should be noted, though, that the conformational changes in EphA2 on ligand binding are relatively minor, much smaller than the conformational changes observed in the B-class receptors (discussed further below).
Figure 5.
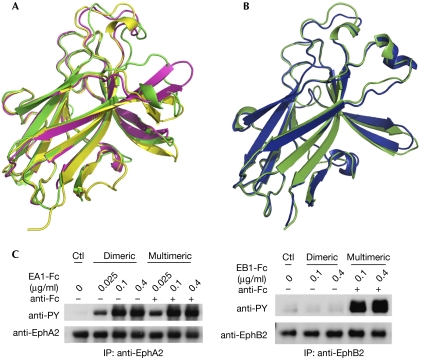
Conformational changes in A- and B-class Eph receptors on ephrin binding. (A) Comparison of the structures of unligated EphB2 (yellow), ephrin-bound EphB2 (green), and a 12-mer antagonistic peptide-bound EphB2 (magenta; Chrencik et al, 2006a). (B) Comparison of the structures of unligated (green) and ephrin-A1 bound (blue) EphA2. (C) Differential requirements for ligand oligomerization for activation of endogenous EphA2 and EphB2 in PC-3 cells. Dimeric ephrin-A1-Fc or ephrin-B1-Fc was clustered with anti-human Fc to generate multimeric ephrins. PC-3 cells were stimulated with the dimeric or multimeric ephrins for 10 min. EphA2 and EphB2 were immunoprecipitated (IP) and blotted with the indicated antibodies. Ctl, control.
Structure of ephrin-A1
The folding topology of ephrin-A1 (Fig 3A,C) is a variation of the Greek key β-barrel with eight mixed β-strands, three short α-helices and two buried disulphide bonds. Ephrin-A1 has significant sequence homology with other ephrins (30–40% amino acid identity; Fig 2B) and thus, the overall structure of ephrin-A1 can be superimposed with an r.m.s.d. value between the equivalent Cα positions of 1.2 Å on ephrin-A5 (Fig 3D; Himanen et al, 2004) and 1.8 Å on ephrin-B2 (Fig 3C; Himanen et al, 2001). The most structurally distinct region of the ephrins is their Eph receptor-binding (G–H) loop, which, interestingly, is also used by ephrin-B1 and -B3 to bind the attachment proteins of the henipaviruses, acting as their cellular receptors (Xu et al, 2008). The different conformations of the G–H loop suggest that it could undergo structural readjustments to accommodate different binding partners. Interestingly, ephrin-A1 has an extra helix (E′) that is absent in the other ephrin structures. Other structural differences between the A- and B-class ephrins include the conformation of the exposed A–B and C–D regions that are proposed to participate in higher-order clustering interactions (Himanen et al, 2001). In our crystals, ephrin-A1 is glycosylated at Asn 28, whereas no glycosylation is visible on EphA2.
Ephrin recognition by A-class Eph receptors
As in the B-class complexes, the EphA2/ephrin-A1 high-affinity interface contains two regions: one involving the ephrin G–H loop that inserts into a hydrophobic Eph receptor channel providing most of the thermodynamic driving force for complex formation, and a separate, mostly polar docking site. In the EphA2/ephrin-A1 case, though, this second, peripheral region is smaller and the overall EphA2/ephrin-A1 architecture is closer to that of the cross-class EphB2/ephrin-A5 complex than to the B-class complexes (Fig 3B). Nevertheless, the EphA2/ephrin-A1 dimerization interface (∼2,350 Å2) is similar in size to those observed in the B-class complexes, including EphB2/ephrin-B2 (1KGY: ∼2,400 Å2; Himanen et al, 2001) and EphB4/ephrin-B2 (2HLE: ∼2,150 Å2; Chrencik et al, 2006b), and significantly larger than the EphB2/ephrin-A5 cross-class complex (1SHW: ∼1,200 Å2; Himanen et al, 2004). This is mostly because of the more intimate Van der Waals interactions of the ephrin G–H loop with the surface receptor channel in the A-class complexes. The B factors of the interacting residues at the Eph receptor/ephrin interface (average value of 14.1 Å2) are significantly lower than the average for the entire complex (20.1 Å2), indicating a high degree of order at the area of contact.
Although the total buried surface area in the A- and B-class Eph receptor/ephrin interfaces is similar, the A-class molecules have been reported to interact, in general, with higher binding affinities than their B-class counterparts (Himanen et al, 2004; Pabbisetty et al, 2007). One possible explanation for this discrepancy is that B-class Eph receptor/ephrin recognition proceeds through an ‘induced-fit' mechanism, whereas A-class recognition seems to proceed through more of a ‘lock-and-key'-type mechanism. This, unique for the Eph receptors, use of different binding modes in different receptor subclasses is illustrated in Fig 5, in which the superimposed structures of the unbound, ephrin-bound and antagonist-peptide-bound EphB2 (Fig 5A) are compared with the superimposed structures of the unbound and ephrin-bound EphA2 (Fig 5B). In the case of EphB2, the loops forming the side of the ephrin-binding channel rearrange on ligand binding, thus requiring the energy to generate the extensive interaction surface that is complementary to the ephrin G–H loop. Conversely, during EphA2 binding to ephrin-A1, two relatively rigid molecular surfaces, already complementary to each other both in shape and in chemical nature, interact without need for significant conformational changes in either molecule. Indeed the r.m.s.d. between all Cα positions of ephrin-A1-bound and free EphA2 is only ∼0.7 Å, whereas it is ∼1.1 Å when comparing ephrin-B2-bound and free EphB2. To further strengthen the A-class binding, EphA2 Arg 103 forms a salt bridge with ephrin-A1 Glu 119. Fig 4B illustrates the two complementary interacting surfaces in EphA2 and ephrin-A1, highlighting the ‘key' (the ephrin G–H loop), the ‘lock' (the Eph receptor surface channel formed by the D–E and J–K loops) and the ‘latch' (EphA2 Arg 103–ephrin-A1 Glu 119). Thus, EphA2/ephrin-A1 binding could be described as a ‘lock, key and latch'-type binding in contrast to the clear ‘induced fit' mechanism used by the B-class receptors.
We evaluated directly the contribution of the ‘latch' to EphA2 receptor activation using structure-based mutagenesis. Compared with wild-type EphA2, an R103E mutant, expressed in human embryonic kidney (HEK)293 cells by retroviral gene transfer, showed a significantly reduced response to ephrin-A1 (Fig 4C). In a dose–response experiment, wild-type EphA2 was activated fully by 0.1 μg/ml ephrinA1-Fc, a dose that did not significantly activate R103E-EphA2 (Fig 4D). These data clearly show the unique importance of the ‘latch' (R103–E119) salt bridge in supporting optimal ligand binding and receptor activation.
The ‘lock and key' versus ‘induced-fit' mechanisms suggest that EphA2 can be activated more readily by its ligands than EphB2. To test this, we examined how EphA2 and EphB2 respond to ephrins that are in different states of oligomerization. It is known that effective activation of the Eph kinase domain requires multimerization of Eph receptors and ephrins, which is often achieved by pre-clustering of ephrin-Fc with, for example, Fc antibodies (Davis et al, 1994). PC-3 cells were used because they express endogenously both EphA2 and EphB2 at similar levels. Fig 5C shows that stimulation of PC-3 cells with pre-clustered ephrin-B1-Fc effectively activates EphB2, whereas un-clustered, dimeric ephrin-B1-Fc fails to do so. By contrast, EphA2 can be fully activated by dimeric ephrin-A1-Fc and pre-clustering does not promote significant further activation. The differential requirements for ligand pre-clustering are consistent with the prediction of our model that the B-class ephrins need more energy to induce the conformational changes necessary for EphB receptor binding, multimerization and activation.
Structural bases for the Eph-class specificity
Comparison of the EphA2/ephrin-A1 structure with the B-class complexes yields an insight into the molecular basis for the observed Eph receptor/ephrin subclass specificity. Indeed, Phe 114 from the G–H loop of ephrin-A1 makes a close Van der Waals contact with Ala 190 from EphA2, whereas in the EphB2/ephrin-B2 complex, the structurally equivalent residues are polar with Asn 123 from ephrin-B2 hydrogen bonding to EphB2 Ser 194 (supplementary Fig S2 online). In the peripheral docking region, EphA2 Met 55 makes a Van der Waals contact with Ser 105 from ephrin-A1, whereas in the B-class receptors, the methionine is substituted with a residue with a smaller side chain (Val 54 in the case of EphB2), which makes contact with a larger side-chain residue (Thr 114 in ephrin-B2; Fig 2). These types of arrangement maintain favourable contacts in A- or B-subclass complexes, but not in mixed (A+B) complexes.
In addition to the individual interacting residues, Eph receptor-subclass specificity is probably also maintained in part by the fact that the differences in the structures of the A- and B-class molecules result in different architectural arrangements of ligands and receptors in the A- and B-subclass complexes. Fig 3B illustrates the fact that the B-class complexes adopt a more ‘compact' conformation with intimate interactions between the Eph receptor B–C region and the juxtaposing C, F and G ephrin strands, whereas the A-class complex is more ‘open' with a smaller number of interactions in the above-mentioned region, but with a more intimate interaction network between the ephrin G–H loop and the Eph receptor D–E, J–K and G–H loops. Finally, the so-called ‘class-specificity' Eph receptor H–I loop might also contribute to the subclass binding and signalling preferences through its participation in tetramerization or higher-order interactions in the Eph receptor/ephrin signalling clusters at the sites of cell–cell contact (Himanen & Nikolov, 2003).
It should be noted that many individual interactions and other structural factors combine to define the specificity of the Eph receptor/ephrin recognition, and substitutions of individual residues would hardly affect ligand–receptor binding and activation. Indeed, we mutated three of the EphA2 residues discussed above (Arg 103, Met 55 and Ala 190) and only the alteration of Arg 103 (the latch) resulted in a significant decrease of EphA2 activation in response to ephrin-A1 (Fig 4C).
Structural insights into drug design
The Eph receptors are the largest RTK family and represent attractive targets for the development of anti-tumour and neuronal regeneration drugs. As the majority of the Eph receptor/ephrin interactions involves the extended G–H ephrin loop interacting with the Eph receptor surface channel, it has been proposed that small peptides and chemical compounds could be identified that bind to the Eph receptor channel and block Eph receptor signalling by preventing ephrin binding (Koolpe et al, 2002; Chrencik et al, 2006a). The EphA2/ephrin-A1 structure suggests that it would be easier to develop inhibitors of the A-class Eph receptor interactions because the ephrin-binding channel in the A-class receptors is already pre-formed in the unbound molecule, whereas in the EphB2 receptors it forms only subsequent to ligand binding. Indeed, small-molecule signalling inhibitors were reported recently that bind to the EphA2 and EphA4 receptors (Noberini et al, 2008; Qin et al, 2008). The antagonizing benzoic acid derivatives occupy a cavity in the ephrin-binding EphA channel by interacting with residues Ile 31–Met 32 in the D–E loop, Gln 43 in the E strand, and Ile 131–Gly 132 in the J–K loop. So far, no small-molecule antagonists have been found for any B-class receptors, highlighting the biological relevance of their different binding modes.
Methods
The human EphA2-LBD (residues 28–206) and the human ephrin-A1 ectodomain (residues 16–153) were expressed in HEK293 cells and purified as described (Himanen et al, 2004). EphA2 and EphA2/ephrin-A1 complex were concentrated to 5 mg/ml in a buffer containing 5 mM HEPES pH 7.0, 10 mM KCl and 2 mM CaCl2. The proteins were crystallized by hanging-drop vapour diffusion at 21°C against a well solution of either 25% PEG 4,000, 100 mM Tris (pH 8.5), 100 mM Na-acetate trihydrate, and 3% ethylene glycol (EphA2) or 20% PEG 8,000, 100 mM Tris (pH 8.5) and 200 mM MgCl2 (EphA2/ephrin-A1 complex). Native crystals were transferred into a cryo-buffer consisting of the mother liquor with an additional 20% glycerol. For structure determination, single-wavelength data sets were used, collected at National Synchrotron Light Source Brookhaven beamline X9A, at Cornell High Energy Synchrotron Source beamline A1, or at Advanced Photon Source beamline ID-24. The structures were determined (supplementary Table S1 online) by molecular replacement with either EphB2 (for EphA2) or ephrin-A5 (for EphA2/ephrin-A1) as models (Himanen et al, 2004; Goldgur et al, 2009), using the CCP4 program Amore (Project CCP4, 1994) and were refined by rigid body fitting in Refmac, followed by an iterative process of model improvement and Cartesian molecular dynamics and energy minimization in the CNS (Brunger et al, 1998). Stereochemical analysis of the final refined model with PROCHECK (Project CCP4, 1994) revealed side-chain parameters better than or within the typical range of values for protein structures. Site-directed mutagenesis and the cell-based Eph receptor activation assays were performed as described previously (Miao et al, 2003; supplementary information online).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank Ms Chen Li for expert technical assistance. This work was supported by National Institutes of Health (NIH) grants NS38486 to D.B.N., GM75886 to J.P.H., and CA96533, CA92259, DK077876 to B.W. The NECAT beamlines are supported by award RR-15301 from the National Center for Research Resources at the NIH. APS use is supported by the U.S. Department of Energy under contract no. DE-AC02-06CH11357.
Footnotes
The authors declare that they have no conflict of interest.
References
- Brunger AT et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Chrencik JE et al. (2006a) Structure and thermodynamic characterization of the EphB4/Ephrin-B2 antagonist peptide complex reveals the determinants for receptor specificity. Structure 14: 321–330 [DOI] [PubMed] [Google Scholar]
- Chrencik JE, Brooun A, Kraus ML, Recht MI, Kolatkar AR, Han GW, Seifert JM, Widmer H, Auer M, Kuhn P (2006b) Structural and biophysical characterization of the EphB4*ephrinB2 protein–protein interaction and receptor specificity. J Biol Chem 281: 28185–28192 [DOI] [PubMed] [Google Scholar]
- Davis S, Gale NW, Aldrich TH, Maisonpierre PC, Lhotak V, Pawson T, Goldfarb M, Yancopoulos GD (1994) Ligands for EPH-related receptor tyrosine kinases that require membrane attachment or clustering for activity. Science 266: 816–819 [DOI] [PubMed] [Google Scholar]
- Goldgur Y, Paavilainen S, Nikolov D, Himanen JP (2009) Structure of the ligand-binding domain of the EphB2 receptor at 2 Å resolution. Acta Crystallogr Sect F Struct Biol Cryst Commun 63: 969–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Miao H, Gerber L, Singh J, Denning MF, Gilliam AC, Wang B (2006) Disruption of EphA2 receptor tyrosine kinase leads to increased susceptibility to carcinogenesis in mouse skin. Cancer Res 66: 7050–7058 [DOI] [PubMed] [Google Scholar]
- Himanen JP, Nikolov DB (2003) Eph signaling: a structural view. Trends Neurosci 26: 46–51 [DOI] [PubMed] [Google Scholar]
- Himanen JP, Rajashankar KR, Lackmann M, Cowan CA, Henkemeyer M, Nikolov DB (2001) Crystal structure of an Eph receptor–ephrin complex. Nature 414: 933–938 [DOI] [PubMed] [Google Scholar]
- Himanen JP et al. (2004) Repelling class discrimination: ephrin-A5 binds to and activates EphB2 receptor signaling. Nat Neurosci 7: 501–509 [DOI] [PubMed] [Google Scholar]
- Koolpe M, Dail M, Pasquale EB (2002) An ephrin mimetic peptide that selectively targets the EphA2 receptor. J Biol Chem 277: 46974–46979 [DOI] [PubMed] [Google Scholar]
- Lackmann M, Oates AC, Dottori M, Smith FM, Do C, Power M, Kravets L, Boyd AW (1998) Distinct subdomains of the EphA3 receptor mediate ligand binding and receptor dimerization. J Biol Chem 273: 20228–20237 [DOI] [PubMed] [Google Scholar]
- Miao H, Nickel CH, Cantley LG, Bruggeman LA, Bennardo LN, Wang B (2003) EphA kinase activation regulates HGF-induced epithelial branching morphogenesis. J Cell Biol 162: 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noberini R, Koolpe M, Peddibhotla S, Dahl R, Su Y, Cosford ND, Roth GP, Pasquale EB (2008) Small molecules can selectively inhibit ephrin binding to the EphA4 and EphA2 receptors. J Biol Chem 283: 29461–29472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabbisetty KB, Yue X, Li C, Himanen JP, Zhou R, Nikolov DB, Hu L (2007) Kinetic analysis of the binding of monomeric and dimeric ephrins to Eph receptors: correlation to function in a growth cone collapse assay. Protein Sci 16: 355–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquale EB (2005) Eph receptor signalling casts a wide net on cell behaviour. Nat Rev Mol Cell Biol 6: 462–475 [DOI] [PubMed] [Google Scholar]
- Project CCP4 (1994) The CCP4 suite: programs for X-ray crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Qin H, Shi J, Noberini R, Pasquale EB, Song J (2008) Crystal structure and NMR binding reveal that two small molecule antagonists target the high-affinity ephrin-binding channel of the EphA4 receptor. J Biol Chem 283: 29473–29484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Rajashankar KR, Chan YP, Himanen JP, Broder CC, Nikolov DB (2008) Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc Natl Acad Sci USA 105: 9953–9958 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information