Abstract
Pancreatic cancer (PC) is the fourth most common cause of cancer death in the United States and the aggressiveness of PC is in part due to its intrinsic and extrinsic drug resistance characteristics, which is also associated with the acquisition of epithelial-to-mesenchymal transition (EMT). Emerging evidence also suggest that the processes of EMT is regulated by the expression status of many microRNAs (miRNAs), which are believed to function as key regulators of various biological and pathological processes during tumor development and progression. In the present study, we compared the expression of miRNAs between gemcitabine-sensitive and gemcitabine-resistant PC cells, and investigated whether the treatment of cells with “natural agents” [3,3′-diinodolylmethane (DIM) or isoflavone] could affect the expression of miRNAs. We found that the expression of miR-200b, miR-200c, let-7b, let-7c, let-7d, and let-7e was significantly down-regulated in gemcitabine-resistant cells that showed EMT characteristics such as elongated fibroblastoid morphology, lower expression of epithelial marker E-cadherin, and higher expression of mesenchymal markers such as vimentin and ZEB1. Moreover, we found that re-expression of miR-200 by transfection studies or treatment of gemcitabine-resistant cells with either DIM or isoflavone resulted in the down-regulation of ZEB1, slug, and vimentin, which was consistent with morphological reversal of EMT phenotype leading to epithelial morphology. These results provide experimental evidence, for the first time, that DIM and isoflavone could function as miRNA regulators leading to the reversal of EMT phenotype, which is likely to be important for designing novel therapies for PC.
Keywords: microRNA, EMT, drug-resistance, pancreatic cancer
Introduction
Pancreatic cancer (PC) is the fourth most common cause of cancer death in the United States with an estimated 37,680 new cases and 34,290 deaths in 2008 (1). For all stages combined, the 5-year relative survival rate is only 5%. The aggressiveness of pancreatic cancer could, in part, be due to their intrinsic and extrinsic drug resistance characteristics, resulting in their invasive behavior during early stages of carcinogenesis. Most patients diagnosed with PC are un-resectable and usually die from metastatic disease even after treatment with existing therapies. Therefore, there is a dire need for understanding the molecular mechanism involved in drug resistance and developing novel therapeutic strategies to overcome drug resistance for the successful treatment of PC.
Recent studies have shown that epithelial-to-mesenchymal transition (EMT) is associated with drug resistance and cancer cell metastasis (2–4). For most epithelial tumors, progression toward malignancy is accompanied by a loss of epithelial differentiation and a shift towards mesenchymal phenotype (5). During the acquisition of EMT characteristics, cancer cells lose the expression of proteins that promote cell-cell contact such as E-cadherin and γ-catenin, and gain the expression of mesenchymal markers such as vimentin, fibronectin, and N-cadherin, leading to enhanced cancer cell migration and invasion (5). Studies have also shown that cancer cells in the tumor center remain positive for the expression of E-cadherin and cytoplasmic β-catenin; however, the tumor cells in the periphery with propensity for invasion display loss of surface E-cadherin and up-regulation of vimentin, the typical characteristics of EMT phenotype (6). EMT has been shown to be important on conferring drug resistance characteristics to cancer cells against conventional therapeutics including taxol, vincristine, oxaliplatin, or epidermal growth factor receptor (EGFR) targeted therapy (4). Therefore, novel inhibitors of EMT or agents that could either reverse the EMT phenotype or kill EMT-type cells would be a novel strategy for the treatment of most PC. Moreover, agents that could either reduce drug resistance or reverse it would likely be useful for increasing therapeutic activity of conventional therapeutics.
Alteration in the expression of critical molecules has been observed during the acquisition of EMT phenotype, consistent with their association in cellular signal transduction pathways. The down-regulation and relocation of E-cadherin and zonula occludens-1, the translocation of β-catenin from cell membrane to nucleus, and the activation of Slug, Twist, and ZEB1 transcription factors result in the induction of EMT phenotype (7–12). Moreover, emerging evidence implicated the critical role of microRNAs (miRNAs) because they are key regulatory molecules in various biological and pathological processes including EMT. These small, non-coding molecules elicit their regulatory effects by imperfectly binding to the 3′ untranslated region (3′UTR) of target mRNA, causing either degradation of mRNA or inhibition of their translation to functional proteins (13, 14). The expression of miRNAs have been recognized as integral components of many normal biological processes, such as those involving cell proliferation, differentiation, apoptosis, and stress resistance (15). Moreover, it has been recently suggested that aberrant expression of miRNAs is associated with the development and progression cancer. Many studies have established this concept by discovering the up-regulation or down-regulation of specific miRNAs in various types of cancer and identifying some of their molecular targets (16–18). More importantly, miRNAs have been found to be the regulators of EMT (19), and thus miRNA appears to play important roles in cancer development and progression (18, 20).
Although the role of miRNAs in cancer has been documented, there are very few studies documenting the cellular consequence due to targeted inactivation or re-expression of specific miRNA. Therefore, we hypothesized that alteration in the expression of miRNAs could be achievable by treating cancer cells with “natural agents” [3,3′-diinodolylmethane (DIM) or isoflavone] and that the “natural agents” could cause reversal of EMT phenotype leading to cancer cell death. It has been well known that dietary chemopreventive agents inhibits cancer cell growth in vitro and in vivo through the regulation of cell signaling transduction (21, 22). Importantly, we have previously found that the dietary compounds including isoflavone genistein and 3,3′-diindolylmethane (DIM) could enhance the anti-tumor activity of chemotherapeutic agents through different mechanisms (23, 24), suggesting that these compounds could exert their pleiotropic effects on cancer treatment through regulation of various cellular signaling pathways. In this study, we assessed the expression pattern of miRNAs between gemcitabine sensitive and gemcitabine resistant pancreatic cancer cells. Moreover, we investigated whether the treatment of gemcitabine resistant cells with either B-DIM (BioResponse formulated DIM with greater bioavailability) or G2535 (a mixture of genistein and other isoflavones) could alter the expression profile of miRNAs and also assessed the cellular consequence. We found that the miRNA expression pattern was different between gemcitabine sensitive and resistant cells. The gemcitabine resistant cells showed EMT characteristics and the treatment of gemcitabine resistant cells with either of the natural agents altered the expression of miRNAs causing partial reversal of EMT phenotype, which was also achieved by re-expression of specific miRNA by transfection studies. These results suggest that re-expression of specific miRNA by non-toxic “natural agents” could be useful strategy for the reversal of EMT phenotype, which could lead to the reversal of drug resistance and the sensitization of PC cells to conventional therapeutic agents for the successful treatment of PC.
Materials and Methods
Cell lines, reagents, and antibodies
The reported gemcitabine-resistant human pancreatic cancer cell lines, MiaPaCa-2, Panc-1, and Aspc-1 (25), were maintained in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin in a 5% CO2 atmosphere at 37°C. The reported gemcitabine-sensitive human pancreatic cancer cell lines, L3.6pl, Colo357, BxPC-3, and HPAC (25), were cultured in DMEM or RPMI1640 (for BxPC-3) supplemented with FBS and antibiotics. B-DIM (BioResponse, Boulder, CO; known as BR-DIM and referred as B-DIM with higher bioavailability in vivo) was generously provided by Dr. Michael Zeligs (26) and was dissolved in DMSO to make a 50 mM stock solution. Isoflavone mixture G2535 (70.54% genistein, 26.34% diadzin and 0.31% glycitein manufactured by Organic Technologies and obtained from NIH) was dissolved in DMSO to make a stock solution containing 50 mM genistein. The concentration of isoflavone we described in this article all refer to the concentration of genistein in isoflavone mixture. Anti-ZEB1 (Santa Cruz, Santa Cruz, CA), antiE-cadherin (Santa Cruz), anti-vimentin (Dako), and anti-GAPDH (Sigma) primary antibodies were used for Western Blot analysis and immunofluorescent staining.
RNA isolation
Total RNA was extracted by using the mirVana™ miRNA Isolation Kit (Ambion Inc, Austin, TX) following the protocol provided by the manufacturer. Briefly, MiaPaCa-2, Panc-1, Aspc-1, L3.6pl, Colo357, BxPC-3, and HPAC cells were treated with 25 μM B-DIM or 25 μM G2535 for 48 hours. Control cells received 0.05% DMSO. Then, the cells from each condition were exposed to Lysis/Binding Solution and vortexed vigorously in order to obtain a homogenous lysate. miRNA Homogenate Additive was added to the lysate and incubated on ice for 10 minutes. After incubation, Acid-Phenol:Chloroform was added and the lysate was subsequently vortexed and centrifuged for 5 min at 10,000 × g. The aqueous phase was recovered and transferred to a fresh tube. After addition of 100% ethanol, each lysate/ethanol mixture was added to a filter cartridge and pulled through via centrifugation at 10,000 × g. Two wash solutions were added to the filter and pulled through via the same method. The total RNA was obtained by adding pre-heated (95°C) Elution Solution to the filter cartridge and centrifuging.
miRNA microarray and data analysis
Five micrograms of each total RNA sample from MiaPaCa-2, Panc-1, L3.6pl, and Colo357 cells were sent to a service provider for completion of the miRNA microarrays (LCSciences, Houston, TX). In LCSciences, the total RNA samples were enriched for microRNAs and the miRNA microarrays were performed on μParaFlo™ microfluidic chips (version 10.0), each of which has a miRNA probe region with multiple repeat regions that detect 711 miRNAs. Multiple control probes are also included on the microarrays for assessing various chip and assay qualities such as uniformity and specificity. Chips were scanned and the data were normalized. The t-tests were performed and the predicting target genes for various miRNAs were also analyzed.
miRNA Real-time RT-PCR
To verify the alterations in the expression of specific miRNAs that were found to be altered in miRNA microarray analysis, we chose representative miRNA (miR-200a, miR-200b, miR-200c, let-7b, let-7e, miR-106b, and miRNA-221) with varying expression profiles for real-time miRNA RT-PCR analysis using TaqMan MicroRNA Assay Kit (Applied Biosystems, Foster City, CA) following manufacturer’s protocol. Briefly, five nanograms of total RNA from each sample were subjected to reverse transcription with a specific miRNA primer (Applied Biosystems). Real-time PCR reactions were then carried out in a total of 25 μl reaction mixture (1.66 μl of RT product, 1.25 μl of 20X TaqMan miRNA Assay mix, 12.5 μl of 2× TaqMan PCR Master Mix, and 9.59 μl of dH2O) in SmartCycler II (Cepheid, Sunnyvale, CA). The PCR program was initiated by 10 min at 95°C before 40 thermal cycles, each of 15 s at 95°C and 1 min at 60°C. Data were analyzed according to the comparative Ct method and were normalized by RNU6B expression in each sample.
Real-time RT-PCR using mRNA
Real-time RT-PCR analysis was also conducted to measure the expression of genes (E-cadherin, vimentin, slug) which are critically related to EMT in pancreatic cancer cells with and without 25 μM B-DIM and 25 μM G2535 treatment. Briefly, one micrograms of total RNA from each sample were subjected to reverse transcription using the High Capacity RNA-to-cDNA Kit (Applied Biosystems) according to the manufacturer’s protocol. Real-time PCR reactions were then carried out in a total of 25 μl reaction mixture (2 μl of cDNA, 12.5 μl of 2X SYBR Green PCR Master Mix from Applied Biosystems, 1.5 μl of each 5 μM forward and reverse primers, and 7.5 μl of dH2O) in SmartCycler II (Cepheid). The PCR program was initiated by 10 min at 95°C before 40 thermal cycles, each of 15 s at 95°C and 1 min at 60°C. Data were analyzed according to the comparative Ct method and were normalized by GAPDH expression in each sample.
Western Blot analysis
We also conducted Western Blot analysis to verify the alterations in the protein expression of genes, which are targets of miRNA or critically involved in EMT. The gemcitabine-sensitive and –resistant pancreatic cancer cells were treated with or without 25 μM B-DIM or 25 μM G2535 for 48 hours. After treatment, the cells were lysed in 62.5 mM Tris-HCl and 2% SDS, and protein concentration was measured using BCA protein assay (PIERCE, Rockford, IL). The proteins were subjected to 10% or 14% SDS-PAGE, and electrophoretically transferred to nitrocellulose membrane. The membranes were incubated with specific primary antibodies, and subsequently incubated with secondary antibody conjugated with peroxidase (Bio-rad, Hercules, CA). The signal was then detected using the chemiluminescent detection system (PIERCE).
Re-expression of miR-200 in gemcitabine-resistant cells
MiaPaCa-2 and Panc-1 cells were seeded in 6 well plates and transfected with miR-200a, miR-200b, miR-200c, or miRNA negative control (Ambion) at a final concentration of 20 nM using DharmaFact Transfection Reagent (Dharmacon, Lafayette. CO). After 3 days of transfection, the cells were split and transfected repeatedly with the miRNA every 3–4 days for indicated times. Total RNA from each samples were then extracted using the Trizol reagent (Invitrogen). One microgram of RNA was subject to RT-PCR using the High Capacity RNA-to-cDNA Kit (Applied Biosystems) and SYBR Green PCR Master Mix ((Applied Biosystems) as described earlier. Total proteins from each sample were also extracted and subject to Western Blot analysis as described earlier.
Immunofluorescence staining
Immunofluorescence staining was performed following standard protocol. Briefly, B-DIM or G2535 treated MiaPaCa-2 cells were fixed with 4% paraformaldehyde, permeabilized in 0.5% Triton X-100, and blocked with 10% goat serum. The cells were then incubated overnight with specific primary antibodies. After washing with PBS, the cells were incubated with fluorescence conjugated secondary antibody for 1 hour. The slides were then washed with PBS and mounted with mounting medium containing anti-fade reagent and DAPI. Cells were viewed under fluorescence microscope and images were analyzed using Advanced Sport software (Diagnostic Instruments, Sterling Heights, MI).
Exposure of gemcitabine-resistant cells to miR-200, B-DIM or G2535 and WST-1 assay for testing drug sensitivity
The gemcitabine-resistant MiaPaCa-2 pancreatic cancer cells were transfected with miR-200b or control miRNA for 7 days or treated with lower concentration of B-DIM (5 μM) and G2535 (10 μM) for 21 days. The morphological changes in MiaPaCa-2 cells were captured under a light microscopy. The cells were then seeded in 96-well plates. After 24 hours, the cells were treated with different concentrations of gemcitabine for 48 hours. After treatment, the cells were subjected to cell proliferation assay using Cell Proliferation Reagent WST-1 assay (Roche, Indianapolis, IN) following the manufacturer’s protocol. Briefly, 10 μl of WST-1 reagent were added to the cells in each well and incubated at 37°Cfor 2 hours. After shaking for 1 min, the spectrophotometric absorbance of the samples was determined by using Ultra Multifunctional Microplate Reader (Tecan, Durham, NC) at 450 nm.
Results
Gemcitabine-resistant PC cells show EMT characteristics documented by miRNA, mRNA, and protein expression
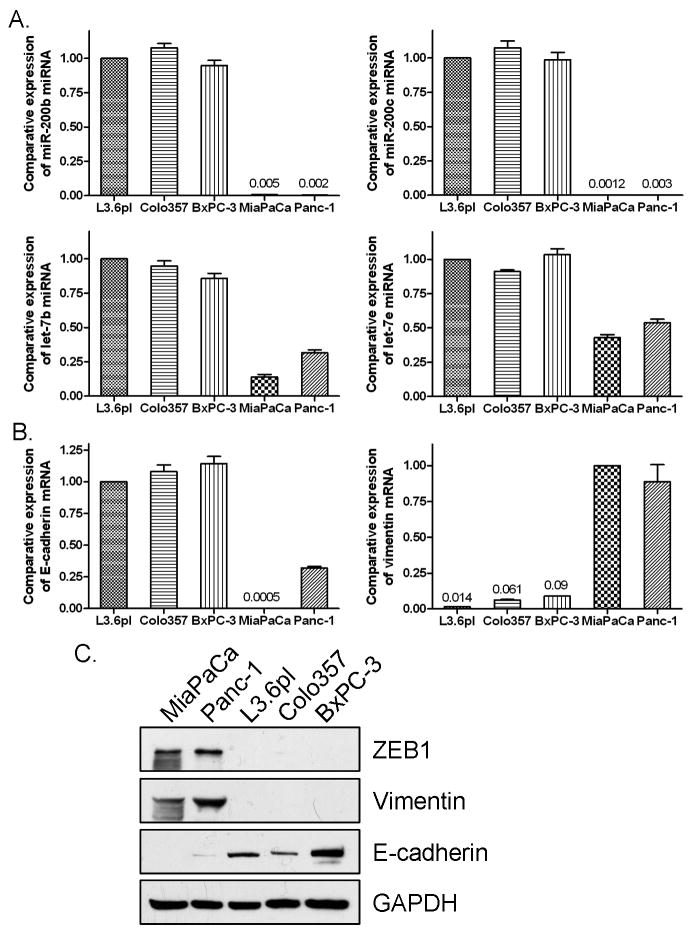
To investigate the difference in miRNA expression between gemcitabine sensitive and resistant PC cells, we conducted miRNA microarray. The results from miRNA microarray were validated by miRNA RT-PCR analysis for representative miRNA with varying expression profiles. The similar alternations of miRNA expression were observed by real-time RT-PCR analysis, although the fold change in the expression level was not exactly the same between these two different analytical methods. By miRNA microarray and RT-PCR, we found differential expression pattern of miRNA between gemcitabine-sensitive and -resistant PC cells. The known EMT regulators such as miR-200b, miR-200c, and miR-200a were down-regulated in gemcitabine-resistant cells (Table 1, Fig. 1A, Supplementary Fig. 1A), which is consistent with the EMT phenotype of gemcitabine-resistant cells. We also found that many members of the tumor suppressor let-7 family were down-regulated in gemcitabine-resistant cells (Table 1, Fig. 1A), and these findings are consistent with the aggressiveness of gemcitabine-resistant PC cells. More importantly, we found that gemcitabine-sensitive PC cells including L3.6pl, Colo357, BxPC-3, and HPAC cells showed strong expression of epithelial marker E-cadherin while gemcitabine-resistant PC cells, MiaPaCa-2, Panc-1, and Aspc-1 exhibited strong expression of mesenchymal makers including vimentin and ZEB-1 at mRNA and protein levels (Fig. 1B and 1C, Supplementary Fig. 1B and 1C). These results suggest that the gemcitabine-resistant cells showing EMT characteristics could be defined by the expression of specific miRNA and its target genes and proteins. Based on these results on the differential expression of miRNA, we hypothesized that “natural agents” that are known to cause cell growth inhibition and induction of apoptosis could regulate the EMT status by the alterations in the miRNA expression of gemcitabine-resistant cells. Therefore, we investigated whether B-DIM or G2535 (isoflavone) treatment could alter miRNA expression of gemcitabine-resistant cells.
Table 1.
Differential expression of let-7 and miR-200 in gemcitabine-sensitive and resistant pancreatic cancer cells treated with B-DIM or G2535 tested by miRNA array
| miRNA | Gemcitabine-sensitive |
Gemcitabine-resistant |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L3.6pl |
Colo357 |
MiaPaCa2 |
Panc-1 |
|||||||||
| Control | B-DIM | G2535 | Control | B-DIM | G2535 | Control | B-DIM | G2535 | Control | B-DIM | G2535 | |
| hsa-let-7a | 15048.0 | 16837.0 | 22667.8 | 14213.1 | 15750.5 | 21823.0 | 7535.6 | 6700.9 | 8243.3 | 11009.4 | 11333.5 | 14812.4 |
| hsa-let-7b | 8118.1 | 9753.4 | 12240.1 | 7554.4 | 9243.4 | 12668.5 | 1490.0 | 1316.9 | 2635.5 | 3582.8 | 3862.0 | 3895.1 |
| hsa-let-7c | 11295.6 | 13596.5 | 17622.9 | 9387.2 | 11167.2 | 14446.9 | 3811.3 | 3265.6 | 4998.7 | 4242.2 | 5799.9 | 8028.4 |
| hsa-let-7d | 10132.5 | 12494.8 | 15597.5 | 9237.1 | 10822.3 | 15712.6 | 4251.8 | 3806.0 | 5806.1 | 6190.5 | 6171.1 | 10086.7 |
| hsa-let-7e | 3973.3 | 7079.5 | 8637.5 | 2718.0 | 3788.0 | 3505.7 | 1728.6 | 2082.5 | 3963.9 | 2442.7 | 2277.6 | 4016.5 |
| hsa-let-7f | 11602.4 | 13645.9 | 17989.5 | 10789.1 | 12277.5 | 17104.3 | 5649.4 | 5375.5 | 7496.4 | 7771.1 | 7679.7 | 12454.1 |
| hsa-miR-200b | 4510.1 | 4520.4 | 5425.1 | 5202.5 | 6822.2 | 4132.7 | 5.93 | 7.04 | 11.60 | 3.96 | 4.87 | 4.79 |
| hsa-miR-200c | 11078.7 | 9360.9 | 14072.4 | 11949.8 | 13942.6 | 18571.2 | 7.57 | 8.02 | 6.61 | 8.51 | 11.10 | 15.54 |
Figure 1.
Real-time RT-PCR showed that miR-200 family (A), let-7 family (A), and E-cadherin (B) were significantly down-regulated and vimentin (B) was up-regulated in gemcitabine-resistant PC cells. Western Blot analysis (C) showed that ZEB1 and vimentin protein expression was up-regulated and E-cadherin was down-regulated in gemcitabine-resistant PC cells.
Regulation of miRNA expression by B-DIM and G2535 treatment in PC cells
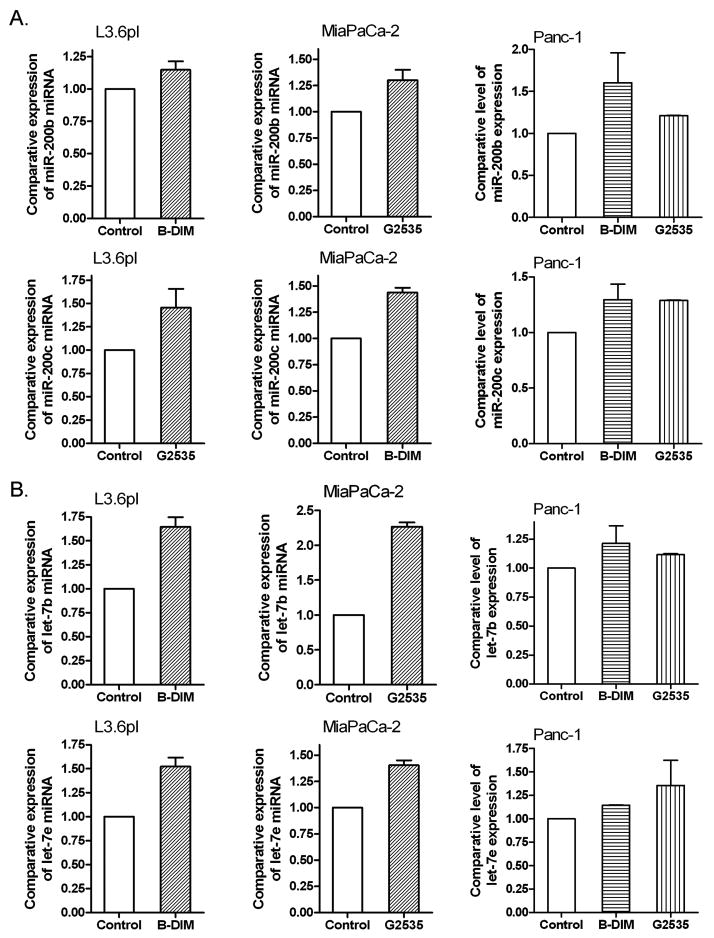
The miRNA microarrays were also used to determine whether treatment of cells with B-DIM or G2535 could alter the expression of miRNA compared to untreated cells. The miRNA RT-PCR was also conducted to confirm the data from miRNA array. By miRNA microarray and RT-PCR, we found that B-DIM treatments caused alterations in the expression of 28 miRNAs while G2535 treatment altered the expression of 67 miRNAs out of a total of 711 miRNA in the chip. Importantly, we found that the members of miR-200 and let-7 families were up-regulated upon G2535 or B-DIM treatment (Table 1, Fig. 2, Supplementary Fig. 1A), suggesting that these agents could cause reversal of EMT characteristics and, in turn, could inhibit tumor aggressiveness. G2535 showed stronger effects on miR-200 and let-7 expression than that of B-DIM as documented by miRNA microarray analysis (Table 1). Based on these novel findings, especially the loss of expression of miR-200 family in gemcitabine-resistant cells and most importantly the up-regulation in the expression of miR-200 family, we assessed the mechanistic role of miR-200 family in our experimental system.
Figure 2.
Real-time miRNA RT-PCR showed that B-DIM and isoflavone (G2535) increased the expression of miR-200 family (A) and let-7 family (B) in gemcitabine-resistant and sensitive PC cells.
Re-expression of miR-200 family leads to the alteration of EMT markers
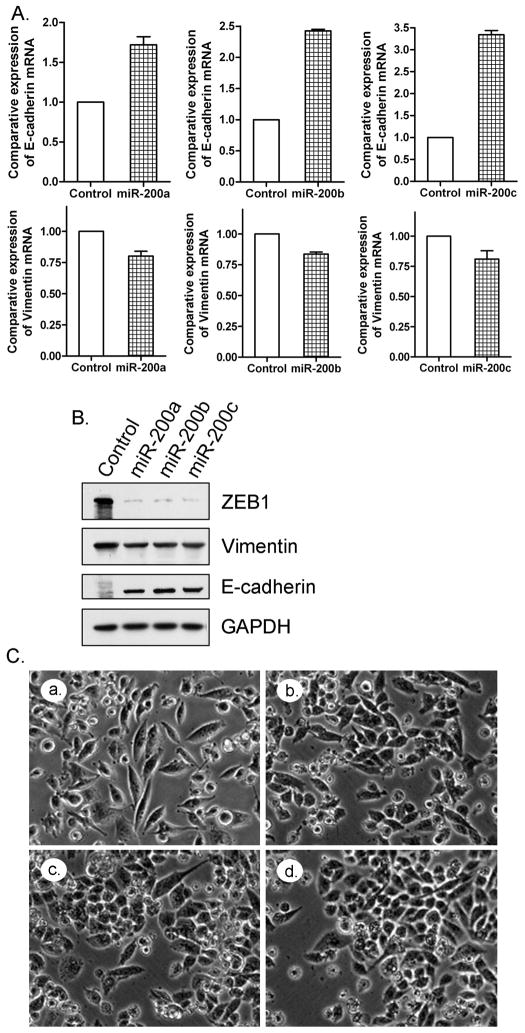
In order to investigate the role of miR-200 family in the reversal of EMT phenotype of gemcitabine-resistant cells, we transfected miR-200a, miR-200b, and miR-200c pre-microRNAs into MiaPaCa-2 cells (gemcitabine resistant) for up to 14 days. We found that the re-expression of miR-200 family in the MiaPaCa-2 cells resulted in the up-regulation of epithelial marker E-cadherin and down-regulation of mesenchymal markers including ZEB1 and vimentin both at the mRNA and protein levels (Fig. 3A and 3B). After 14 days of transfection, the morphology of miR-200 transfected MiaPaCa-2 cells was partially changed from elongated fibroblastoid to epithelial cobblestone-like appearance, and the cells appeared to grow in close contact with each other (Fig. 3C). These results suggest that the loss of miR-200 family is critical for the acquisition of EMT characteristics and that the re-expression of miR-200 could reverse the EMT phenotype of gemcitabine-resistant cells. Since re-expression of miRNA by transfection is not a practically useful strategy at the present time in clinical setting, we investigated whether “natural agents” could be useful for such a purpose and, if so, then the natural agents could be easily tested for their effects in the clinical setting.
Figure 3.
Re-expression of miR-200 family in MiaPaCa-2 resulted in the up-regulation in the expression of E-cadherin and the down-regulated expression of ZEB1 and vimentin as assessed by real-time RT-PCR (A) and Western Blot analysis (B). The morphology of MiaPaCa-2 cells changed from fibroblastoid to epithelial-like appearance (C) after miR-200 re-expression by transfection (a: control; b: miR-200a transfection; c: miR-200b transfection; d: miR-200c transfection).
B-DIM and G2535 treatment leads to the regulation of EMT marker expression and the reversal of EMT
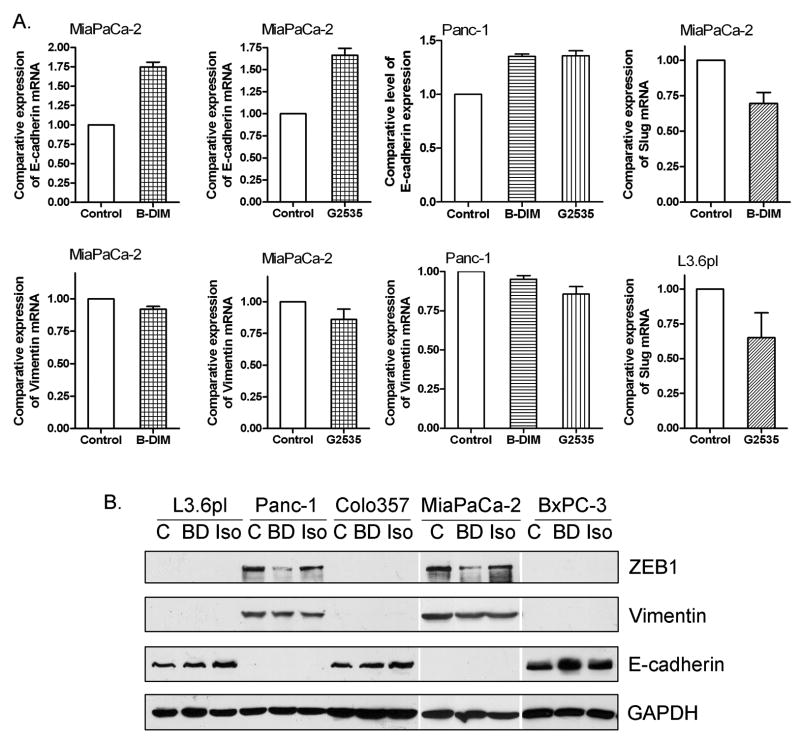
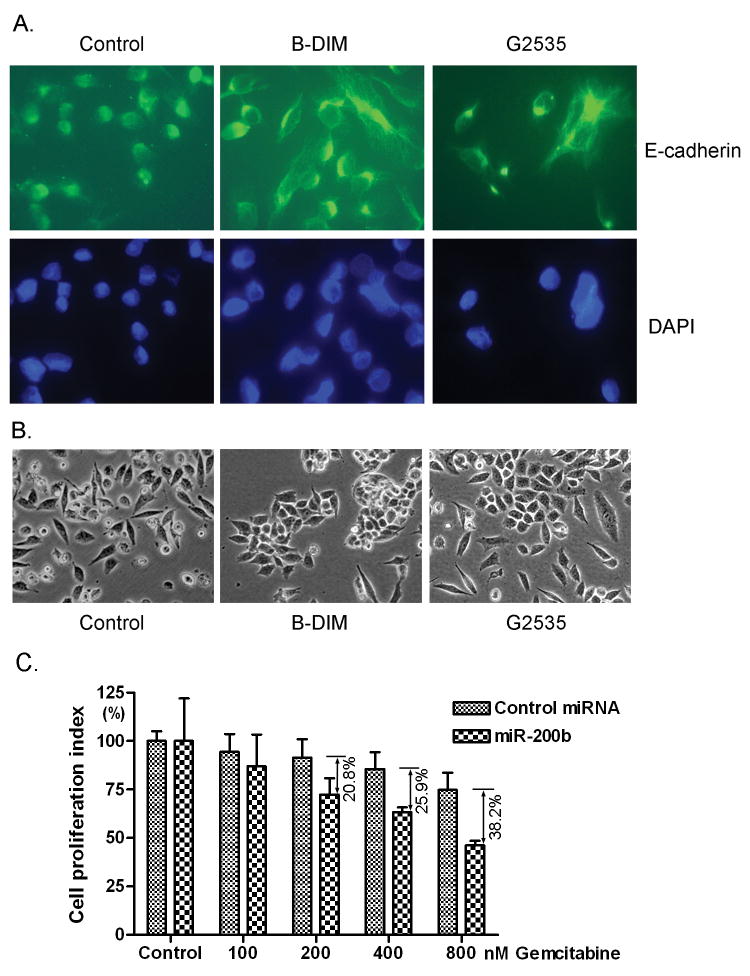
Since we observed that B-DIM and G2535 treatments increased the level of miR-200 family in MiaPaCa-2 cells, we further investigated whether B-DIM and G2535 treatment could show similar effects that were observed by re-expression of miR-200 family using transfection approach. After the treatment of PC cells with low concentration of B-DIM (5 μM) and G2535 (10 μM) for 21 days, we found up-regulation of epithelial marker E-cadherin and down-regulation of mesenchymal markers, ZEB1, vimentin and slug, at different levels tested by real-time RT-PCR (Fig. 4A, Supplementary Fig. 1B) and Western blot analysis (Fig. 4B, Supplementary Fig. 1C). Immunofluorescence staining showed that E-cadherin was redistributed in the cytoplasm closer to cell membrane after B-DIM and G2535 treatments (Fig. 5A). Importantly, we also observed that the morphology of MiaPaCa-2 cells changed from elongated fibroblastoid to epithelial cobblestone-like appearance, and it seems that the cell-cell contact was increased after the treatments with B-DIM and G2535 (Fig. 5B). These results are consistent with the changes in the expression of miRNAs and EMT markers. We also noted that some PC cells treated with G2535 showed more elongated shape which was commonly seen in genistein treated cells; however, G2535 has stronger effect on E-cadherin expression while B-DIM exerted stronger effect on ZEB1 expression as shown by Western Blot analysis (Fig. 4B). Nevertheless, these results suggest that both B-DIM and isoflavone could regulate the expression of EMT markers and indeed could be useful for the reversal of EMT phenotype, which could lead to the sensitization of gemcitabine-resistant cells to gemcitabine. Therefore, we assessed the sensitivity of gemcitabine-resistant cells to gemcitabine after being transfected with miR-200b or treated with B-DIM or G2535.
Figure 4.
B-DIM and isoflavone (G2535) increased the expression of E-cadherin and decreased the expression of ZEB1, slug, and vimentin in PC cells as assessed by real-time RT-PCR (A) and Western Blot analysis (B).
Figure 5.
E-cadherin appears to distribute in the cytoplasm closer to the cell membrane after B-DIM or isoflavone treatment (A). The morphology of MiaPaCa-2 cells changed from fibroblastoid to epithelial-like appearance after B-DIM or isoflavone treatment (B). Cell proliferation assay showed that re-expression of miR-200b increased the sensitivity of MiaPaCa-2 cells to gemcitabine (C).
miR-200 re-expression or B-DIM and G2535 treatment increased sensitivity of gemcitabine-resistant PC cells to gemcitabine
We transfected MiaPaCa-2 cells with miR-200b or control miRNA for a period of 7 days. We also treated MiaPaCa-2 cells with or without low concentration of B-DIM or G2535 for 21 days. Then, these cells were exposed to different concentrations of gemcitabine for 48 hours. Cell proliferation index was accessed by WST-1 assay. We found that the sensitivity to gemcitabine was significantly increased after miR-200 re-expression compared to control cells (Fig. 5C). The cells transfected with miR-200b showed 20.8% to 38.2% more inhibition compared to control. The pre-treatment of MiaPaCa-2 cells with B-DIM or G2535 also increased the sensitivity of MiaPaCa-2 cells to gemcitabine. The cell proliferation rate of MiaPaCa-2 cells treated with 500 nM gemcitabine was 79.05% compared to untreated control, whereas the rate was 67.39% or 65.56% upon B-DIM or G2535 pre-treatment, respectively (Supplementary Table 1). The MiaPaCa-2 cells pre-treated with B-DIM showed 14.8% to 17.4% more inhibition compared to control while the MiaPaCa-2 cells pre-treated with G2535 showed 15.4% to 17.1% more inhibition (Supplementary Table 2). These results suggest that miR-200 re-expression, and B-DIM or isoflavone treatment could partially increase the sensitivity of gemcitabine-resistant cells to gemcitabine possibly through miR-200 mediated reversal of EMT status.
Discussion
In recent years, it has become increasingly clear that EMT plays important roles in the progression of cancer and also responsible for resistant phenotype of cancer cells to conventional chemotherapeutics. It is believed that the sensitivity of therapeutic agents correlates with cell phenotype. The mesenchymal-type cancer cells showed increased expression of genes that are involved in the processes of invasion and metastasis, which are resistant to conventional therapeutics whereas the epithelial-type cancer cells are less invasive and metastatic and found to be significantly more susceptible to chemotherapeutics, suggesting that EMT decreases the sensitivity of cancer cells to therapeutics and promotes cancer cell invasion and metastasis (2). Further studies have shown that EMT marker proteins including E-cadherin and vimentin could serve as a determinant of EGFR activity in non-small cell lung cancer (NSCLC) cells and that E-cadherin expression was a novel biomarker predicting clinical positive activity of the EGFR inhibitor erlotinib in NSCLC patients (27). In our study, we found that gemcitabine-resistant cells showed EMT characteristics including elongated fibroblastoid shape, high expression of vimentin, and low expression of E-cadherin. Taken together, these results suggest that EMT plays critical roles in drug resistance to most conventional therapeutics. It has been reported that EMT is associated with poor survival in surgically resected pancreatic adenocarcinoma (28). Therefore, the presence of EMT phenotypic cells in PC could be one of the reasons that patients with PC are typically drug resistance, which contributes to high mortality.
It has been shown that down-regulation or the loss in the expression of miR-200 family is associated with EMT. Gregory et al have reported that all five members of the miR-200 family (miR-200a, miR-200b, miR-200c, miR-141 and miR-429) and miR-205 were significantly down-regulated in cells that had undergone EMT process in response to TGF-β(29). They also found that re-expression of the miR-200 family alone was sufficient to prevent TGF-β-induced EMT. In our study, we found similar phenomenon in gemcitabine resistant pancreatic cancer cells. The gemcitabine-resistant cells having EMT characteristics showed low expression of miR-200 family. Interestingly, re-expression of miR-200 in gemcitabine-resistant cells showed partial reversal of EMT characteristics as documented by increased expression of E-cadherin and decreased expression of vimentin, ZEB1, and slug. These results suggest that miR-200 family regulates the expression of ZEB1, slug, E-cadherin, and vimentin and that the re-expression of miR-200 could be useful for the reversal of EMT phenotype to mesenchymal-epithelial transition (MET).
Both ZEB1 and slug are transcription factors which repress the expression of E-cadherin (30, 31). It has also been shown that ZEB1 is the target gene of miR-200 family (29, 31, 32). By binding to 3′UTR of ZEB1 mRNA, miR-200 inhibits the translation of ZEB1. Consistent with these findings, our results showed decreased levels of ZEB1 and increased levels of E-cadherin expression in miR-200 transfected gemcitabine-resistant cells, suggesting that novel approaches by which miR-200 family could be up-regulated would be an useful approach for the reversal of EMT and thus would be useful for the reversal of the drug resistance phenotype of PC cells. It has been found that the expression of miR-200 is lost in invasive breast cancer cell lines with mesenchymal phenotype and in regions of metaplastic breast cancer specimens lacking E-cadherin (29), suggesting that the loss of miR-200 expression could be a critical step in cancer progression. Further studies have shown that the ectopic expression of miR-200 increased E-cadherin expression, altered cell morphology to an epithelial phenotype, and reduced the in vitro motility of 4TO7 cells in migration assays (31), suggesting that the increase in miR-200 could reverse EMT and inhibit cancer cell invasion and metastasis. In our present study, we found that B-DIM and isoflavone could induce miR-200 expression, resulting in altered cellular morphology from mesenchymal to epithelial appearance, and induced E-cadherin distribution that is typically found in epithelial-like cells. These alterations by B-DIM and isoflavone could be responsible for the inhibition of cancer progression, invasion, and metastasis that was previously observed by our laboratory (33, 34).
The let-7 miRNA has been found to regulate cell proliferation and differentiation. Importantly, recent studies have shown that decreased let-7 expression is linked to increased tumorigenesis and poor patient prognosis (18, 35). Since let-7 inhibits the expression of multiple oncogenes including ras, myc, HMGA2, etc, let-7 has been recognized as a tumor suppressor (20, 35). Moreover, it has been found that let-7 could regulate “stemness” by repressing self-renewal and promoting differentiation in both normal development and cancer and that let-7 miRNAs are markedly reduced in breast cancer initiating cells and increased with differentiation (35, 36), suggesting that let-7 could regulate cancer stem cell and EMT. In our present study, we found that B-DIM and isoflavone could significantly up-regulate the expression of let-7 family, suggesting that B-DIM and isoflavone could inhibit pancreatic tumor progression by reversing EMT characteristics in part due to the up-regulation of let-7; however, further mechanistic studies are warranted for assessing the role of let-7 in the processes of EMT in PC.
The successful treatment of pancreatic cancer is dependent on the sensitivity of cancer cells to conventional therapeutic agents. Since mesenchymal-type cancer cells are more resistant to chemotherapeutic agents than epithelial-type cancer cells (2, 37), the status of EMT characteristics must be reversed to overcome drug resistance, which could lead to the sensitization of drug-resistant cancer cells to conventional chemotherapeutic agents. The results presented in this manuscript showed, for the first time, that the non-toxic “natural agents” such as B-DIM and isoflavone could up-regulate the expression of miR-200 and let-7, which in turn down-regulate the expression of ZEB1 resulting in the reversal of EMT characteristics of gemcitabine-resistant cells. We also found that the sensitivity of PC cells to gemcitabine could be partly increased after re-expression of miR-200 or prolong exposure of gemcitabine-resistant cells to lower concentrations of either B-DIM or isoflavone. It is important to note that B-DIM and isoflavone showed differential effects on miRNA and EMT marker expression; therefore, further studies could be done by using the combination of B-DIM and isoflavone to reverse EMT and increase the sensitivity of PC cells to gemcitabine. In conclusion, these exciting results should provide incentives for further investigation and optimization in establishing the mechanistic role of B-DIM and isoflavone in the reversal of EMT characteristics and drug resistance, and their utility in the clinical setting for the treatment of PC for which there is no effective and curative therapy.
Supplementary Material
Acknowledgments
Grant support: National Cancer Institute, NIH (5R01CA083695, 5R01CA108535, and 5R01CA101870 to FHS). We also thank Puschelberg Foundation for their generous contribution for the completion of this study.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Fuchs BC, Fujii T, Dorfman JD, et al. Epithelial-to-mesenchymal transition and integrin-linked kinase mediate sensitivity to epidermal growth factor receptor inhibition in human hepatoma cells. Cancer Res. 2008;68:2391–9. doi: 10.1158/0008-5472.CAN-07-2460. [DOI] [PubMed] [Google Scholar]
- 3.Cheng GZ, Chan J, Wang Q, Zhang W, Sun CD, Wang LH. Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to paclitaxel. Cancer Res. 2007;67:1979–87. doi: 10.1158/0008-5472.CAN-06-1479. [DOI] [PubMed] [Google Scholar]
- 4.Sabbah M, Emami S, Redeuilh G, et al. Molecular signature and therapeutic perspective of the epithelial-to-mesenchymal transitions in epithelial cancers. Drug Resist Updat. 2008 doi: 10.1016/j.drup.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–54. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 6.Brabletz T, Jung A, Reu S, et al. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci U S A. 2001;98:10356–61. doi: 10.1073/pnas.171610498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stemmer V, de CB, Berx G, Behrens J. Snail promotes Wnt target gene expression and interacts with beta-catenin. Oncogene. 2008;27:5075–80. doi: 10.1038/onc.2008.140. [DOI] [PubMed] [Google Scholar]
- 8.Graham TR, Zhau HE, Odero-Marah VA, et al. Insulin-like growth factor-I-dependent up-regulation of ZEB1 drives epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2008;68:2479–88. doi: 10.1158/0008-5472.CAN-07-2559. [DOI] [PubMed] [Google Scholar]
- 9.Yang MH, Wu MZ, Chiou SH, et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 10.Spaderna S, Schmalhofer O, Wahlbuhl M, et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 2008;68:537–44. doi: 10.1158/0008-5472.CAN-07-5682. [DOI] [PubMed] [Google Scholar]
- 11.Yang J, Mani SA, Donaher JL, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Muller T, Bain G, Wang X, Papkoff J. Regulation of epithelial cell migration and tumor formation by beta-catenin signaling. Exp Cell Res. 2002;280:119–33. doi: 10.1006/excr.2002.5630. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–23. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saxena S, Jonsson ZO, Dutta A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J Biol Chem. 2003;278:44312–9. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- 15.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–6. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 16.Gironella M, Seux M, Xie MJ, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci U S A. 2007;104:16170–5. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 18.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 19.Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7:3112–8. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 20.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–30. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Surh YJ. Cancer chemoprevention with dietary phytochemicals. Nat Rev Cancer. 2003;3:768–80. doi: 10.1038/nrc1189. [DOI] [PubMed] [Google Scholar]
- 22.Aggarwal BB, Shishodia S. Molecular targets of dietary agents for prevention and therapy of cancer. Biochem Pharmacol. 2006;71:1397–421. doi: 10.1016/j.bcp.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 23.Ali S, Banerjee S, Ahmad A, El-Rayes BF, Philip PA, Sarkar FH. Apoptosis-inducing effect of erlotinib is potentiated by 3,3′-diindolylmethane in vitro and in vivo using an orthotopic model of pancreatic cancer. Mol Cancer Ther. 2008;7:1708–19. doi: 10.1158/1535-7163.MCT-08-0354. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Li Y, Ahmed F, Ali S, Philip PA, Kucuk O, Sarkar FH. Inactivation of nuclear factor kappaB by soy isoflavone genistein contributes to increased apoptosis induced by chemotherapeutic agents in human cancer cells. Cancer Res. 2005;65:6934–42. doi: 10.1158/0008-5472.CAN-04-4604. [DOI] [PubMed] [Google Scholar]
- 25.Pan X, Arumugam T, Yamamoto T, et al. Nuclear factor-kappaB p65/relA silencing induces apoptosis and increases gemcitabine effectiveness in a subset of pancreatic cancer cells. Clin Cancer Res. 2008;14:8143–51. doi: 10.1158/1078-0432.CCR-08-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderton MJ, Manson MM, Verschoyle R, et al. Physiological modeling of formulated and crystalline 3,3′-diindolylmethane pharmacokinetics following oral administration in mice. Drug Metab Dispos. 2004;32:632–8. doi: 10.1124/dmd.32.6.632. [DOI] [PubMed] [Google Scholar]
- 27.Thomson S, Buck E, Petti F, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005;65:9455–62. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- 28.Javle MM, Gibbs JF, Iwata KK, et al. Epithelial-mesenchymal transition (EMT) and activated extracellular signal-regulated kinase (p-Erk) in surgically resected pancreatic cancer. Ann Surg Oncol. 2007;14:3527–33. doi: 10.1245/s10434-007-9540-3. [DOI] [PubMed] [Google Scholar]
- 29.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 30.Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 31.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–4. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong D, Li Y, Wang Z, Banerjee S, Sarkar FH. Inhibition of angiogenesis and invasion by 3,3′-diindolylmethane is mediated by the nuclear factor-kappaB downstream target genes MMP-9 and uPA that regulated bioavailability of vascular endothelial growth factor in prostate cancer. Cancer Res. 2007;67:3310–9. doi: 10.1158/0008-5472.CAN-06-4277. [DOI] [PubMed] [Google Scholar]
- 34.Li Y, Kucuk O, Hussain M, Abrams J, Cher ML, Sarkar FH. Antitumor and antimetastatic activities of docetaxel are enhanced by genistein through regulation of osteoprotegerin/receptor activator of nuclear factor-kappaB (RANK)/RANK ligand/MMP-9 signaling in prostate cancer. Cancer Res. 2006;66:4816–25. doi: 10.1158/0008-5472.CAN-05-3752. [DOI] [PubMed] [Google Scholar]
- 35.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–9. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Yu F, Yao H, Zhu P, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 37.Black PC, Brown GA, Inamoto T, et al. Sensitivity to epidermal growth factor receptor inhibitor requires E-cadherin expression in urothelial carcinoma cells. Clin Cancer Res. 2008;14:1478–86. doi: 10.1158/1078-0432.CCR-07-1593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.