The Keystone Symposium on Chromatin Dynamics and Higher Order Organization took place between 25 February and 2 March 2009, in Coeur d'Alene, Idaho, USA, and was organized by T.Kohwi-Shigematsu & P.D. Varga-Weisz. Credit: Patrick Varga-Weisz.
Glossary
Introduction
At the recent Keystone Symposium on Chromatin Dynamics and Higher Order Organization, an emerging and expanded view of the chromatin landscape was apparent. Many reports connected the organization of individual nucleosomes and histone-modification domains with the spatial co-localization of genes and regulatory elements in three dimensions. It seems, now more than ever, that the nucleus is a lively place. Genes interact with one another at multiple levels during development and disease, and R. Ohlsson (Stockholm, Sweden) introduced the meeting attendees to useful monikers for the various levels of interaction: pairwise (‘dates'), small groups (‘parties') or large ensembles (‘galas'), all of which occur in a highly regulated manner. This report conveys several of the main themes of this active area of research, and highlights some of the advances and insights discussed at the meeting; we apologize to those speakers whose work we could not mention owing to space limitations.
The connected genome: chromatin-interaction networks
The co-localization of similarly regulated genes was a main topic of the meeting and the keynote lecture by M. Rosenfeld (San Diego, CA, USA) touched on many of the emerging themes. He presented several examples of how steroid hormone-induced transcriptional activation affects the nuclear positioning of target genes, as well as their spatial relationships. For example, he showed oestrogen-induced co-localization of many oestrogen receptor-responsive genes in a few foci, and presented experiments that indicated androgen-induced co-localization of androgen receptor-responsive genes in prostate cells. He also presented an intriguing series of experiments which suggested that co-localized genes might engage in preferred translocation events through a mechanism involving local DNA-break formation by the action of LINE-derived enzymes and subsequent non-homologous end joining. This observation might be related to the occurrence of specific translocation events in particular tissues and cancers in which LINE elements are derepressed. The idea sparked lively discussions during the session, as well as later in the bar.
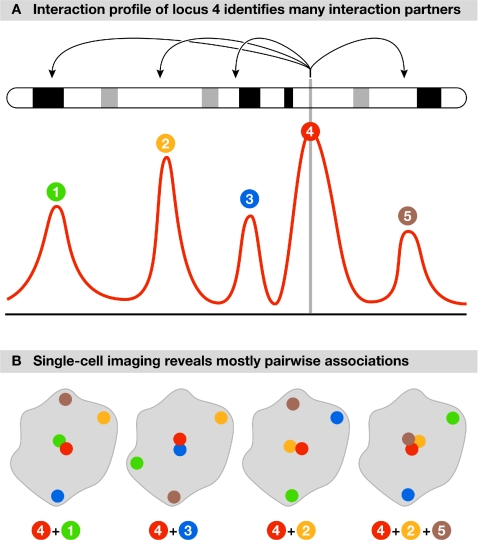
Long-range interactions are not only abundant, but also have roles in a wide range of processes (Dekker, 2008). Interactions such as those between genes and distant enhancers, as well as spatial clustering of genes, have long been thought to have a function in genome regulation (Fig 1). The ability to detect long-range interactions between genomic elements has been greatly facilitated by the development of 3C and related methods (4C, 5C and so on), which are used to detect the physical associations between single or multiple genomic loci, both along (in cis) and between (in trans) chromosomes (Dekker et al, 2002). The large number of these analyses presented at the meeting demonstrated that during the past few years 3C-based approaches have been widely and successfully used to study higher-order chromosome structure and, with the advent of high-throughput variants, nuclear organization in general.
Figure 1.
3C identifies which loci interact, whereas single-cell imaging reveals the topology of the interaction network. (A) The hypothetical 4C profile of a locus (numbered 4) across a chromosome identifies long-range associations with four other loci. (B) When analysed at the single-cell level, many associations occur in a pairwise manner, and only rarely do multiple loci interact simultaneously. 3C, chromosome conformation capture; 4C, chromosome conformation capture-on-chip or circular chromosome conformation capture.
Of particular interest is 5C—a high-throughput variant of 3C specifically designed to map networks of interactions among many loci (Dostie et al, 2006)—which can yield extremely dense interaction maps for genomic regions of up to tens of megabases. This is particularly suitable for making high-resolution three-dimensional models of higher-order chromatin organization, as shown in a presentation by J. Dekker (Worcester, MA, USA) in collaboration with M. Marti-Renom (Valencia, Spain).
Several new variations of high-throughput 3C methods were also reported. The laboratories of T. Kohwi-Shigematsu (Berkeley, CA, USA), P. Fraser (Cambridge, UK) and G. Cavalli (Montpellier, France) all presented clever variations of the previously developed 4C methods, which they used to identify loci throughout the genome that are spatially close to a single locus of interest. The Fraser laboratory, for example, applied their ‘enhanced' 4C method to the analysis of genome-wide interaction profiles involving the α-globin and β-globin loci in mice. Interestingly, they found that the active globin loci interact with many other transcribed genes in the genome, several of which are regulated by the transcription factor KLF1, which is a known regulator of globin loci. The interactions between these genes were shown to be dependent upon KLF1, leading Fraser to propose that genes sharing regulatory factors might congregate at a limited number of specialized transcription factories that are enriched in these factors. Subsequent FISH and immunofluorescence studies confirmed that many of the interacting genes found by enhanced 4C indeed co-localized at transcription factories enriched in KLF1.
Kohwi-Shigematsu briefly highlighted a study from her laboratory indicating that in thymocytes, the MYC gene interacts with a range of other genes throughout the genome at high frequencies, with many of the interactions apparently being mediated by the chromosome architectural protein SATB1. Interestingly, the set of genes that interact with the MYC gene was enriched for loci involved in MYC biology and MYC-regulated processes, again suggesting that biologically related genes tend to interact, perhaps at specialized subnuclear locations.
The R. Ohlsson (Stockholm, Sweden) laboratory used their 4C approach to search the genome for loci that interact with the ICR that is involved in regulating imprinting at the H19 gene locus in the mouse. Interestingly, they found that this locus frequently associates with sites that are near to other imprinted domains throughout the genome. Ohlsson suggested that the H19 ICR, which is one of the first evolved imprinted domains, has additional roles in organizing imprinted loci throughout the genome.
Taken together, these analyses suggest that the interaction profiles of loci and the resulting interaction networks reflect a spatial organization of the nucleus in which the co-regulated expression of genes occurs at a limited number of positions. However, 3C-based mapping approaches sample large cell populations and do not provide insight into the number of interactions in a single cell nucleus in which a particular locus is engaged. To address this problem, the groups mentioned above all used FISH to confirm their observations. Importantly, their FISH studies showed that, in most cases, interactions occur in a pairwise manner, with various pairs interacting in different cells. These observations emphasize the importance of combining 3C-based methods with single-cell observations. From this work, a picture emerges of the nucleus as an intricate and highly dynamic network of interacting loci. 3C-based methods uncover the many potential interaction partners throughout the genome, whereas single-cell FISH studies reveal significant cell-to-cell variation in which pairs of loci are co-localized (Fig 1).
Nuclear organization: inside out
Although nuclear organization is generally thought to be dynamic and variable, one aspect is typically considered to be constant: heterochromatin is located at the nuclear periphery, whereas euchromatic regions are localized at more internal positions. I. Solovei (Munich, Germany) presented a fascinating analysis of rod cells of the mouse retina, which was the result of a collaborative study between the laboratories of T. Cremer (Munich, Germany), L. Peichel (Frankfurt, Germany) and J. Gluck (Cambridge, UK). These investigators found that heterochromatin is located at the centre of the nucleus and that euchromatin is located at the periphery in these rod cells, inverting the usual global organization of the nucleus, although heterochromatic and euchromatic loci remain spatially separated (Solovei et al, 2009). Interestingly, the central location of heterochromatin reduces light scatter and makes rod cells more light sensitive, potentially explaining why this inverted organization is found specifically in nocturnal mammals.
The Polycomb connection
The role of PcG proteins in mediating chromatin dynamics and long-range interactions was another prominent theme at the meeting. The Polycomb genes were first identified in Drosophila as being required to maintain the silent state of Hox genes that control body-segment identity (Schuettengruber et al, 2007). The PRC2, which includes Ezh2, Suz12 and Eed, methylates histone H3 on lysine 27. Ezh2 is the SET domain-containing lysine methyltransferase, but the other proteins in the complex are required to reconstitute activity. H3K27me3 is then recognized and bound by PRC1. Binding of PRC1 is thought to cause gene silencing by several mechanisms, including chromatin compaction and the ubiquitination of histone H2A. PcG proteins bind to thousands of sites in the genome in a dynamic and cell type-specific manner. As PcG proteins cluster in the nucleus into 30–50 Polycomb bodies in fly cells, interaction with PcG can, in principle, bring several gene loci into close proximity (Schuettengruber et al, 2007).
How important are long-range interactions among target genes for Polycomb-mediated silencing? S. Baylin (Baltimore, MD, USA) showed that gene silencing in embryonic carcinoma cells is associated with H3K27me3 and PRC2 occupancy at promoters and distal elements, and that these PRC2-occupied sites come into physical proximity, as shown by 3C experiments (Tiwari et al, 2008). Depletion of PRC2 or the differentiation of embryonic carcinoma cells led to a cessation of the looping, as well as the derepression of target genes. Adding a new twist, Baylin showed that colon cancer cells exhibit enhanced chromosomal looping and DNA methylation, and that the loss of DNA methylation causes the chromosomal looping pattern to revert to one more similar to that in embryonic cells. Clearly, PRC2 occupancy might induce the close spatial proximity between the silenced target genes; however, whether the clustering is actually necessary for gene silencing is still unclear. The spatial clustering of Polycomb target genes might facilitate the recruitment of genes into nuclear territories that are inhospitable to transcription, as shown by A. Peters (Basel, Switzerland; Terranova et al, 2008).
Noncoding RNAs and repeats
The genome is pervasively transcribed, and both lncRNAs and sncRNAs are emerging as potential interfaces between DNA and specific enzymatic activities that programme chromatin states. Rosenfeld described a DNA damage-inducible ncRNA upstream of the cyclin D1 gene that recruited silencing factors in cis (Wang et al, 2008). J. Lee (Boston, MA, USA) highlighted her recent discovery of a specific RNA motif—known as ‘repeat A'—in the long ncRNA Xist, which interacts directly with Ezh2 and allows Xist to recruit PRC2 to the inactive X chromosome (Zhao et al, 2008). H. Chang (Stanford, CA, USA) discussed an lncRNA known as HOTAIR that can target the PRC2 to distant gene loci, thereby regulating their chromatin states (Rinn et al, 2007). Chang showed that HOTAIR not only regulates genes in the HOX loci, but might also regulate diverse classes of genes targeted by PRC2. Given the recent identification of a large number of evolutionarily conserved and highly regulated lncRNAs (Guttman et al, 2009), these types of mechanism will become evident in many other genes and biological states.
Repetitive elements are often thought of as junk DNA; however, the presence of similar sequences in multiple locations in the genome can be an important force that shapes long-range chromosomal interactions. S. Grewal (Bethesda, MD, USA) showed that many types of repeat in Saccharomyces pombe generate siRNAs, which, in turn, recruit the RNAi machinery and associated chromatin-modification enzymes to create distinct chromatin states, such as at centromeres and other types of repeat. Rosenfeld also showed that AR-induced gene clustering and translocation depends, in part, on homologous half AR sites on both partners of the kissing genes. It is likely that repeat elements have active regulatory roles in higher-order nuclear structure.
Insulators: a soiree of loci and factors
Chromatin insulators are elements that can either block enhancer–promoter communication or prevent heterochromatin spreading to delimit the genome into distinct functional chromatin domains. Insulators are bound by specific insulator proteins and their function might rely on their ability to form ‘insulator bodies' in which distant insulators might coalesce into discrete nuclear foci. As such, insulator bodies are the sites where long-range interactions occur between distant loci (reviewed in Gaszner & Felsenfeld, 2006; Valenzuela & Kamakaka, 2006).
M. Merkenschlager, (London, UK) and K. Yokomori (Irvine, CA, USA) reported the cohesin complex as a new component in mediating long-range interactions between sites bound by CTCF insulators. Cohesins form a ring-shaped protein complex that tethers sister chromatids together until mitosis (reviewed in Onn et al, 2008). Mutation in cohesin genes cause Cornelia de Lange syndrome, which leads to multiple developmental abnormalities (reviewed in Dorsett & Krantz, 2009). A role in gene expression was further illustrated in a presentation by D. Dorsett (St Louis, MO, USA), who found that Nipped-B—a cohesin loading factor in Drosophila—and cohesin are associated specifically with active genes, as well as with an interesting set of genes including the Enhancer of Split gene complex that is simultaneously bound by PcG proteins. At these sites, cohesin seems to repress transcription, and might therefore be a crucial orchestrator of the assembly/disassembly of insulator bodies and other types of higher-order chromosome dynamics. G. Felsenfeld (Bethesda, MA, USA) showed that BGP1, which is the chicken homologue of the mouse transcription factor Vezf1, contributes to the full function of a barrier insulator through a distinct mechanism controlling DNA methylation (Gowher et al, 2008).
Several families of insulators have been identified in organisms ranging from yeast to humans. In the case of Drosophila, each family contains a distinct set of DNA-binding motifs that are specifically recognized by one of the insulator proteins CTCF, Su(Hw) or BEAF. V. Corces (Atlanta, GA, USA) presented evidence that these insulator proteins might operate through some shared cofactors including CP190. As CP190 is known to be important for the formation of Su(Hw) or dCTCF insulator bodies (Gerasimova et al, 2007), it might provide an important conserved mechanism for insulator clustering and function.
O. Cuvier (Toulouse, France) showed that when BEAF binds to its characteristic ‘dual-cores' binding sites (Emberly et al, 2008)—which encompass positioned nucleosomes—it might in fact regulate nucleosome occupancy. A recent study also showed that CP190 can be independently detected at active promoter sites that are marked by the loss of nucleosome occupancy (Lefevre et al, 2008; Bartkuhn et al, 2009). Therefore, nucleosome occupancy might be regulated at all insulator bodies, including CTCF or Su(Hw), through the involvement of CP190. The modification of nucleosome occupancy at insulators might also account both for the presence of DHS in their sequences, which were used to identify the first insulator elements in Drosophila (reviewed in Gaszner & Felsenfeld, 2006), and for the phenomenon of ‘general Dnase I sensitivity', as suggested by C. Crane-Robinson (Portsmouth, UK).
Illustrating the links among insulators, chromatin domains and/or nuclear organization, J. van Bemmel (Amsterdam, the Netherlands) presented a genome-wide analysis of nuclear LADs in Drosophila conducted in the B. van Steensel laboratory. LADs define domains that are located at the nuclear periphery, where genes are generally repressed. Human LADs are known to be demarcated by the insulator protein CTCF, promoters or CpG islands, suggesting that there are various mechanisms to delimit them. In Drosophila, LADs are well defined by sharp transitions between chromatin domains, which coincide with increased Su(Hw) occupancy. It remains to be seen whether BEAF and CTCF might also delimit the repressed domains that are tagged by H3K27me3. The relationship between LADs and other chromosomal features is likely to provide a range of clues about the functions of insulators and chromosome organization.
Felsenfeld also reported that boundary elements of the chicken β-globin locus are marked by the histone isoforms H3.3 and H2AZ, which form unstable nucleosomes (Jin & Felsenfeld, 2007). Nucleosomes that are turned over in such a rapid manner might be used by the cell, together with epigenetic regulators, to demarcate chromatin domains by preventing the progressive spread of histone modifications from adjacent domains (Dion et al, 2007; Mito et al, 2007).
Emerging questions
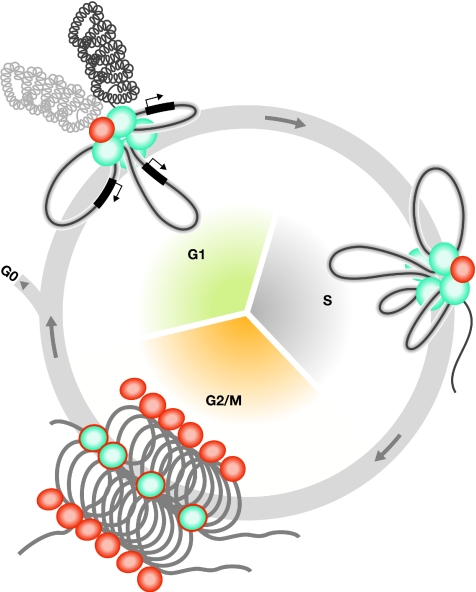
From the diverse presentations at the meeting, some of which we have related here, it became clear that local chromatin states and the three-dimensional organization of the genome are both dynamic and highly regulated. Going forward, it is important to determine how best to visualize and understand multiparameter chromosomal interaction maps, and how to overcome the limitation that many of the measurements we make are single snapshots in time. The field must also address how the interactions between various loci are coordinated with the cell cycle—against the competing needs of DNA replication and mitosis—and how they are controlled during development (Fig 2). Finally, it will be important to establish whether long-range chromosomal interactions can be targeted for use in therapeutic strategies against disease, which is certainly a worthy topic for future studies.
Figure 2.
Orchestration of higher-order chromatin dynamics during the cell cycle. Several crucial processes are regulated through the formation of factories/bodies such as transcription, Polycomb, insulator or SATB1 bodies/factories in the G1 phase, or replication, repair or recombination factories during the S phase. Their assembly/disassembly must be coordinated in a timely manner during the cell cycle, possibly involving common partners (perhaps cohesin) or mechanisms (such as DNA looping) that are also involved in the higher-order organization of chromosomes during G2/M phases. SATB1, special AT-rich sequence-binding protein 1.
3C chromosome conformation capture
4C chromosome conformation capture-on-chip or circular chromosome conformation capture
5C chromosome conformation capture-carbon copy
AR androgen receptor
BEAF boundary element-associated factor
BGP1 β-globin protein 1
CP190 centrosomal protein 190
CTCF CCCTC-binding factor
dCTCF Drosophila homologue of CTCF
DHS DNase I ‘hypersensitive sites'
Eed embryonic ectoderm development gene
Ezh2 enhancer of zeste homologue 2
FISH fluorescent in situ hybridization
H3K27me3 trimethylated lysine 27 on histone H3
HOTAIR Hox transcript antisense intergenic RNA
Hox homeobox
ICR imprinting control region
KLF Kruppel-like factor
LAD lamina-associated domain
LINE long-interspersed nuclear element
lncRNA long-noncoding RNA
ncRNA noncoding RNA
PcG polycomb group
PRC polycomb repressive complex
RNAi RNA interference
SATB1 special AT-rich sequence-binding protein 1
SET suppressor of variegation-enhancer of zeste-trithorax domain; typically encodes protein lysine methylase activity
siRNA short interfering RNA
sncRNA short-noncoding RNA
Su(Hw) suppressor of Hairy wing
Suz12 suppressor of zeste 12 homologue
Vezf1 vascular endothelial zing-finger 1
Xist X inactivation-specific transcript

Howard Y. Chang

Olivier Cuvier

Job Dekker
References
- Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, Saumweber H, Gilfillan GD, Becker PB, Renkawitz R (2009) Active promoters and insulators are marked by the centrosomal protein 190. EMBO J 28: 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J (2008) Gene regulation in the third dimension. Science 319: 1793–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N (2002) Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ (2007) Dynamics of replication-independent histone turnover in budding yeast. Science 315: 1405–1408 [DOI] [PubMed] [Google Scholar]
- Dorsett D, Krantz ID (2009) On the molecular etiology of Cornelia de Lange syndrome. Ann NY Acad Sci 1151: 22–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J et al. (2006) Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res 16: 1299–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emberly E, Blattes R, Schuettengruber B, Hennion M, Jiang N, Hart CM, Kas E, Cuvier O (2008) BEAF regulates cell-cycle genes through the controlled deposition of H3K9 methylation marks into its conserved dual-core binding sites. PLoS Biol 6: 2896–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G (2006) Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 7: 703–713 [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Lei EP, Bushey AM, Corces VG (2007) Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol Cell 28: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowher H, Stuhlmann H, Felsenfeld G (2008) Vezf1 regulates genomic DNA methylation through its effects on expression of DNA methyltransferase Dnmt3b. Genes Dev 22: 2075–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M et al. (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G (2007) Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev 21: 1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre P, Witham J, Lacroix CE, Cockerill PN, Bonifer C (2008) The LPS-induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol Cell 32: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S (2007) Histone replacement marks the boundaries of cis-regulatory domains. Science 315: 1408–1411 [DOI] [PubMed] [Google Scholar]
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE (2008) Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol 24: 105–129 [DOI] [PubMed] [Google Scholar]
- Rinn JL et al. (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G (2007) Genome regulation by polycomb and trithorax proteins. Cell 128: 735–745 [DOI] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, Guck J, Joffe B (2009) Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137: 356–368 [DOI] [PubMed] [Google Scholar]
- Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, Orkin SH, Peters AH (2008) Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell 15: 668–679 [DOI] [PubMed] [Google Scholar]
- Tiwari VK, McGarvey KM, Licchesi JD, Ohm JE, Herman JG, Schubeler D, Baylin SB (2008) PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol 6: 2911–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela L, Kamakaka RT (2006) Chromatin insulators. Annu Rev Genet 40: 107–138 [DOI] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R (2008) Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454: 126–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]