Abstract
Cellular survival from radiation induced DNA damage requires access to sites of damage for the assembly of repair complexes and the subsequent repair, particularly the repair of DNA double strand breaks (DSBs). Hyperthermia causes changes in protein-protein/DNA interactions in the nucleus that block access to sites of DNA damage. Studies presented here indicate that the nucleolar protein, nucleophosmin (NPM), redistributes from the nucleolus following hyperthermia, increases its association with DNA, and blocks access to DNA DSBs. Reduction of NPM significantly reduces heat-induced radiosensitization, but reduced NPM level does not alter radiation sensitivity per se. NPM knock down reduces heat-induced inhibition of DNA DSB repair. Also, these results suggest that NPM associates with nuclear matrix attachment region (MAR) DNA in heat shocked cells.
Keywords: Nucleophosmin, Hyperthermia, Radiosensitization
INTRODUCTION
The potential for using hyperthermia as a radiosensitizing adjuvant for radiation therapy has long been recognized. Recent meta-analysis shows that hyperthermia significantly improves radiation therapy (1). Further, hyperthermia is a potent radiosensitizer, producing thermal enhancement ratios (TERs) from 3 to 5 (1, 2). Nevertheless its clinical use remains limited, in part, because the mechanism of hyperthermia induced radiosensitization remains poorly understood. Reasonably, inhibition of DNA double strand break (DSB) repair should be involved in heat-induced radiosensitization. However, the literature indicates a more complex situation. Cells defective in the non-homologous end joining pathway or in homologous recombination show similar TERs but are more radiosensitive than isogenic wild type cells (2). One interpretation of these results is that neither of these pathways is a critical target for heat-induced radiosensitization. However, since the mutants studied are defective in the effector mechanisms of these pathways, an alternative interpretation is that hyperthermia affects an upstream step such as the sensing of DNA damage which would impact both DNA DSB repair pathways. One explanation (3-6) suggests that masking of DNA strand breaks by heat-induced protein-DNA complexes would sterically inhibit access of the sensing proteins to the damage site. However, little work has been done toward determining what protein(s) involved in DNA damage masking and the DNA regions masked in heat shocked cells. Here we present evidence that heat-induced masking of DNA damage is due (in part) to the heat-induced association of the nucleolar protein, nucleophosmin (NPM) with MAR DNA.
NPM is an abundant phosphoprotein, localized to the granular regions of the nucleolus (7). NPM trafficking contributes to its essential functions (8). Thus, changing the ability of NPM to mobilize to distinct cellular locales should alter NPM function (8). Additionally, loss of NPM function results in attenuation of cellular proliferation and increased apoptosis (9-16). The nucleolus acts as repository of stress responsive proteins (17-20). Specifically, the nucleolar protein, nucleolin (NCL or C23), redistributes from the nucleolus and interacts with replication protein A resulting in inhibition of DNA synthesis (20-22). The nucleolus is a heat sensitive organelle, undergoing detectable changes after 1 hr at 41°C and general changes after 15 min at 42°C, which leave cytoplasmic organelles largely unaffected (23-25). Further, HSP70 and HSP110, which modulate heat-induced radiosensitization localize in the nucleolus in cells during and immediately after heat shock (26-27). Therefore, we determined if NPM could play a key role in heat radiosensitization.
MATERIALS AND METHODS
Cell Lines and Cell Culture
HeLaS3 cells (ATCC) were maintained in alpha-MEM media with 10% calf serum, 100 U/ml penicillin and 100 mg/ml streptomycin. Human breast cancer cells, HCC1806, were maintained in RPMI 1640 media with 10% Bovine Growth Serum and antibiotics. For survival experiments, control and treated cells were plated in replicate T-25 flasks. After 10 days (HeLaS3) or 19 days (HCC1806), flasks were stained with Crystal Violet and viable colonies (≥ 50 cells) counted.
Hyperthermia and Irradiation
For acute hyperthermia, flasks were immersed in a temperature controlled water bath. Irradiation was performed using a PANTAK pmc1000 X-ray machine with a 0.1 Cu+ 2.5 AL filter at a dose rate of 88.7 cGy/min.
Transfection with siRNA
HeLaS3 cell transfections were accomplished using a Nucleofector II electroporation apparatus (Amaxa) following the manufacturer’s instructions. NPM specific siRNA sequences (28) and siCONTROL Non-Targeting siRNA #1 were obtained from Dharmacon. GFP-NPM (29) images were captured with a Spot Pursuit digital camera (Diagnostic Instruments) on an Olympus fluorescent microscope.
Assay for DNA Double Strand Breaks
The Neutral Comet Assay was performed using the Comet Assay Kit from Trevigen following manufacturer’s instructions. Proteinase K (Sigma), when used, was added to the lysis solution (0.27 U/ml). Lysis occurred at 4°C for 30 min. Comets were analyzed by CometScore software (TriTek).
Assay for DNA Supercoiling Ability
The halo assay was done as described in (30).
Chromatin Immunoprecipitation (CHiP) Assay
The enzymatic shearing Chip-IT kit (Active Motif) was used following the manufacturer’s instructions. NPM was immunoprecipitated with mouse anti-NPM antibodies (Zymed Invitrogen). Co-immunoprecipitated DNA fragments were PCR amplified using primers to Topoisomerase I; MAR II, Exon 13, Exon 20; c-jun and the GAPDH promoters (31).
Immunoblots, Antibodies and Other Reagents
SDS-PAGE and immunoblotting were done following standard procedures using a BioRad Mini-Protean II gel and blotting system. Immunoblots were probed with mouse anti-NPM or HSP70/HSC70 antibodies and alkaline phosphatase conjugated Affinipure Goat anti-Mouse IgG+IgM secondary antibodies. The targeted bands were visualized colorimetrically by BCIP/NBT.
RESULTS
To confirm that heat increased NPM-DNA binding we crosslinked DNA-associated proteins to DNA with cisplatin, then isolated the DNA with hydroxyapatite, and resolved the DNA associated proteins by 2D gel electrophoresis. Hyperthermia increased the DNA association of NPM isoform, B23.1, more than 6-fold. Several NPM isoforms became associated with DNA in heated cells (Fig. 1A). These results are consistent with previous reports showing that hyperthermia increases NPM association with DNA in vivo (5) binds DNA in vitro (32).
Figure 1. Nucleophosmin redistribution following heat shock.
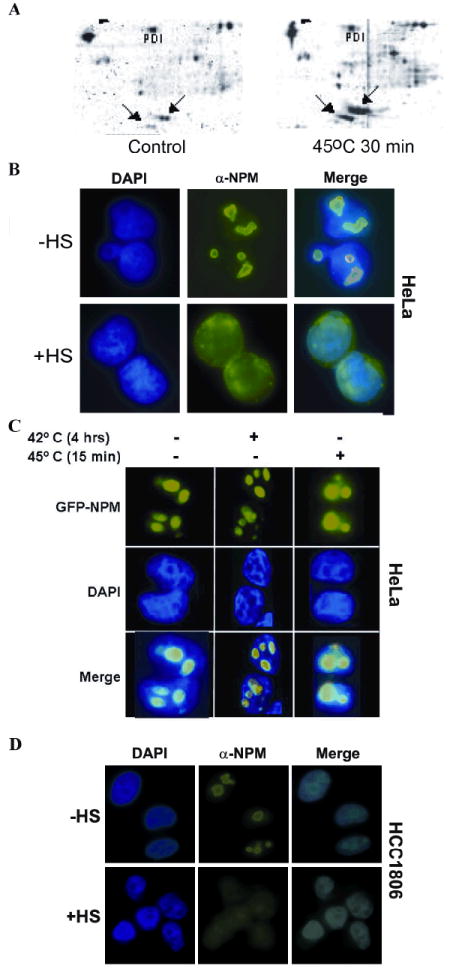
(A) Proteins were crosslinked to DNA by cisplatin from control and heat shocked HeLaS3 cells. Two forms of NPM, B23.1 and B23.2, are denoted by arrows. (B) HeLaS3 cells were incubated at 45°C for 45’, fixed and stained with antibodies recognizing NPM. DNA was visualized with Hoechst dye and NPM with FITC filters. (C) Cells expressing a GFP-NPM fusion protein and counter stained with DAPI. (D) Redistribution of NPM following heat shock in HCC1806 cells.
To determine the effects of hyperthermia on the sub-nuclear localization of NPM we exposed HeLaS3 cells and HCC1806 cells to increasing heat shocks and visualized the localization of NPM using fluorescently labeled antibodies or transiently expressed GFP-tagged NPM fusion protein. NPM readily re-localizes from the nucleolus to the nucleoplasm after heat shocks of 15-45 min at 45°C in HeLaS3 and HCC1806 cells (Fig. 1). Thus, NPM redistributes into the nucleoplasm after hyperthermia and becomes associated with DNA.
Since NPM becomes associated with DNA, we determined if NPM could be involved in radiation sensitivity. We measured the TER after a 90-95% NPM knockdown, using siRNA. Cells were X-irradiated with or without prior hyperthermia (30 min at 45°C) and their colony forming ability measured (Fig. 2A). For HeLaS3 and HCC1806 cells, TER values were calculated at the control survival level after 2 and 4 Gy which were chosen because these doses are commonly used in clinical radiation therapy and averaged. Hyperthermia significantly increased the non-transfected cell’s sensitivity to X-rays, TER = 2.22 and 2.08 for HeLaS3 and HCC1806, respectively (Fig. 2D). Transfection with the nonspecific siRNA did not significantly change the TER (2.22 and 2.16). However, transfection with the siRNA specific for NPM significantly lowered the TER to 1.74 and 1.39 (p<0.05; Students t-test) in HeLaS3 and HCC1806 cells, respectively. Also, NPM reduction made a statistically significant increase in survival at each dose level by the Student’s t-test. Thus, heat induced radiosensitization is due, in part, to the presence of NPM. Without heat shock, NPM knockdown had no effect on survival after 2, 4 or 6 Gy (Fig. 2A).
Figure 2. Reducing NPM reduces the TER.
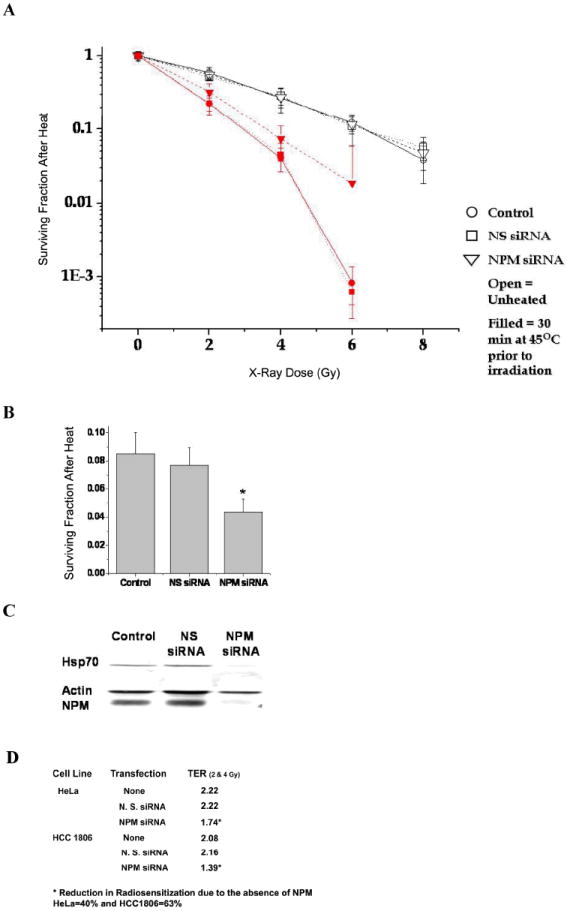
(A) HeLaS3 cells were transfected with a NPM specific siRNA or a nonspecific siRNA, then irradiated with or without prior heat shock (30 min @ 45°C). The symbols are the average of 3-6 experiments ± 1 SD (n ≤ 3). The curves are normalized for cell killing by heat shock alone. (B) Heat shocked HeLaS3 cells were plated for cell survival and cell extracts prepared for protein analysis. (C) HeLaS3 cell extracts were probed for HSP70 and NPM. The average cell survival, after heat shock, is significantly lower for cells with lower NPM levels (p < 0.05, t-test). (D) Effect of NPM knockdown on the TER in the HeLaS3 cells and HCC1806 cells. See text for TER calculation.
Transient NPM knockdown and transfection with a non-specific siRNA did not significantly alter the plating efficiency in HeLaS3 cells (80 ± 3%) or in HCC1806 cells (30 ± 3%). However, NPM knockdown sensitized HeLaS3 cells to heat shock alone. To understand this observation, we measured HSC70/HSP70 levels. NPM knockdown reduced the endogenous levels of HSC70/HSP70 (Fig. 2C), which confers resistance to heat-induced cell killing (33-34). This result also supports the hypothesis that NPM specifically contributes to heat induced radiosensitization; because its knockdown increased thermal sensitivity while reducing the TER without affecting radiation sensitivity per se. In contrast, nonspecific thermal sensitizers and protectors have been shown to alter both heat sensitivity and the TER in concert (35).
Since NPM knockdown reduced the TER from heat shock, we determined if heat induced a NPM dependent masking of DNA DSB and the inhibition of DNA DSB repair. Using the neutral comet assay with PK (0.27 U/ml) the level of DNA damage immediately after 8 Gy, was the same between cells heated at 45°C for 30 min and unheated. However, a significant inhibition of subsequent DNA DSB repair could be observed (Fig. 3A, left). In control cells 75% of the DNA DSBs were repaired after 90 min, but in heated cells only 20-25% was repaired. After 150 min, more than 90% of the DNA DSBs were repaired in unheated cells, while ~ 50% remained unrepaired in the heated cells. In the absence of PK there is a 60% reduction in the measured comet moment (Fig. 3A, right) for cells, heated at 45°C for 30 min, compared with unheated cells. Since the comet moment is the same in heated versus non-heated cells in the presence of PK, we conclude that 60% of the DNA DSBs, following 8 Gy, are masked (Fig. 3A, right), by a heat-induced, PK digestible component i.e. a protein or proteins. We define masking as the inability of an assay (or DNA repair pathway) to detect DNA damage in the presence of protein-DNA interactions even though the damage would be detectable in the absence of such protein-DNA interactions. Without PK, there appeared to be no repair of DNA DSB for 150 min (Fig 3A left), which may be due to a combination of unmasking the DSBs and their subsequent repair.
Figure 3. NPM contributes to DNA DSB repair inhibition and DSB masking.
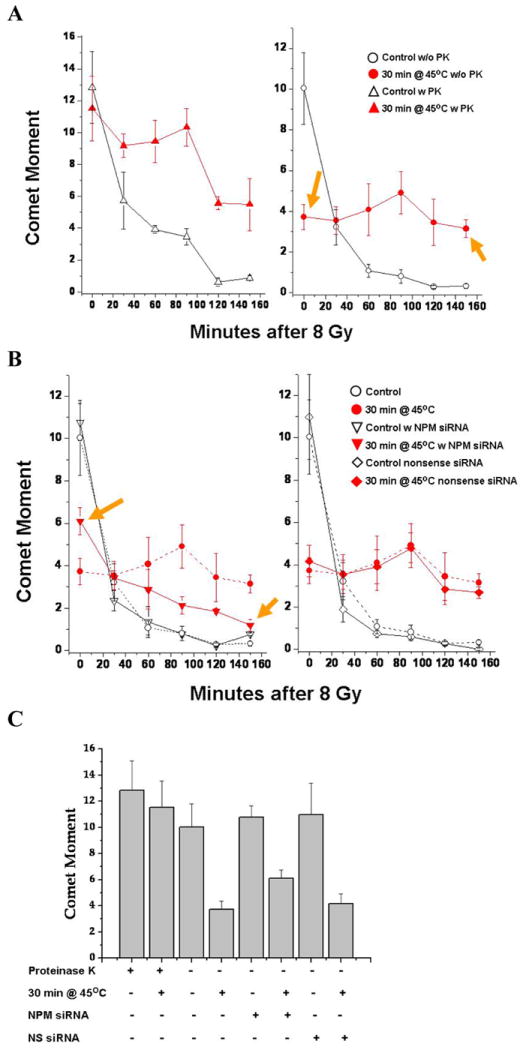
(A) DNA DSBs were detected by the neutral comet assay. Left Panel; comet moment, measured after 30 min incubation with 0.27 U/ml PK. The initial damage was the same but a repair inhibition was evident in the heated sample. Right Panel; without PK digestion. the apparent reduction in initial damage shows that DNA DSBs are masked by a PK digestible component in heated cells (left arrow). Heat induced inhibition of repair can be seen as the increase in residual damage at 150 min. of repair (right arrow). (B) Left Panel; NPM knockdown reduces DNA DSB masking (right arrow) and repair inhibition (left arrow). The residual unrepaired DSBs are reduced by 50%. NPM knockdown had no effect on DNA DSB repair in the absence of heat shock. The Right Panel; the lack of effects for cells transfected with a nonsense siRNA. (C) Masking of DNA DSB after 8 Gy is PK labile and partially reduced in cells with transient NPM knockdown as shown by reduction in comet moment. With versus without PK (p<0.05); NPM siRNA versus control or nonsense (NS) siRNA (p<0.05 experiments).
We determined the effects of NPM knockdown on the heat-induced masking of DNA damage and repair inhibition. The masking of DNA DSB is reduced by 40% in cells treated with the NPM siRNA (Fig. 3B right). To illustrate the effects of NPM knockdown on the masking of DNA damage, the comet moment values immediately after irradiation are plotted (Fig. 3C). Hyperthermia reduced the post 8 Gy comet moment ~3-fold. The comet moment was reduced ~2-fold after NPM knockdown (p < 0.05). Heated NPM knockdown cells, 90 min after 8 Gy, had 22% of the initial DNA DSB remaining for a 5-fold reduction in repair inhibition. Further the residual unrepaired DSB was reduced from 50% unrepaired to 12% unrepaired. Unrepaired DSBs are critical in hyperthermic radiosensitization (36-37). In contrast, in cells treated with a nonsense siRNA the heat-induced masking of DNA DSB and inhibition of their repair were identical to the untreated control (Fig. 3B right). Without heat shock, NPM knockdown had no effect on DNA DSB repair (Fig. 3B left) consistent with the observation that NPM knockdown had no effect on radiosensitivity in the absence of heat shock.
Previous studies of the masking effect using the halo assay, a DNA supercoiling assay that depends on DNA-nuclear matrix anchoring (38) found that it correlated with the TER in cell lines having various levels of resistance to heat-induced radiosensitization (39). Therefore, we determined the effects of NPM on DNA damage masking using this assay. In the halo assay DNA supercoil unwinding/rewinding is driven by increasing the concentration of the intercalating dye, propidium iodide. DNA strand breaks induced by ionizing radiation inhibit the ability of the loops to rewind supercoils in a dose dependent manner (40). The inability of DNA loops to be rewound is quantified as the difference between rewinding in the absence vs the presence of DNA damage (38, 40). This difference referred to as the excess halo diameter (EHD) is estimated for PI concentrations between 10 and 50 μg/ml (38). When cells are heated prior to irradiation, the EHD is reduced even though the amount of DNA damage per cell or per unit of DNA remains the same (4). Cells were untreated, irradiated with 10 Gy or heat shocked for 30 min at 45°C and then irradiated with 10 Gy prior to running the halo assay (Fig. 4A). Cells with normal NPM levels showed a typical heat-induced reduction in the inhibition of DNA supercoil rewinding following irradiation. In contrast, in cells with reduced NPM protein levels, hyperthermia had minimal effect on DNA supercoil rewinding after irradiation. Thus the masking of DNA damage, as detected by the halo assay, is reduced in cells with 80-90% lower NPM protein levels, suggesting that NPM plays a significant role in masking of damage when detected by the halo assay. The EHD (Fig. 4B, calculated as described above) for heated and irradiated cells with normal NPM levels was significantly reduced compared with radiation alone. In contrast, the heat-induced reduction of EHD, when NPM was knocked down, was ~ 80% less than that for normal NPM levels (Fig. 4B). Similar results were obtained in HCC1806 cells (Fig. 4C). In summary a significant fraction (80%) of the masking effect, detected by the halo assay depends on the presence of NPM.
Figure 4. Effect of NPM knockdown on DNA supercoil unwinding/rewinding after irradiation and/or heat shock.
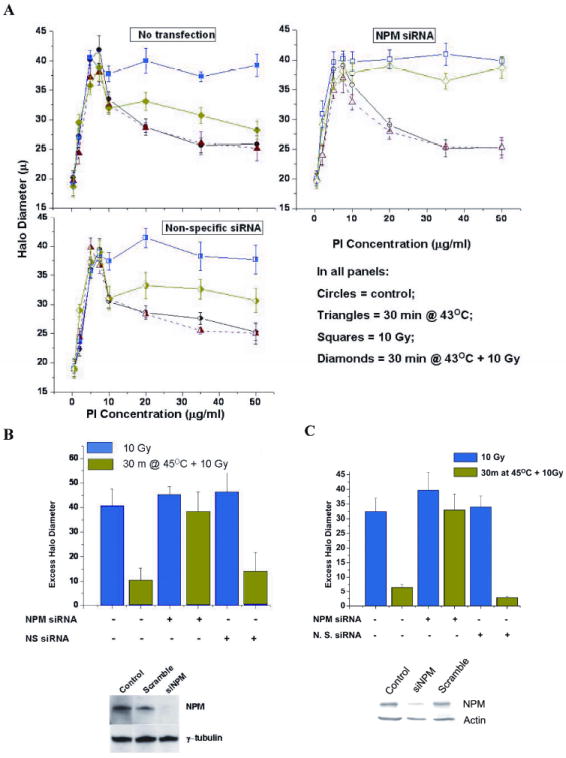
(A) The halo assay was used to determine the effects of NPM knockdown on the ability of nuclear DNA to undergo supercoiling changes after irradiation with or without prior heat shock. The points are a mean of 4-5 experiments. (B) Excess halo diameter (EHD) values were calculated from the rewinding portion of the 4-5 individual experiments and averaged (bars) ± the SD. The only the non-transfected and the non-specific transfected showed a significant EHD reduction (p < 0.05, t-test) after heat shock. Inset: typical NPM levels. (C) EHD values for irradiated and heat plus irradiated HCC1806 cells. The non-transfected and the non-specific transfected cells showed a significant EHD reduction after heat shock.
We used the ChIP assay to determine if NPM is associated with MAR, exon, or promoter DNA. NPM was immunoprecipitated with a NPM specific antibody from heated and control cells and the associated DNA analyzed by PCR using primers specific for MAR DNA, promoter DNA and exon DNA (31). Detection of specific DNA sequences (Fig. 5A) showed that NPM is associated with Topo I-MAR1, which increased more than 3-fold following hyperthermia (Fig. 5A & C). There appeared to be no association between NPM and the promoter DNA of c-jun or GAPDH, nor to Topo I exons 13 and 20 (Fig. 5C & D). Thus, only MAR DNA sequences were amplified by the PCR reactions on the DNA that co-precipitated with NPM, suggesting (albeit with limited sample size) that NPM binds MAR DNA, but not promoter or exon DNA. Further, the enhanced association between NPM and MAR DNA was reversed in a time dependent manner during post heat incubation (Fig. 5D), typical for post heat recovery from the masking effect (5). Since the ability to rewind DNA supercoils depends upon DNA-nuclear matrix anchoring, the association of NPM with MAR DNA is consistent with the observation that the masking effect detected by the halo assay depends on NPM.
Figure 5. PCR reaction products from DNA associated with NPM.
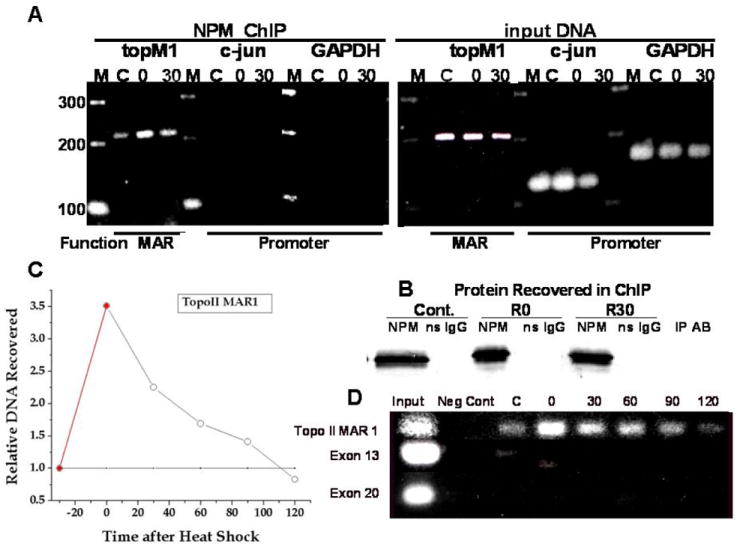
(A) PCR results for the input DNA and that for the NPM-associated DNA recovered in the ChIP assay are indicated. Amplification of promoter sequences was detected in the input DNA but not in the DNA sequences co-immunoprecipitated with NPM. Control is marked “C”’ heated at 45°C for 30 min is “O” and 30 min after heat is “30”. NPM recovery is shown in (B). Immunoprecipitates from unheated control cells are marked C. Immunoprecipitates from heat shocked (30 min at 45°C) cells are indicated by numbers, i.e. 0 = 0 min and 30 =30 min after heat shock. After cells were heat shocked (45°C for 30 min) the CHIP assay was performed at the indicated time (D). Exon DNA was also analyzed. The relative change in NPM-Topo I MAR1 binding (open circles) and NPM-Topo I exon DNA (dotted line) is quantified (C).
DISCUSSION
Here, we show that heat-induced NPM redistribution and DNA association contributes to the radiosensitization of cells, because multiple radiation sensitivity related endpoints (i.e., TER, inhibition of DSB repair and reduction of the EHD) are modulated in heated cells after NPM knockdown. The results also indicate that in heated cells NPM associates with MAR DNA, shown by modulation of the masking effect and the increased association of MAR DNA sequences with NPM. A model illustrating the effect of masking on DNA supercoil rewinding and the masking of DSBs is shown in Fig. 6. The hypothesized effects of DSB masking on downstream pathways and endpoints are illustrated in Panel C, assuming that if DNA damage is masked from a particular assay, then that damage will also be masked from (at least some of) the sensor steps in DNA repair. This assumption is supported by the observation that the duration of the initial DSB repair inhibition, 90 min, measured in the presence of PK corresponded to the duration of masking (reported previously, (6)) and the association of the TOPO I MAR with NPM.
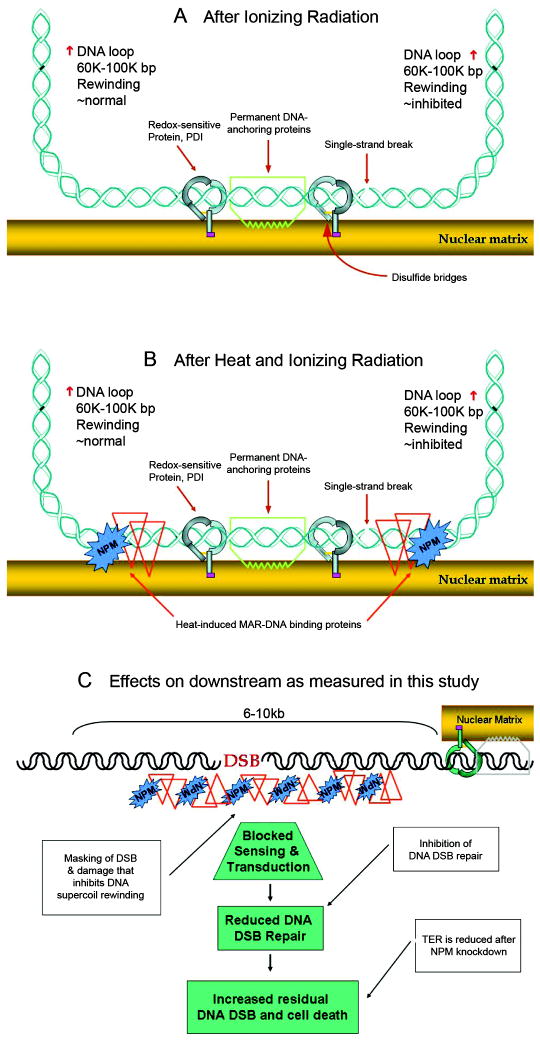
Figure 6. Model of the role of heat-induced NPM-DNA association in increased radiation sensitivity.
(A) Illustration of the MAR region in an unheated cell with a radiation induced strand break that inhibits the rewinding of the right DNA loop. (B) Illustration of the MAR region in a heated cell with a radiation induced strand break that is buried in a protein-DNA aggregate and cannot inhibit the rewinding of the right DNA loop. (C) The downstream effects of the heat-induced masking of DNA damage which include inhibition of repair and increased radiation induced cell killing. Note that the 6-10 Kb range beyond the MAR is suggested by previous studies.
It has been proposed that the nucleolus can act as repository of regulatory proteins that are released as needed in response to stress (17-19). Recent reports show that NPM binds chromatin in response to DNA double-strand breaks (41-42). These results suggest the importance of the nucleolus in modulating the cell’s response to stress. The current work further supports the hypothesis that the nucleolus is a key element in the response to stress conditions by showing a role for NPM redistribution in heat-induced radiosensitization.
Since most clinical hyperthermia delivers 41-42°C temperatures, and NPM redistribution is induced by 45°C, the NPM radiosensitizing mechanism is not contributing to the TER achieved by current clinical hyperthermia. Therefore if the thermal effectiveness of a given heat exposure could be enhanced to induce NPM redistribution and DNA association, then a significant increase in TER would be expected. Thus, NPM redistribution and DNA association will be an excellent target for the development of thermal enhancing agents, reported previously (43-44).
In summary, after heat shock the nucleolar protein, NPM or B23 re-localizes from the nucleolus into the nucleoplasm increasing its association with MAR DNA, masks a fraction of X-ray induced DNA damage including DSB, inhibits their repair and contributes to heat-induced radiosensitization in two cell lines. This observation builds on previous reports that suggest the nucleolus is a repository of stress responsive proteins. Thus NPM redistribution and MAR DNA binding should make a reasonable target for enhancing radiosensitization via thermal enhancers or mimetics.
Acknowledgments
Supported by grant 5PO1CA10445705 from the NIH US Department of HHS. J.D.W. is supported by the Pew Charitable Foundations and the Komen Race for the Cure.
References
- 1.Horsman MR, Overgaard J. Hyperthermia: a potent enhancer of radiotherapy. Clinical oncology (Royal College of Radiologists (Great Britain)) 2007;19(6):418–26. doi: 10.1016/j.clon.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 2.Kampinga HH, Dikomey E. Hyperthermic radiosensitization: mode of action and clinical relevance. International journal of radiation biology. 2001;77(4):399–408. doi: 10.1080/09553000010024687. [DOI] [PubMed] [Google Scholar]
- 3.Kampinga HH, Dynlacht JR, Dikomey E. Mechanism of radiosensitization by hyperthermia (> or = 43 degrees C) as derived from studies with DNA repair defective mutant cell lines. Int J Hyperthermia. 2004;20(2):131–9. doi: 10.1080/02656730310001627713. [DOI] [PubMed] [Google Scholar]
- 4.Kampinga HH, Wright WD, Konings AW, Roti Roti JL. The interaction of heat and radiation affecting the ability of nuclear DNA to undergo supercoiling changes. Radiation research. 1988;116(1):114–23. [PubMed] [Google Scholar]
- 5.Vanderwaal RP, Roti Roti JL. Heat induced ‘masking’ of redox sensitive component(s) of the DNA-nuclear matrix anchoring complex. Int J Hyperthermia. 2004;20(2):234–9. doi: 10.1080/02656730310001627704. [DOI] [PubMed] [Google Scholar]
- 6.Wynstra JH, Wright WD, Roti Roti JL. Repair of radiation-induced DNA damage in thermotolerant and nonthermotolerant HeLa cells. Radiation research. 1990;124(1):85–9. [PubMed] [Google Scholar]
- 7.Spector DL, Ochs RL, Busch H. Silver staining, immunofluorescence, and immunoelectron microscopic localization of nucleolar phosphoproteins B23 and C23. 1Chromosoma. 1984;90(2):139–48. doi: 10.1007/BF00292451. [DOI] [PubMed] [Google Scholar]
- 8.Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56(3):379–90. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 9.Chan WY, Liu QR, Borjigin J, et al. Characterization of the cDNA encoding human nucleophosmin and studies of its role in normal and abnormal growth. Biochemistry. 1989;28(3):1033–9. doi: 10.1021/bi00429a017. [DOI] [PubMed] [Google Scholar]
- 10.Feuerstein N, Chan PK, Mond JJ. Identification of numatrin, the nuclear matrix protein associated with induction of mitogenesis, as the nucleolar protein B23. Implication for the role of the nucleolus in early transduction of mitogenic signals. J Biol Chem. 1988;263(22):10608–12. [PubMed] [Google Scholar]
- 11.Kondo T, Minamino N, Nagamura-Inoue T, Matsumoto M, Taniguchi T, Tanaka N. Identification and characterization of nucleophosmin/B23/numatrin which binds the anti-oncogenic transcription factor IRF-1 and manifests oncogenic activity. Oncogene. 1997;15(11):1275–81. doi: 10.1038/sj.onc.1201286. [DOI] [PubMed] [Google Scholar]
- 12.Okuwaki M, Matsumoto K, Tsujimoto M, Nagata K. Function of nucleophosmin/B23, a nucleolar acidic protein, as a histone chaperone. FEBS Lett. 2001;506(3):272–6. doi: 10.1016/s0014-5793(01)02939-8. [DOI] [PubMed] [Google Scholar]
- 13.Yung BY, Chan PK. Identification and characterization of a hexameric form of nucleolar phosphoprotein B23. Biochim Biophys Acta. 1987;925(1):74–82. doi: 10.1016/0304-4165(87)90149-8. [DOI] [PubMed] [Google Scholar]
- 14.Brady SN, Yu Y, Maggi LB, Weber JD. ARF impedes NPM/B23 shuttling in an Mdm2-sensitive tumor suppressor pathway. Mol Cell Biol. 2004;24(21) doi: 10.1128/MCB.24.21.9327-9338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertwistle D, Sugimoto M, Sherr CJ. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol Cell Biol. 2004;24(3):985–96. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itahana K, Bhat KP, Jin A, et al. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol Cell. 2003;12(5):1151–64. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- 17.Mayer C, Bierhoff H, Grummt I. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes & development. 2005;19(8):933–41. doi: 10.1101/gad.333205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olson MO, Dundr M, Szebeni A. The nucleolus: an old factory with unexpected capabilities. Trends in cell biology. 2000;10(5):189–96. doi: 10.1016/s0962-8924(00)01738-4. [DOI] [PubMed] [Google Scholar]
- 19.Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. The EMBO journal. 2003;22(22):6068–77. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Guan J, Wang H, Wang Y, Leeper D, Iliakis G. Regulation of dna replication after heat shock by replication protein a-nucleolin interactions. The Journal of biological chemistry. 2001;276(23):20579–88. doi: 10.1074/jbc.M100874200. [DOI] [PubMed] [Google Scholar]
- 21.Daniely Y, Borowiec JA. Formation of a complex between nucleolin and replication protein A after cell stress prevents initiation of DNA replication. The Journal of cell biology. 2000;149(4):799–810. doi: 10.1083/jcb.149.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iliakis G, Krieg T, Guan J, Wang Y, Leeper D. Evidence for an S-phase checkpoint regulating DNA replication after heat shock: a review. Int J Hyperthermia. 2004;20(2):240–9. doi: 10.1080/02656730310001656379. [DOI] [PubMed] [Google Scholar]
- 23.Simard R, Bernhard W. A heat-sensitive cellular function located in the nucleolus. The Journal of cell biology. 1967;34(1):61–76. doi: 10.1083/jcb.34.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amalric F, Simard R, Zalta JP. Effect of supra-optimal temperature on nucleolar ribonucleoproteins and RNA. II. Biochemical study. Experimental cell research. 1969;55(3):370–7. doi: 10.1016/0014-4827(69)90571-0. [DOI] [PubMed] [Google Scholar]
- 25.Simard R, Amalric F, Zalta JP. Effect of supra-optimal temperature on nucleolar ribonucleoproteins and RNA. 1. Ultrastructural study. Experimental cell research. 1969;55(3):359–69. doi: 10.1016/0014-4827(69)90570-9. [DOI] [PubMed] [Google Scholar]
- 26.Subjeck JR, Shyy T, Shen J, Johnson RJ. Association between the mammalian 110,000-dalton heat-shock protein and nucleoli. The Journal of cell biology. 1983;97(5 Pt 1):1389–95. doi: 10.1083/jcb.97.5.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welch WJ, Feramisco JR. Nuclear and nucleolar localization of the 72,000-dalton heat shock protein in heat-shocked mammalian cells. The Journal of biological chemistry. 1984;259(7):4501–13. [PubMed] [Google Scholar]
- 28.Yu Y, Maggi LB, Jr, Brady SN, et al. Nucleophosmin is essential for ribosomal protein L5 nuclear export. Molecular and cellular biology. 2006;26(10):3798–809. doi: 10.1128/MCB.26.10.3798-3809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maggi LB, Jr, Kuchenruether M, Dadey DY, et al. Nucleophosmin serves as a rate-limiting nuclear export chaperone for the Mammalian ribosome. Molecular and cellular biology. 2008;28(23):7050–65. doi: 10.1128/MCB.01548-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.VanderWaal RP, Spitz DR, Griffith CL, Higashikubo R, Roti Roti JL. Evidence that protein disulfide isomerase (PDI) is involved in DNA-nuclear matrix anchoring. Journal of cellular biochemistry. 2002;85(4):689–702. doi: 10.1002/jcb.10169. [DOI] [PubMed] [Google Scholar]
- 31.Martens JHA, Verlaan M, Kalkhoven E, Dorsman JC, Zantema A. Scaffold/Matrix Attachment Region Elements Interact with a p300-Scaffold Attachment Factor A Complex and Are Bound by Acetylated Nucleosomes. Mol Cell Biol. 2002;22(8):2598–606. doi: 10.1128/MCB.22.8.2598-2606.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feuerstein N, Mond JJ, Kinchington PR, et al. Evidence for DNA binding activity of numatrin (B23), a cell cycle-regulated nuclear matrix protein. Biochimica et biophysica acta. 1990;1087(2):127–36. doi: 10.1016/0167-4781(90)90196-9. [DOI] [PubMed] [Google Scholar]
- 33.Huang L, Mivechi NF, Moskophidis D. Insights into regulation and function of the major stress-induced hsp70 molecular chaperone in vivo: analysis of mice with targeted gene disruption of the hsp70.1 or hsp70.3 gene. Molecular and cellular biology. 2001;21(24):8575–91. doi: 10.1128/MCB.21.24.8575-8591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yogev O, Saadon K, Anzi S, Inoue K, Shaulian E. DNA damage-dependent translocation of B23 and p19 ARF is regulated by the Jun N-terminal kinase pathway. Cancer research. 2008;68(5):1398–406. doi: 10.1158/0008-5472.CAN-07-2865. [DOI] [PubMed] [Google Scholar]
- 35.Dewey WC. Failla memorial lecture. The search for critical cellular targets damaged by heat. Radiation research. 1989;120(2):191–204. [PubMed] [Google Scholar]
- 36.Mills MD, Meyn RE. Effects of hyperthermia on repair of radiation-induced DNA strand breaks. Radiation research. 1981;87(2):314–28. [PubMed] [Google Scholar]
- 37.Mills MD, Meyn RE. Hyperthermic potentiation of unrejoined DNA strand breaks following irradiation. Radiation research. 1983;95(2):327–38. [PubMed] [Google Scholar]
- 38.Wright WD, Lagroye I, Zhang P, Malyapa RS, Roti Roti JL, Zbigniew Darzynkiewicz HACJPR. Methods in Cell Biology. Academic Press; 2001. Cytometric methods to analyze ionizing-radiation effects; pp. 251–68. [DOI] [PubMed] [Google Scholar]
- 39.Laszlo A, Davidson T, Harvey A, et al. Alterations in heat-induced radiosensitization accompanied by nuclear structure alterations in Chinese hamster cells. Int J Hyperthermia. 2006;22(1):43–60. doi: 10.1080/02656730500394296. [DOI] [PubMed] [Google Scholar]
- 40.Roti Roti JL, Wright WD. Visualization of DNA loops in nucleoids from HeLa cells: assays for DNA damage and repair. Cytometry. 1987;8(5):461–7. doi: 10.1002/cyto.990080505. [DOI] [PubMed] [Google Scholar]
- 41.Lee SY, Park JH, Kim S, Park EJ, Yun Y, Kwon J. A proteomics approach for the identification of nucleophosmin and heterogeneous nuclear ribonucleoprotein C1/C2 as chromatin-binding proteins in response to DNA double-strand breaks. The Biochemical journal. 2005;388(Pt 1):7–15. doi: 10.1042/BJ20042033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wardleworth BN, Downs JA. Spotting new DNA damage-responsive chromatin-binding proteins. The Biochemical journal. 2005;388(Pt 1):e1–2. doi: 10.1042/BJ20050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sekhar KR, Sonar VN, Muthusamy V, et al. Novel chemical enhancers of heat shock increase thermal radiosensitization through a mitotic catastrophe pathway. Cancer research. 2007;67(2):695–701. doi: 10.1158/0008-5472.CAN-06-3212. [DOI] [PubMed] [Google Scholar]
- 44.Sonar VN, Thirupathi Reddy Y, Sekhar KR, Sasi S, Freeman ML, Crooks PA. Novel substituted (Z)-2-(N-benzylindol-3-ylmethylene)quinuclidin-3-one and (Z)-(±)-2-(N-benzylindol-3-ylmethylene)quinuclidin-3-ol derivatives as potent thermal sensitizing agents. Bioorganic & Medicinal Chemistry Letters. 2007;17(24):6821–4. doi: 10.1016/j.bmcl.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]