Summary
The basic helix-loop-helix (bHLH) transcription factor Hand2 is required for growth and development of the heart, branchial arches and limb buds. To determine whether DNA binding is required for Hand2 to regulate the growth and development of these different embryonic tissues, we generated mutant mice in which the Hand2 locus was modified by a mutation (referred to as Hand2EDE) that abolished the DNA-binding activity of Hand2, leaving the remainder of the protein intact. In contrast to Hand2 null embryos, which display right ventricular hypoplasia and vascular abnormalities, causing severe growth retardation by E9.5 and death by E10.5, early development of the heart appeared remarkably normal in homozygous Hand2EDE mutant embryos. These mutant embryos also lacked the early defects in growth of the branchial arches seen in Hand2 null embryos and survived up to 2 to 3 days longer than did Hand2 null embryos. However, Hand2EDE mutant embryos exhibited growth defects in the limb buds similar to those of Hand2 null embryos. These findings suggest that Hand2 regulates tissue growth and development in vivo through DNA binding-dependent and -independent mechanisms.
Keywords: bHLH, Hand2, Heart development, Limb development, Craniofacial development
INTRODUCTION
Basic helix-loop-helix (bHLH) transcription factors control the cell fate specification, growth and differentiation of numerous cell types during embryonic development (Massari and Murre, 2000). The HLH motif mediates the dimerization of tissue-specific class B bHLH factors with ubiquitous class A bHLH proteins, called E-proteins, juxtaposing their basic regions to form a bipartite DNA-binding domain that recognizes the E box consensus sequence (CANNTG). Binding of bHLH dimers to DNA results in either transcriptional activation or repression, depending on the components of the dimer and the target gene. The basic regions of bHLH proteins also participate in protein-protein interactions and, in the case of the myogenic bHLH proteins of the MyoD family, have been proposed to constitute a transcriptional code for muscle-specific gene activation (Brennan et al., 1991; Davis et al., 1990).
Hand1 (eHAND/Thing1/Hxt) and Hand2 (dHAND/Thing2/Hed) are bHLH transcription factors expressed in a variety of embryonic tissues, including the heart, branchial arches and limb buds (Angelo et al., 2000; Charite et al., 2000; Cross et al., 1995; Cserjesi et al., 1995; Hollenberg et al., 1995; Srivastava et al., 1995; Yelon et al., 2000). During mouse development, Hand1 and Hand2 are both expressed in the cardiac crescent. As the heart tube forms, Hand1 expression becomes localized to the outer curvature of the left ventricle (LV), derived from the primary heart field, whereas Hand2 expression becomes restricted to the right ventricle (RV) and derivatives of the secondary heart field. Mice lacking Hand1 die at E8.5 of placental and extra-embryonic defects, precluding analysis of its potential functions in the heart (Firulli et al., 1998; Riley et al., 1998). However, cardiac-specific deletion of the Hand1 gene in mice causes perinatal lethality with a spectrum of congenital heart defects that reflect abnormalities in ventricular growth (McFadden et al., 2005). Mice homozygous for a Hand2 null mutation die between E9.5 and E10.5 from right ventricular hypoplasia and defects in vascular development (Srivastava et al., 1997; Yamagishi et al., 2000). The phenotypes of Hand1/Hand2 double mutant mice indicate dose-sensitive functions of these transcription factors in the control of cardiac morphogenesis (McFadden et al., 2005). Further support for the essential role of Hand genes in ventricular growth comes from the phenotype of the zebrafish mutant hands off, which lacks the single Hand gene in that organism, and fails to form a ventricular chamber (Yelon et al., 2000).
Hand1 and Hand2 are expressed in the neural crest-derived mesenchyme of the developing branchial arches, where their expression is regulated by endothelin 1 (ET-1; Edn1 – Mouse Genome Informatics) signaling (Clouthier et al., 2000; Cserjesi et al., 1995; Thomas et al., 1998). Hand2 null embryos display hypoplastic first and second branchial arches, due to apoptosis, and fail to form the third and fourth arches before lethality at E10.5 (Srivastava et al., 1997; Thomas et al., 1998). Hand2 is also expressed in the posterior region of the limb mesenchyme that encompasses the zone of polarizing activity (ZPA) (Charite et al., 2000), where it is necessary to induce the expression of the morphogen sonic hedgehog (Shh) (Charite et al., 2000). Misexpression of Hand2 throughout the limb bud results in ectopic expression of Shh and its target genes in the anterior compartment of the limb bud with consequent preaxial polydactyly and mirror image duplications of posterior digits (Charite et al., 2000; Fernandez-Teran et al., 2000). It was also shown that the transcriptional repressor Gli3 restricts Hand2 expression from the anterior mesenchyme, and Hand2 in turn excludes Gli3 and Alx4 from posterior mesenchyme (te Welscher et al., 2002). Hand2 also positively regulates posterior expression of the BMP antagonist gremlin. These genetic interactions between Gli3 and Hand2 polarize the nascent limb bud mesenchyme prior to Shh signaling (te Welscher et al., 2002).
Despite detailed analysis of the consequences of loss-of-function Hand gene mutations in mutant mice, relatively little is known of the mechanism of action of Hand proteins during development. Like other bHLH proteins, Hand1 and Hand2 form heterodimers with E-proteins and can activate the transcription of E-box-dependent reporter genes when overexpressed in vitro (Firulli, 2003). However, mutant forms of Hand proteins defective in DNA binding can also activate transcription in overexpression assays (Xu et al., 2003; Rychlik et al., 2003). Similarly, a DNA binding-defective mutant of Hand2 was as effective as the wild-type protein in inducing preaxial polydactyly and digit duplications when misexpressed in the developing limb bud of transgenic mice (McFadden et al., 2002). These studies raise questions about the precise mechanism of action of Hand proteins and whether they can function at physiological levels in vivo without binding DNA.
In the present study, we used homologous recombination to introduce a mutation into the endogenous mouse Hand2 gene that abolished the DNA-binding activity of Hand2, but left the Hand2 protein otherwise intact. In contrast to a Hand2 null allele, which causes severe cardiac defects by E9.5 and lethality by 10.5, mice homozygous for the DNA-binding mutation, referred to as Hand2EDE, showed remarkably normal development of the heart and branchial arches until at least E11.5, when right ventricular growth became impaired and embryonic lethality ensued. However, similar to Hand2 null embryos, Hand2EDE/EDE embryos also showed underdeveloped limb buds and failed to express Shh and to restrict anterior gene expression, These findings suggest that Hand2 exerts at least a subset of its embryonic functions independently of DNA binding during the development of the heart and branchial arches, whereas during limb development the DNA-binding activity of Hand2 is indispensable for its function.
MATERIALS AND METHODS
Gene targeting
A previously described 8 kb Hand2 genomic clone (from BamHI site to BamHI site) was used to generate the targeting vector. A 4.6-kb fragment was used as the 5′ long arm for homologous recombination, and a 2-kb fragment was used as the 3′ short arm. Site-directed PCR mutagenesis was used to introduce mutations in six nucleic acids that changed the amino acid sequence RRR to EDE from amino acid 109-111 of the mouse Hand2 protein. Mutations were verified by sequencing of the entire PCR fragment. A Neomycin-resistance gene cassette driven by the pGK promoter and flanked by two Frt sites was inserted into the BstBI site within the intron sequence of the Hand2 gene. A diphtheria toxin A gene (DTA gene cassette from plasmid pGKDTAbpA) was inserted downstream of the Hand2 genomic fragment for natural negative selection. Integrity of the targeting vector was confirmed by restriction mapping and DNA sequencing. PCR primer sequences are available upon request.
The completed targeting vector was linearized with AhdI and electroporated into SM-1 ES cells. Following positive selection with G418 and natural negative selection of the DTA cassette, resistant colonies were screened by Southern blot analysis of NcoI-digested genomic DNA using a probe from the 3′ flanking region (Fig. 1B). Homologous recombination of the 5′ arm was confirmed by Southern blotting of NdeI-digested genomic DNA using a probe from the 5′ flanking region (Fig. 1B). Genomic DNA from positive clones was also subjected to PCR and sequencing around the exon 1 region of the Hand2 gene to confirm the introduced mutations. Three correctly targeted clones were expanded and injected into C57BL/6 blastocysts, and transferred into the uteri of pseudopregnant female mice. Chimeric males were bred to C57BL/6 mice to obtain germline transmission of the Hand2EDE-neo allele. In order to remove the neomycin resistance cassette in the germline, Hand2EDE-neo/+ mutant mice were bred to hACTB::FLPe transgenic mice (Rodriguez et al., 2000). Removal of the neomycin resistance cassette was confirmed by PCR genotyping (data not shown). The resulting Hand2EDE/+ heterozygous mutants were bred to each other to obtain Hand2EDE/EDE homozygous mutants.
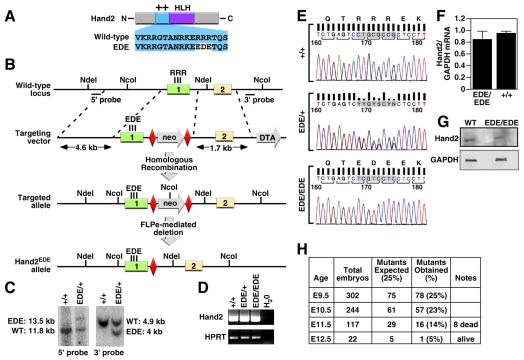
Fig. 1.
Generation of Hand2EDE mutant mice. (A) Schematic of the Hand2 protein. Amino acid sequences of the basic domain (++) of wild-type and EDE mutant protein are shown. The EDE mutant protein is also referred to as Hand2EDE. (B) Targeting strategy. Homologous recombination resulted in replacement of the region of exon 1 encoding the basic region and part of the intron with the exon 1 containing mutations and a neor cassette flanked by FRT sites (red diamond). After FLPe-mediated excision, one FRT site remained in the intron. Mutations representing the amino acid changes within the basic domain in exon 1 are shown. The positions of the 5′ long arm (4.6 kb), the 3′ short arm (1.7 kb), and the 5′ and 3′ probes used for Southern blotting are shown. (C) Southern blot analysis. Genomic DNA from ES cell clones was isolated and analyzed by Southern blotting with the 5′ probe after NdeI digest and the 3′ probe after NcoI digest. Hybridization of NdeI-digested ES cell DNA with the 5′ probe yielded an 11.8-kb fragment for the wild-type allele and a 13.5-kb fragment for the targeted allele because of the insertion of the neor cassette. Hybridization of NcoI-digested ES cell DNA with a 3′ probe yielded a 4.9-kb DNA fragment for the wild-type allele and a 4-kb fragment for the targeted allele due to an additional NcoI site in the neor cassette. The positions of wild-type and mutant bands are shown. Genotypes are shown at the top. (D,E) Analysis of Hand2 transcripts by RT-PCR and sequencing. RNA was isolated from wild-type, Hand2EDE/+ and Hand2EDE/EDE embryos at E9.5 and analyzed by RT-PCR to detect Hand2 transcripts. Transcripts for HPRT were detected as a control for RNA loading and integrity. The Hand2 PCR products were then gel-isolated and subjected to sequencing. The sequencing traces from wild-type, Hand2EDE/+ and Hand2EDE/EDE embryos are shown in E and the amino acid sequences are indicated. Because the Hand2 locus was sequenced with a reverse primer, the antisense sequences are shown and amino acid sequences of Hand2 are shown from right to left. (F) The Hand2 mRNA level was analyzed by quantitative real-time PCR. RNA isolated from wild-type and Hand2EDE/EDE embryos at E9.5 was analyzed by real-time PCR to detect levels of Hand2 mRNA. Relative expression normalized to GAPDH in wild-type and Hand2EDE/EDE embryos is shown. (G) Hand2 protein expression was detected by western blotting. Protein isolated from pooled E9.5 hearts from wild-type and Hand2EDE/EDE embryos was blotted against anti-Hand2 antibody. GAPDH protein was detected as a loading control. (H) Summary of homozygous Hand2EDE/EDE mutants obtained from heterozygous intercrosses.
PCR genotyping
Genotyping of Hand2EDE loci was performed by PCR with primers flanking the inserted FRT sequences. Tail and yolk sac DNA was isolated as previously described. One μl of tail or yolk sac DNA was used as a template in 25 μl PCR reactions using CLP Taq polymerase and 2 mM MgCl2. Thermal cycle reactions were as follows: 2 minutes at 95°C, 30 cycles of 30 seconds at 95°C, 30 seconds at 60°C, 20 seconds at 72°C and a final 5 minutes extension at 72°C. Reactions were visualized on 2% agarose/TAE gels. The wild-type allele gave rise to a 244-bp fragment, whereas the Hand2EDE allele gave rise to a 390-bp fragment. Primer sequences are available upon request.
RNA isolation, RT-PCR and real-time RT-PCR
Whole embryos or embryonic hearts at E9.5 were dissected and immediately frozen and stored in liquid nitrogen until yolk sac DNA was isolated and genotyped. Hearts of the same genotypes were pooled as one sample before RNA isolation. Total RNA was isolated using Trizol reagent and standard protocols. Total RNA (10 μg) was used as a template for reverse transcription with random hexamer primers. cDNA (25 ng) was used as template for PCR reactions using Promega Taq polymerase to detect Hand2 transcripts and transcripts for hypoxanthine phosphoribosyl transferase (HPRT) as a control. Reactions were visualized on 1% agarose/TAE gels and Hand2 RT-PCR products were gel-isolated and subjected to sequencing. Primer sequences are available upon request.
For quantitative real-time PCR, cDNA (25 ng) was amplified in each reaction by using the TaqMan Universal PCR Master Mix Kit (Applied Biosystems, Foster City, CA). Mean relative gene expression was calculated by using standard curves from serial dilutions of cDNA from wild-type embryos and normalized to GAPDH (n=3 per group).
Western blotting analysis
Hearts from E9.5 embryos were dissected and immediately frozen and stored in liquid nitrogen until yolk sac DNA was isolated and genotyped. Hearts from the same genotypes were pooled as one sample before protein isolation. Equal amounts of protein were loaded onto 4-20% SDS-PAGE gels and western blotting was performed according to standard protocols. The antibody against Hand2 (Santa Cruz, 1:100) was described previously (Zhao et al., 2005).
Whole-mount in situ hybridization
Embryos were harvested at the indicated age and fixed in 4% paraformaldehyde in PBS overnight at 4°C. Whole-mount in situ hybridizations were performed as previously described (Clouthier et al., 2000) using digoxigenin-labeled riboprobes.
Histology and skeletal analysis
Embryos were harvested from timed matings and fixed overnight in 4% paraformaldehyde in phosphate-buffered saline (PBS) overnight at 4°C. Following fixation, embryos were rinsed in PBS and then embedded in paraffin as previously described. Sections were cut and stained with Hematoxylin and Eosin. For skeletal analysis, postnatal day 1 (P1) mice were collected, prepared and stained with Alizarin Red and Alcian Blue for visualization of bone and cartilage formation, respectively (Yanagisawa et al., 1998).
TUNEL and immunohistochemistry
TUNEL staining was performed on paraffin-embedded sections according to the Roche in situ cell death detection kit. Phospho-histone H3 antibody staining was performed as described previously (Xin et al., 2006). Briefly, sections were deparaffinized in xylene, rehydrated through graded ethanol to PBS, and permeabilized in 0.3% Triton X-100 in PBS. Sections were then blocked by 1.5% normal horse serum in PBS followed by incubation with rabbit anti-phosphohistone H3 (Upstate Cell Signaling Solutions, Charlottesville, VA) at a 1:200 dilution in 0.1% BSA in PBS overnight at 4°C. Sections were washed in PBS, and fluorescein-conjugated secondary antibodies (Vector Laboratories, Burlingame, CA) were applied at a 1:200 dilution in 1% normal horse serum for 1 hour.
RESULTS
Generation of mice expressing a DNA-binding-defective mutant form of Hand2
To determine whether the actions of Hand2 in vivo require direct DNA binding, we used homologous recombination to replace three asparagine residues (RRR) in the basic domain with acidic residues, Glu-Asp-Glu (EDE), creating a mutant Hand2 protein, referred to as Hand2EDE (Fig. 1A,B). Prior studies showed that this mutation abolished all detectable DNA-binding activity of Hand2 in vitro (McFadden et al., 2002). The mutant Hand2 protein was also expressed at the same level as the wild-type protein in transfected cells, dimerized with E12, and was localized to the nucleus (McFadden et al., 2002) (data not shown).
The mouse Hand2 gene contains two exons, and the basic domain is encoded by exon 1 (Fig. 1A). Our targeting strategy involved insertion of a neomycin-resistance cassette flanked by sites for the FLP recombinase (Frt) into the intron of the Hand2 gene (Fig. 1B). The targeting vector was electroporated into ES cells and positive clones containing the targeted allele were identified by Southern blot analysis (Fig. 1C). Injection of heterozygous ES cells into mouse blastocysts yielded chimeric male mice, which transmitted the mutant allele through the germline. The heterozygous mice were bred to mice expressing the FLPe recombinase to remove the neomycin-resistance cassette, leaving only a single Frt site in the intron (Fig. 1B). Successful removal of the neomycin-resistance cassette was detected by PCR analysis of tail DNA (data not shown).
To confirm that the Frt site in the intron did not alter Hand2 expression, we performed RT-PCR on RNA isolated from E9.5 embryos using primers that flank the junctions of exons 1 and 2 of the Hand2 gene. As shown in Fig. 1D, RNA from wild-type and Hand2EDE/EDE embryos yielded the expected PCR fragments, and DNA sequencing confirmed the presence of the EDE mutation in homozygous mutant embryos (Fig. 1E). The junction sequences between exons 1 and 2 in the homozygous embryos were also correct in the mutant embryos, indicating that the mutant allele was correctly spliced. Quantitative real-time PCR confirmed that wild-type and mutant Hand2 transcripts were expressed at comparable levels (Fig. 1F). Western blotting on E9.5 embryonic hearts also showed that the mutant Hand2 protein is expressed at a comparable level to the wild-type Hand2 protein (Fig. 1G). These results indicate that the mutations in the basic region did not alter protein stability or subcellular localization.
Prolonged survival of Hand2EDE/EDE mutant embryos
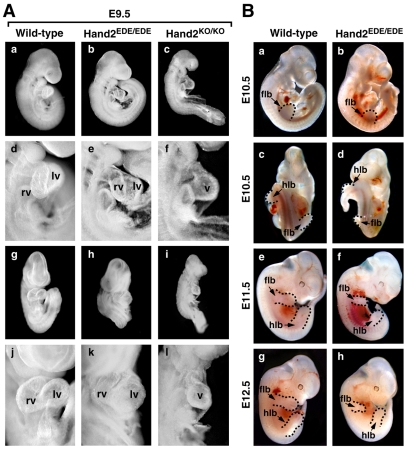
Genotyping of offspring from Hand2EDE/+ heterozygous crosses at postnatal day 10 revealed no viable Hand2EDE/EDE mice among more than 200 offspring examined, suggesting the homozygous mutation resulted in embryonic lethality (Fig. 1H). Homozygous Hand2 null (Hand2KO/KO) embryos show severe cardiac defects and a markedly dilated aortic sac by E9.5 followed by growth retardation and death by E10.5 with complete penetrance (Srivastava et al., 1997). By contrast, homozygous Hand2EDE/EDE embryos were morphologically indistinguishable from wild-type embryos at E9.5, except for a slightly smaller right ventricle and outflow tract (Fig. 2A). At E10.5, we obtained 57 viable Hand2EDE/EDE embryos out of 244 embryos examined from Hand2EDE/+ heterozygous crosses, representing Mendelian ratios (Fig. 1H). At this stage, these mutant embryos appeared grossly normal compared with wild-type embryos (Fig. 2B, parts a-d). We have never observed viable Hand2KO/KO embryos at E10.5 from several hundred embryos analyzed from heterozygous Hand2KO/+ intercrosses. At E11.5, 16 Hand2EDE/EDE embryos were obtained out of 117 embryos collected from heterozygous crosses and many showed multifocal hemorrhages, indicative of cardiovascular defects (Fig. 2B). The forelimb and hindlimb buds of Hand2EDE/EDE embryos at this stage were hypoplastic, and the face and jaws were also obviously abnormal (Fig. 2B, parts e,f).
Fig. 2.
Hand2EDE/EDE embryos from E9.5 to E12.5. (A) Wild-type, Hand2EDE/EDE and Hand2KO/KO embryos at E9.5 are shown. (a-c) Right view; (d-f) higher magnification shows hearts from the right view; (g-i) ventral view; (j-l) higher magnification shows hearts from the ventral view. Hand2EDE/EDE embryos were morphologically indistinguishable from wild-type embryos and showed significant formation of the RV at E9.5. By contrast, Hand2KO/KO embryos showed growth retardation and a hypoplastic RV. Lv, left ventricle; rv, right ventricle; v, single ventricular chamber. (B) Embryos at E10.5, E11.5 and E12.5. Hand2EDE/EDE embryos appear relatively normal but show multifocal hemorrhages at E11.5. The surviving Hand2EDE/EDE embryo at E12.5 showed no obvious heart defects. Delayed growth of the limb buds of Hand2EDE/EDE embryos was apparent at E10.5 and became more severe at E11.5 and E12.5. (a,b,e-h) Right view of the embryos; (c,d) ventral view. Limb buds are highlighted by dashed lines. flb, forelimb bud; hlb, hindlimb bud.
At E12.5, we obtained one viable Hand2EDE/EDE embryo out of 22 embryos collected from heterozygous crosses. This mutant embryo showed no signs of hemorrhage, but its limb buds were underdeveloped relative to those of wild-type embryos (Fig. 2B). No viable Hand2EDE/EDE embryos were obtained after E12.5. A summary of homozygous Hand2EDE/EDE embryos obtained from timed matings is shown in Fig. 1H. We conclude that the homozygous Hand2EDE/EDE mutation results in lethality between E10.5 and E12.5 due to cardiovascular defects, one to three days later than does the knockout of the Hand2 gene in Hand2KO/KO embryos.
Cardiac abnormalities of Hand2EDE/EDE embryos
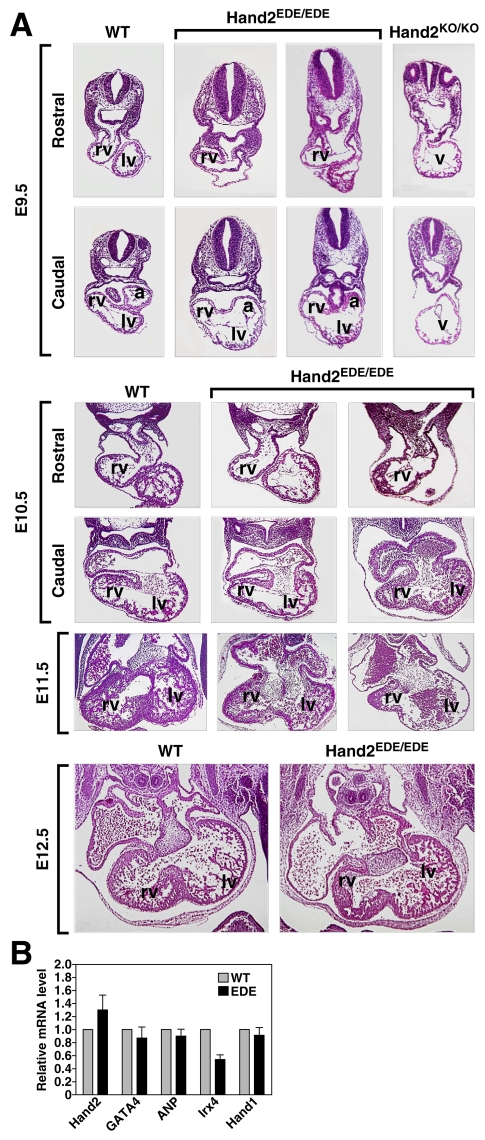
At E9.5, Hand2KO/KO embryos have an extremely hypoplastic right ventricle and outflow tract (Fig. 2A, Fig. 3A) (Srivastava et al., 1997). By contrast, the RV and outflow tract appeared to develop nearly normally in the Hand2EDE/EDE embryos at this stage, although they were slightly smaller relative to those of wild-type littermates (Fig. 3A). At E10.5, the RV of Hand2EDE/EDE embryos continued to develop and contained trabeculations (Fig. 3A). The endocardial cushions appeared slightly disorganized in the Hand2EDE/EDE mutant embryos at this stage, indicative of a potential delay in their formation. At E11.5, the Hand2EDE/EDE mutant embryos showed enlarged and disorganized endocardial cushions, and the RV showed reduced trabeculation compared with wild type (Fig. 3A). The LV of the Hand2EDE/EDE embryos was also abnormally thin with few trabeculations. The interventricular septum (IVS) in wild-type embryos was apparent, as evidenced by the thickening and alignment of cardiomyocytes in this region. In Hand2EDE/EDE mutants, the interventricular groove began to form but its development was delayed and myocytes failed to thicken (Fig. 3A). The cushion defect together with the underdeveloped RV and dilated LV probably cause embryonic lethality in Hand2EDE/EDE mutants. At E12.5, the heart of the surviving Hand2EDE/EDE embryo appeared normal, except for a slightly hypoplastic RV and IVS, and less trabeculation in the RV (Fig. 3A).
Fig. 3.
Heart development in Hand2EDE/EDE mutant embryos. (A) Hearts from wild-type, Hand2EDE/EDE and Hand2KO/KO mutant embryos at different time points were sectioned and stained with Hematoxylin and Eosin. Note that Hand2EDE/EDE hearts show significant formation of the RV compared with the lack of RV in the Hand2KO/KO embryos at E9.5. a, atrium; lv, left ventricle; rv, right ventricle; v, single ventricular chamber. (B) Expression of Hand2 and its target genes in the heart as detected by real-time PCR. RNA isolated from pooled E9.5 hearts from wild-type and Hand2EDE/EDE embryos was analyzed by quantitative real-time PCR. Relative expression normalized to GAPDH in wild-type and Hand2EDE/EDE embryos is shown.
Hand2 has been shown to regulate the expression of Gata4, Irx4 and ANP (Nppa – Mouse Genome Informatics) in the developing heart (Bruneau et al., 2000; Srivastava et al., 1997; Thattaliyath et al., 2002; Yamagishi et al., 2001). To further define the functions of the Hand2EDE protein, we analyzed the expression of these genes by quantitative real-time PCR in E9.5 hearts. As shown in Fig. 3B, expression of Gata4, ANP and Hand1 was unchanged in the hearts of mutant embryos compared with wild type, but expression of Irx4 was slightly decreased. These results suggest that Hand2 regulates expression of Gata4, ANP and Hand1 in a DNA-binding-independent manner, whereas regulation of Irx4 expression by Hand2 requires DNA binding.
Limb development in Hand2EDE/EDE embryos
During limb outgrowth and patterning, Hand2 is expressed in the posterior region of the limb bud, and it is necessary and sufficient to induce the expression of Shh (Charite et al., 2000). Hand2 was shown to be repressed by the transcriptional repressor Gli3 in the anterior mesenchyme of the limb buds, and Hand2 in turn represses the anterior genes Gli3 and Alx4 from being expressed in the posterior region (te Welscher et al., 2002). Hand2 is necessary for expression of the posterior gene gremlin (te Welscher et al., 2002). The repressive interactions between Gli3 and Hand2 prepattern the limb bud mesenchyme prior to Shh signaling (te Welscher et al., 2002). In Hand2 null mice, the forelimbs are hypoplastic, variably malformed and fail to upregulate Shh (Charite et al., 2000). Expression of Gli3 and Alx4 is no longer restricted to the anterior-most mesenchyme, but is expanded posteriorly in Hand2 null limb buds (te Welscher et al., 2002).
A developmental delay in the growth of the limb buds was apparent in the majority of Hand2EDE/EDE mutant embryos by E10.5 and became more extreme by E11.5 (Fig. 2B; Fig. 4A,B). The growth defects were apparent in both the anteroposterior (AP) and proximodistal dimensions of the fore- and hind-limbs at E11.5 (Fig. 4A,B). Additionally, the anterior margin of the forelimb buds appeared to be shifted posteriorly compared with that of wild-type embryos (Fig. 2B; Fig. 4A,C), a phenotype also seen in Hand2 null limb buds (Charite et al., 2000).
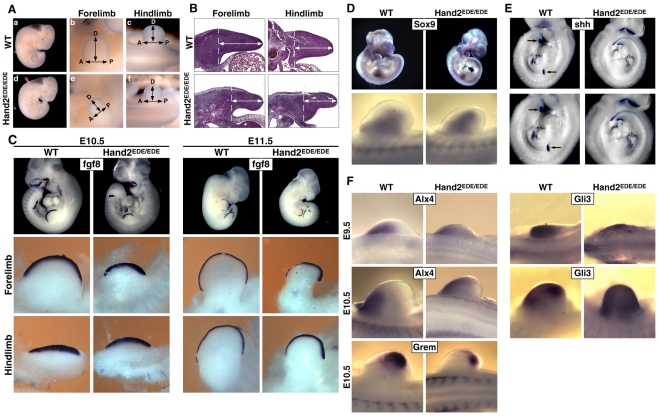
Fig. 4.
Limb development in Hand2EDE/EDE embryos. (A) At E11.5, Hand2EDE/EDE embryos have underdeveloped forelimb and hindlimb buds. Embryos were stained in 0.2% CoCl2 for visualization of the limb buds. (Left) Right view of the whole embryos; (middle and right) dorsal views of the forelimb and hindlimb buds with distal on the top and anterior to the left. (B) Transverse section of wild-type and mutant limb buds at E11.5 with distal on the top and anterior to the left. (C) Fgf8 expression in wild-type and Hand2EDE/EDE embryos detected by whole-mount in situ hybridization at E10.5 and E11.5. (Top) Views of the whole embryos; (middle and bottom) dorsal views of the forelimb and hindlimb buds with distal on the top and anterior to the left. (D) Sox9 expression in wild-type and Hand2EDE/EDE embryos detected by whole-mount in situ hybridization at E10.5. Bottom panel shows dorsal view of the forelimb buds. (E) Shh expression in wild-type and Hand2EDE/EDE embryos at E10.5 analyzed by whole-mount in situ hybridization. Arrows point to the Shh expression domain at the posterior margin of the forelimb and hindlimb buds of wild-type embryos. Expression of Shh was absent in the limb buds of Hand2EDE/EDE embryos. Upper images show the entire embryo, and lower images show higher magnification of the limb buds. (F) Expression of Alx4, Gli3 and gremlin (Grem) in wild-type and Hand2EDE/EDE embryos at the stages indicated. Dorsal views of the forelimb buds were shown with distal on the top and anterior to the left.
Using Fgf8 expression as a marker for the apical ectodermal ridge (AER) (Lewandoski et al., 2000; Moon and Capecchi, 2000), we examined the length of the AER in Hand2EDE/EDE mutant limb buds. We found that, at E10.5, the AER of Hand2EDE/EDE forelimbs was shorter along the AP axis, but the AER of the hindlimbs was comparable to that of the wild-type hindlimbs (Fig. 4C). At E11.5, the AER was significantly shorter in both forelimbs and hindlimbs of Hand2EDE/EDE mutant embryos (Fig. 4C). The shortened AERs in Hand2EDE/EDE mutant limb buds are likely to be due to disruption of the Shh/gremlin/Fgf loop that is important for maintenance of the AER (Panman et al., 2006; Sun et al., 2000; Zuniga et al., 1999). We also analyzed early chondrocyte condensation by Sox9 expression and observed no difference between wild-type and Hand2EDE/EDE mutant embryos at E10.5 (Fig. 4D).
Interestingly, all of the genes that are known to be regulated by Hand2 in limb buds were also affected in Hand2EDE/EDE mutant embryos (Fig. 4E,F). Shh, which was absent in Hand2 null limb buds, was also absent in the Hand2EDE/EDE forelimb and hindlimb buds at E10.5 (Fig. 4E). Similarly, expression of the anterior genes Alx4 and Gli3 was no longer restricted to the anterior-most mensenchyme, but was expanded posteriorly in Hand2EDE/EDE limb buds (Fig. 4F). In addition, gremlin expression was diminished in the posterior region of the Hand2EDE/EDE limb buds (Fig. 4F). Other genes involved in limb development, such as Msx1, Msx2, Prx1 and Tbx5, were expressed at normal levels in Hand2EDE/EDE embryos (data not shown).
Taken together, the result that Hand2EDE/EDE mutant embryos resemble Hand2 null embryos in terms of limb defects suggests that the DNA-binding activity of Hand2 is necessary for the regulation of Hand2 target genes during limb outgrowth and patterning.
Branchial arch development in Hand2EDE/EDE mutant embryos
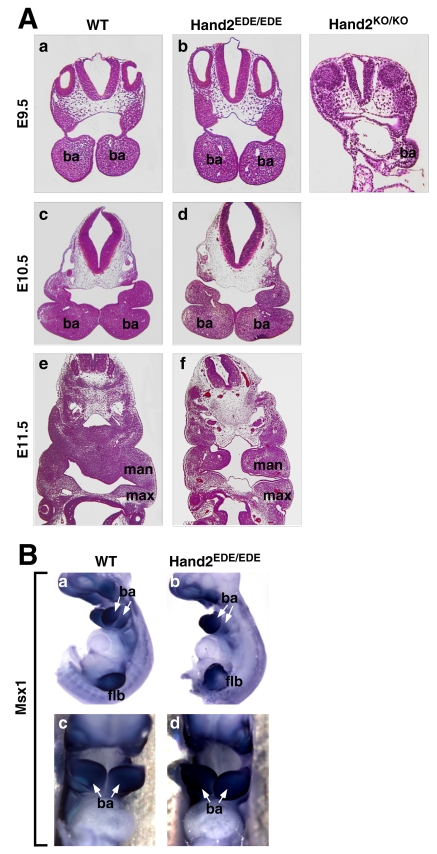
In Hand2 null embryos, the first and second branchial arches are severely hypoplastic at E9.5 due to extensive apoptosis, and the third and fourth arches fail to form (Srivastava et al., 1997; Thomas et al., 1998). At E9.5 and E10.5, the majority of Hand2EDE/EDE embryos had relatively normal branchial arches, with all four (first to fourth) being formed (Fig. 5A). However, at E11.5 in some Hand2EDE/EDE embryos, the left and right mandibular components of the first branchial arch of the Hand2EDE/EDE embryos failed to fuse, leaving a cavity in the center of the mandible (Fig. 5A). Hand2 was shown to regulate Msx1 expression during branchial arch growth, and, in Hand2-null embryos, no expression of Msx1 is detectable in the branchial arches (Thomas et al., 1998). Interestingly, in Hand2EDE/EDE embryos, Msx1 expression was not affected in the branchial arches at E10.5, as detected by whole-mount in situ hybridization (Fig. 5B). Other markers of the neural-crest-derived ectomesenchyme were expressed normally in branchial arches of Hand2EDE/EDE embryos at E10.5 (data not shown).
Fig. 5.
Branchial arch development in Hand2EDE/EDE embryos. (A) Transverse sections of branchial arches are shown from E9.5 to E11.5. Hand2EDE/EDE embryos have normal growth of branchial arches until E10.5. At E11.5, the mandibular components of the first brachial arch (man) of the mutant embryo were not fused (compare with those of the wild type). ba, branchial arch; man, mandibular components of the first branchial arch; max, maxillary components of the first branchial arch. (B) Expression of Msx1 transcripts at E10.5 was analyzed by whole-mount in situ hybridization. There is no significant difference in expression level of Msx1 between wild-type and Hand2EDE/EDE embryos. Arrows indicate branchial arches. Top panel, lateral view; bottom panel, ventral view. ba, branchial arches; flb, forelimb bud.
To determine the molecular basis of the defect in the first branchial arch of Hand2EDE/EDE embryos, we examined apoptosis and cell proliferation in the branchial arches of these embryos. In contrast to the extensive apoptosis in the first and second branchial arches at E9.5 in Hand2 null embryos (Thomas et al., 1998), there was no difference in the number of apoptotic cells observed by TUNEL assay in the branchial arches of the wild-type and Hand2EDE/EDE embryos at E10.5 (data not shown). Analysis of cell proliferation at E10.5 by staining with anti-phosphohistone H3 antibody also failed to reveal differences in the number of proliferating cells (data not shown). These results, together with results from whole-mount in situ hybridization, confirmed that, at least until E10.5, development of branchial arches was normal in the Hand2EDE/EDE embryos, indicating that regulation of the initial events of branchial arch development by Hand2 does not require its DNA-binding activity.
Hand2EDE mutant protein cannot support craniofacial development
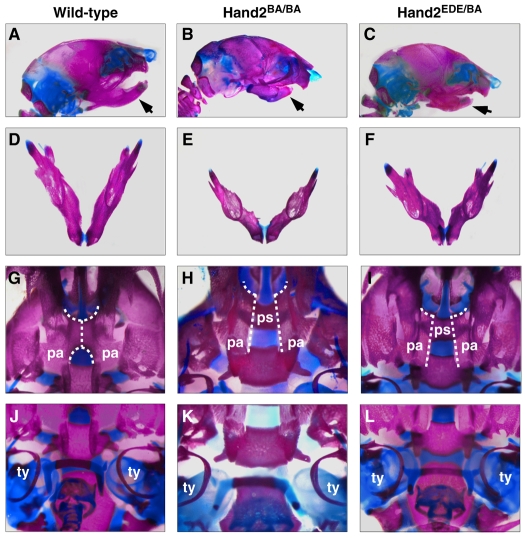
The embryonic lethality of Hand2EDE/EDE mutant mice precluded analysis of the potential requirement of Hand2 DNA binding in later phases of development in these embryos. To test whether DNA-binding activity was required for later steps in craniofacial development, we crossed Hand2EDE/+ mice with mice heterozygous for a deletion of the Hand2 enhancer that directs expression in the first and second branchial arches, referred to as Hand2BA mice (Yanagisawa et al., 2003). Homozygous Hand2BA/BA mice die at postnatal day 1 from failure to suckle and exhibit craniofacial abnormalities, including cleft palate and malformations of the mandible and Meckel's cartilage (Yanagisawa et al., 2003) (see also Fig. 6B,E,H,K). Hand2EDE/BA mice also died at P1 from failure to suckle and exhibited cleft palate. The mandibles of Hand2BA/BA mice (Fig. 6B, arrow), like those of Hand2EDE/BA mice (Fig. 6C, arrow), were hypoplastic and foreshortened compared with wild-type mice (Fig. 6A, arrow). Close examination showed that the angle between the right and left mandibular bones was also wider in both Hand2BA/BA and Hand2EDE/BA mutants than in wild-type mice (Fig. 6D-F). In wild-type mice, bilateral palatine processes extend horizontally and fuse to form the secondary palate (Fig. 6G, dotted vertical line). By contrast, the palatine processes of the Hand2EDE/BA and Hand2BA/BA mice did not appear to be elevated and thus the secondary palate was not formed (Fig. 6H,I, dotted vertical line), causing the underlying presphenoid bone to be visible in ventral view (ps in Fig. 6H,I). In addition, the tympanic rings of the Hand2EDE/BA mouse (Fig. 6L) were shortened and deformed compared with those of the wild-type mouse (Fig. 6J), a phenotype also observed in the Hand2EDE/BA mouse (Fig. 6K). In summary, the Hand2EDE/BA mouse exhibited similar craniofacial defects to the Hand2BA/BA mouse at P1, suggesting that Hand2EDE protein cannot support late craniofacial development. Thus, the DNA-binding activity of Hand2 is essential for its role in patterning and development of the cranial neural crest.
Fig. 6.
Craniofacial analysis of wild-type, Hand2BA/BA and Hand2EDE/BA mice at P1. (A,D,G,J) Wild-type mouse; (B,E,H,K) Hand2BA/BA mutant mouse; (C,F,I,L) Hand2EDE/BA mutant mouse. (A-C) Lateral view; (D-F) ventral view of the isolated jaws; (G-L) ventral view. (A-C) The mandibles (arrows) are smaller and are deformed in Hand2EDE/BA and Hand2BA/BA mice. (D-F) The mandibles in both mutant mice are shorter and are deformed. The angle between the left and right mandible is wider than that in the wild-type mouse. (G-I) In the wild-type mouse, the secondary palate is formed by the fusion of bilateral palatine processes (dotted vertical line). In Hand2BA/BA and Hand2EDE/BA mice, the palatine processes are not formed (dotted vertical lines), so the presphenoid (ps) bone becomes visible. (J-L) Tympanic rings (ty) are shortened and are deformed in the Hand2BA/BA and Hand2EDE/BA mice. pa, palatine; ps, presphenoid; ty, tympanic ring.
DISCUSSION
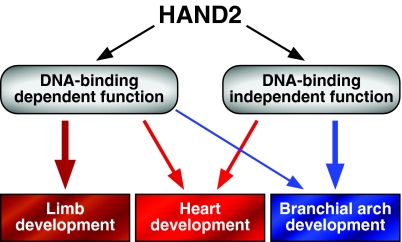
The results of this study demonstrate that a Hand2 mutant protein (Hand2EDE) devoid of DNA-binding activity is capable of supporting a subset of functions of the wild-type Hand2 protein during mouse embryogenesis. Hand2EDE is able to support heart development beyond the stage in which Hand2 null embryos die from ventricular hypoplasia and associated cardiovascular abnormalities, and can also support the early steps of branchial arch growth that require Hand2 function. However, the eventual death of homozygous Hand2EDE embryos indicates that this mutant cannot fully substitute for Hand2. These findings suggest that Hand2 acts through multiple mechanisms, both DNA-binding dependent and independent, to control tissue growth and morphogenesis in vivo (Fig. 7).
Fig. 7.
A model of Hand2 functions during development. Hand2 regulates gene expression by two different mechanisms: DNA-binding dependent and DNA-binding independent. During limb development, Hand2 functions through DNA-binding-dependent mechanisms. During early branchial arch development, Hand2 mainly acts through a DNA-binding-independent mechanism, but DNA binding is required for mandible development later. Both the DNA-binding-dependent and -independent functions of Hand2 are necessary for heart morphogenesis.
DNA-binding-dependent and -independent functions of Hand2
Tissue-specific (class B) bHLH proteins dimerize with ubiquitous bHLH proteins (E proteins) to form a bipartite DNA-binding domain that recognizes the E box consensus sequence (Massari and Murre, 2000). The basic regions of bHLH proteins are required not only for binding to target DNA, but also for protein-protein interactions and tissue-specific gene activation (Brennan et al., 1991; Davis et al., 1990).
It is striking that Hand2 is able to partially function in vivo in the absence of DNA binding. Although we cannot formally rule out the possibility that the Hand2EDE mutant protein retains residual DNA binding activity in vivo, we feel this is unlikely because we have detected no DNA-binding activity of this protein with a high-affinity binding site in vitro and because the replacement of basic residues with acidic residues in the basic regions of other bHLH proteins completely abolishes DNA-binding activity (McFadden et al., 2002).
How might Hand2 function in the absence of DNA binding? We suggest two possibilities. (1) Hand2 might interact with other transcriptional activators that are bound to DNA, thereby establishing a multi-protein transcriptional complex. There is evidence for such a mechanism from transfection assays (Rychlik et al., 2003; Xu et al., 2003). (2) Hand2 might act by titrating out repressor proteins, possibly through the HLH region, that negatively regulate specific gene programs.
SCL (also known as Tal1), a bHLH protein required for hematopoiesis and vascular development, has also been shown to possess DNA-binding-independent functions such that a DNA-binding-defective mutant can rescue hematopoiesis in SCL–/– ES cells, and can restore hematopoiesis and vasculogenesis when expressed in zebrafish (O'Neil et al., 2001; Porcher et al., 1999). Inhibition of the pro-osteogenic activity of the Runx2 transcription factor by the Twist family of bHLH proteins has also been shown to occur independently of DNA binding, at least in vitro (Bialek et al., 2004). Together, these studies suggest that bHLH proteins may operate through DNA-binding-dependent and -independent mechanisms. The present study is the first to analyze the requirement of bHLH protein DNA binding in vivo through the expression of a DNA binding-defective mutant from the endogenous gene locus at physiological levels.
Functions of Hand2 in the heart
The finding that formation and growth of the RV in Hand2EDE/EDE embryos proceed until at least E11.5 suggests that Hand2 may regulate two sets of target genes via different mechanisms during ventricular growth. Genes that control the initial patterning and growth of the RV may be regulated by Hand2 independently of DNA binding, whereas as the ventricle continues to expand, Hand2 may regulate another set of genes that require direct binding of Hand2 to its target DNA.
Hand2EDE/EDE mutant embryos display disorganized and enlarged endocardial cushions and a delay in formation of the IVS. We cannot rule out the possibility that these defects are secondary to delayed RV development, but we favor the interpretation that Hand2 is important in the formation of the endocardial cushions and the IVS, because embryos that lack cardiac expression of Hand1, which shares partially redundant functions with Hand2, also displayed a thickened and disorganized ventricular septum and hyperplastic endocardial cushions (McFadden et al., 2005). It has also been reported that overexpression of Hand2 in the ventricles results in a complete absence of the IVS, indicative of a negative role of Hand2 in the formation of this cardiac structure (Togi et al., 2006).
Functions of Hand2 in the limb buds
Limb bud patterning and outgrowth along the three axes are controlled by distinct but interdependent signaling pathways from both ectoderm and mesodermal mesenchyme (Mariani and Martin, 2003; Niswander, 2002). For example, limb outgrowth along the proximodistal axis is regulated by signals from the Fgf family in the AER (AER-Fgfs) (Boulet et al., 2004; Sun et al., 2002), and AP patterning is controlled by Shh secreted from the ZPA (Riddle et al., 1993). Precise interactions between the AER and the ZPA via the Fgf/gremlin/Shh positive loop and the Fgf/gremlin negative loop determine limb bud size and shape during limb outgrowth and termination (Panman et al., 2006; Sun et al., 2000; Verheyden and Sun, 2008; Zuniga et al., 1999). During initiation of limb bud outgrowth, prepatterning between anterior and posterior mesenchyme through genetic repression between Gli3/Alx4 and Hand2 determines posterior identity, which is essential for differential mesenchymal responsiveness to future Shh signaling (te Welscher et al., 2002).
Although elegant genetic studies have been performed, it remains unclear at the molecular level how Hand2 activates Shh and gremlin and represses Gli3 and Alx4 expression. The finding that Hand2EDE/EDE embryos display growth defects in limb buds suggests that Hand2 regulates limb growth through a DNA-binding-dependent mechanism. Our results also show that the regulation of Shh, Gli3, Alx4 and gremlin by Hand2 requires a functional DNA-binding domain. We showed previously that overexpression of the Hand2EDE mutant in the limb buds of transgenic mice can induce ectopic digit formation as effectively as the wild-type Hand2 protein (McFadden et al., 2002). The disparity between these phenotypes suggests either that regulation of digit patterning and growth by Hand2 are indeed independent of DNA binding, whereas earlier functions in limb bud outgrowth require DNA binding, or that the overexpressed Hand2EDE protein acts through a non-physiological mechanism to induce ectopic digits.
Functions of Hand2 in the branchial arches
Hand2 controls development of the branchial arches via a signaling pathway involving ET-1, Hand2 and Msx1 (Charite et al., 2001; Thomas et al., 1998). Our results suggest that the DNA-binding activity of Hand2 is dispensable during early branchial arch development, but is required for the development of mandibular components of the first branchial arch, as evidenced by failure in fusion of the left and right mandibular components in mutant embryos at E11.5. Interestingly, Hand2EDE/BA mutant mice display craniofacial defects at P1 that include shortened jaw (derived from mandibular components of the first branchial arch) and cleft palate, a phenotype similar to that of the Hand2BA/BA mice (Yanagisawa et al., 2003). These findings indicate that the DNA-binding activity of Hand2 is required for late craniofacial development, especially jaw formation.
Implications
In conclusion, the results of this study demonstrate both DNA-binding-dependent and -independent functions of the Hand2 protein during mouse embryonic development, underscoring the complexity of mechanisms by which this bHLH protein regulates such a diverse spectrum of developmental processes. Thus, the complete set of regulatory influences of Hand2 is likely to reflect not only direct protein-DNA interactions, but also protein-protein interactions, both positive and negative, in different cell types. We anticipate that this level of complexity is shared by other bHLH proteins that regulate the cell growth, specification and differentiation of other cell types.
We thank the Pathology Core Facility at UT Southwestern for histology. We thank John Shelton, Diana Chang, Cheryl Nolen and Gaile Vitug for technical help. We are grateful to Drs X. Sun and G. R. Martin for in situ probes and Dr X. Sun for discussions. We are grateful to Dr Noriko Funato for providing bone staining pictures of Hand2BA/BA pups. We thank Alisha Tizenor and Jose Cabrera for graphics and Jennifer Brown for editorial assistance. This work was supported by grants from the National Institutes of Health and the Donald W. Reynolds Clinical Cardiovascular Research Center. N.L. was supported by a grant from American Heart Association. Deposited in PMC for release after 12 months.
References
- Angelo, S., Lohr, J., Lee, K. H., Ticho, B. S., Breitbart, R. E., Hill, S., Yost, H. J. and Srivastava, D. (2000). Conservation of sequence and expression of Xenopus and zebrafish dHAND during cardiac, branchial arch and lateral mesoderm development. Mech. Dev. 95, 231-237. [DOI] [PubMed] [Google Scholar]
- Bialek, P., Kern, B., Yang, X., Schrock, M., Sosic, D., Hong, N., Wu, H., Yu, K., Ornitz, D. M., Olson, E. N. et al. (2004). A twist code determines the onset of osteoblast differentiation. Dev. Cell 6, 423-435. [DOI] [PubMed] [Google Scholar]
- Boulet, A. M., Moon, A. M., Arenkiel, B. R. and Capecchi, M. R. (2004). The roles of Fgf4 and Fgf8 in limb bud initiation and outgrowth. Dev. Biol. 273, 361-372. [DOI] [PubMed] [Google Scholar]
- Brennan, T. J., Chakraborty, T. and Olson, E. N. (1991). Mutagenesis of the myogenin basic region identifies an ancient protein motif critical for activation of myogenesis. Proc. Natl. Acad. Sci. USA 88, 5675-5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau, B. G., Bao, Z. Z., Tanaka, M., Schott, J. J., Izumo, S., Cepko, C. L., Seidman, J. G. and Seidman, C. E. (2000). Cardiac expression of the ventricle-specific homeobox gene Irx4 is modulated by Nkx2-5 and dHand. Dev. Biol. 217, 266-277. [DOI] [PubMed] [Google Scholar]
- Charite, J., McFadden, D. G. and Olson, E. N. (2000). The bHLH transcription factor dHAND controls Sonic hedgehog expression and establishment of the zone of polarizing activity during limb development. Development 127, 2461-2470. [DOI] [PubMed] [Google Scholar]
- Charite, J., McFadden, D. G., Merlo, G., Levi, G., Clouthier, D. E., Yanagisawa, M., Richardson, J. A. and Olson, E. N. (2001). Role of Dlx6 in regulation of an endothelin-1-dependent, dHAND branchial arch enhancer. Genes Dev. 15, 3039-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier, D. E., Williams, S. C., Yanagisawa, H., Wieduwilt, M., Richardson, J. A. and Yanagisawa, M. (2000). Signaling pathways crucial for craniofacial development revealed by endothelin-A receptor-deficient mice. Dev. Biol. 217, 10-24. [DOI] [PubMed] [Google Scholar]
- Cross, J. C., Flannery, M. L., Blanar, M. A., Steingrimsson, E., Jenkins, N. A., Copeland, N. G., Rutter, W. J. and Werb, Z. (1995). Hxt encodes a basic helix-loop-helix transcription factor that regulates trophoblast cell development. Development 121, 2513-2523. [DOI] [PubMed] [Google Scholar]
- Cserjesi, P., Brown, D., Lyons, G. E. and Olson, E. N. (1995). Expression of the novel basic helix-loop-helix gene eHAND in neural crest derivatives and extraembryonic membranes during mouse development. Dev. Biol. 170, 664-678. [DOI] [PubMed] [Google Scholar]
- Davis, R. L., Cheng, P. F., Lassar, A. B. and Weintraub, H. (1990). The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell 60, 733-746. [DOI] [PubMed] [Google Scholar]
- Fernandez-Teran, M., Piedra, M. E., Kathiriya, I. S., Srivastava, D., Rodriguez-Rey, J. C. and Ros, M. A. (2000). Role of dHAND in the anterior-posterior polarization of the limb bud: implications for the Sonic hedgehog pathway. Development 127, 2133-2142. [DOI] [PubMed] [Google Scholar]
- Firulli, A. B. (2003). A HANDful of questions: the molecular biology of the heart and neural crest derivatives (HAND) – subclass of basic helix-loop-helix transcription factors. Genes 312, 27-40. [DOI] [PubMed] [Google Scholar]
- Firulli, A. B., McFadden, D. G., Lin, Q., Srivastava, D. and Olson, E. N. (1998). Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat. Genet. 18, 266-270. [DOI] [PubMed] [Google Scholar]
- Hollenberg, S. M., Sternglanz, R., Cheng, P. F. and Weintraub, H. (1995). Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 15, 3813-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandoski, M., Sun, X. and Martin, G. R. (2000). Fgf8 signalling from the AER is essential for normal limb development. Nat. Genet. 26, 460-463. [DOI] [PubMed] [Google Scholar]
- Mariani, F. V. and Martin, G. R. (2003). Deciphering skeletal patterning: clues from the limb. Nature 423, 319-325. [DOI] [PubMed] [Google Scholar]
- Massari, M. E. and Murre, C. (2000). Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20, 429-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden, D. G., McAnally, J., Richardson, J. A., Charite, J. and Olson, E. N. (2002). Misexpression of dHAND induces ectopic digits in the developing limb bud in the absence of direct DNA binding. Development 129, 3077-3088. [DOI] [PubMed] [Google Scholar]
- McFadden, D. G., Barbosa, A. C., Richardson, J. A., Schneider, M. D., Srivastava, D. and Olson, E. N. (2005). The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development 132, 189-201. [DOI] [PubMed] [Google Scholar]
- Moon, A. M. and Capecchi, M. R. (2000). Fgf8 is required for outgrowth and patterning of the limbs. Nat. Genet. 26, 455-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswander, L. (2002). Interplay between the molecular signals that control vertebrate limb development. Int. J. Dev. Biol. 46, 877-881. [PubMed] [Google Scholar]
- O'Neil, J., Billa, M., Oikemus, S. and Kelliher, M. (2001). The DNA binding activity of TAL-1 is not required to induce leukemia/lymphoma in mice. Oncogene 20, 3897-3905. [DOI] [PubMed] [Google Scholar]
- Panman, L., Galli, A., Lagarde, N., Michos, O., Soete, G., Zuniga, A. and Zeller, R. (2006). Differential regulation of gene expression in the digit forming area of the mouse limb bud by SHH and gremlin 1/FGF-mediated epithelial-mesenchymal signalling. Development 133, 3419-3428. [DOI] [PubMed] [Google Scholar]
- Porcher, C., Liao, E. C., Fujiwara, Y., Zon, L. I. and Orkin, S. H. (1999). Specification of hematopoietic and vascular development by the bHLH transcription factor SCL without direct DNA binding. Development 126, 4603-4615. [DOI] [PubMed] [Google Scholar]
- Riddle, R. D., Johnson, R. L., Laufer, E. and Tabin, C. (1993). Sonic hedgehog mediates the polarizing activity of the ZPA. Cell 75, 1401-1416. [DOI] [PubMed] [Google Scholar]
- Riley, P., Anson-Cartwright, L. and Cross, J. C. (1998). The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat. Genet. 18, 271-275. [DOI] [PubMed] [Google Scholar]
- Rodriguez, C. I., Buchholz, F., Galloway, J., Sequerra, R., Kasper, J., Ayala, R., Stewart, A. F. and Dymecki, S. M. (2000). High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25, 139-140. [DOI] [PubMed] [Google Scholar]
- Rychlik, J. L., Gerbasi, V. and Lewis, E. J. (2003). The interaction between dHAND and Arix at the dopamine beta-hydroxylase promoter region is independent of direct dHAND binding to DNA. J. Biol. Chem. 278, 49652-49660. [DOI] [PubMed] [Google Scholar]
- Srivastava, D., Cserjesi, P. and Olson, E. N. (1995). A subclass of bHLH proteins required for cardiac morphogenesis. Science 270, 1995-1999. [DOI] [PubMed] [Google Scholar]
- Srivastava, D., Thomas, T., Lin, Q., Kirby, M. L., Brown, D. and Olson, E. N. (1997). Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat. Genet. 16, 154-160. [DOI] [PubMed] [Google Scholar]
- Sun, X., Lewandoski, M., Meyers, E. N., Liu, Y. H., Maxson, R. E., Jr and Martin, G. R. (2000). Conditional inactivation of Fgf4 reveals complexity of signalling during limb bud development. Nat. Genet. 25, 83-86. [DOI] [PubMed] [Google Scholar]
- Sun, X., Mariani, F. V. and Martin, G. R. (2002). Functions of FGF signalling from the apical ectodermal ridge in limb development. Nature 418, 501-508. [DOI] [PubMed] [Google Scholar]
- te Welscher, P., Fernandez-Teran, M., Ros, M. A. and Zeller, R. (2002). Mutual genetic antagonism involving GLI3 and dHAND prepatterns the vertebrate limb bud mesenchyme prior to SHH signaling. Genes Dev. 16, 421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thattaliyath, B. D., Firulli, B. A. and Firulli, A. B. (2002). The basic-helix-loop-helix transcription factor HAND2 directly regulates transcription of the atrial naturetic peptide gene. J. Mol. Cell. Cardiol. 34, 1335-1344. [DOI] [PubMed] [Google Scholar]
- Thomas, T., Kurihara, H., Yamagishi, H., Kurihara, Y., Yazaki, Y., Olson, E. N. and Srivastava, D. (1998). A signaling cascade involving endothelin-1, dHAND and msx1 regulates development of neural-crest-derived branchial arch mesenchyme. Development 125, 3005-3014. [DOI] [PubMed] [Google Scholar]
- Togi, K., Yoshida, Y., Matsumae, H., Nakashima, Y., Kita, T. and Tanaka, M. (2006). Essential role of Hand2 in interventricular septum formation and trabeculation during cardiac development. Biochem. Biophys. Res. Commun. 343, 144-151. [DOI] [PubMed] [Google Scholar]
- Verheyden, J. M. and Sun, X. (2008). An Fgf/Gremlin inhibitory feedback loop triggers termination of limb bud outgrowth. Nature 454, 638-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, M., Davis, C. A., Molkentin, J. D., Lien, C. L., Duncan, S. A., Richardson, J. A. and Olson, E. N. (2006). A threshold of GATA4 and GATA6 expression is required for cardiovascular development. Proc. Natl. Acad. Sci. USA 103, 11189-11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H., Firulli, A. B., Zhang, X. and Howard, M. J. (2003). HAND2 synergistically enhances transcription of dopamine-beta-hydroxylase in the presence of Phox2a. Dev. Biol. 262, 183-193. [DOI] [PubMed] [Google Scholar]
- Yamagishi, H., Olson, E. N. and Srivastava, D. (2000). The basic helix-loop-helix transcription factor, dHAND, is required for vascular development. J. Clin. Invest. 105, 261-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi, H., Yamagishi, C., Nakagawa, O., Harvey, R. P., Olson, E. N. and Srivastava, D. (2001). The combinatorial activities of Nkx2.5 and dHAND are essential for cardiac ventricle formation. Dev. Biol. 239, 190-203. [DOI] [PubMed] [Google Scholar]
- Yanagisawa, H., Yanagisawa, M., Kapur, R. P., Richardson, J. A., Williams, S. C., Clouthier, D. E., de Wit, D., Emoto, N. and Hammer, R. E. (1998). Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development 125, 825-836. [DOI] [PubMed] [Google Scholar]
- Yanagisawa, H., Clouthier, D. E., Richardson, J. A., Charite, J. and Olson, E. N. (2003). Targeted deletion of a branchial arch-specific enhancer reveals a role of dHAND in craniofacial development. Development 130, 1069-1078. [DOI] [PubMed] [Google Scholar]
- Yelon, D., Ticho, B., Halpern, M. E., Ruvinsky, I., Ho, R. K., Silver, L. M. and Stainier, D. Y. (2000). The bHLH transcription factor hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development 127, 2573-2582. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Samal, E. and Srivastava, D. (2005). Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436, 214-220. [DOI] [PubMed] [Google Scholar]
- Zuniga, A., Haramis, A. P., McMahon, A. P. and Zeller, R. (1999). Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature 401, 598-602. [DOI] [PubMed] [Google Scholar]