Abstract
Technical advances have seen the rapid adoption of genomics and multiplex genetic polymorphism identification to research on vascular diseases. The utilization of proteomics for the study of vascular diseases has been limited by comparison. In this review we outline currently available proteomics techniques, the challenges to using these approaches and modifications which may improve the utilization of proteomics in the study of vascular diseases.
Introduction
Common vascular diseases, including atherosclerosis and aortic aneurysm appear to result from interactions between environmental risk factors and genetic predisposition which exacerbate normal aging processes. Mechanisms underlying these diseases include inflammation, alterations of the vascular extracellular matrix and dyslipidaemia. Changes in protein expression are a feature of pathological progression and drive such alterations. Inflammation, for example, is determined by the expression of adhesion molecules, chemokines and pro-inflammatory cytokines. Understanding the protein profiles associated with different vascular diseases may help to improve disease management in a number of areas including diagnosis, prognosis and treatment.
Traditionally, identification of new disease-associated proteins relied on hypothesis-led approaches, using techniques such as ELISA and western blotting to investigate expression of individual candidates. However, the relatively recent development of techniques, including two-dimensional electrophoresis in 1975,1 which enable more complete protein profiling have given rise to a discipline known as “proteomics”, with potential to investigate protein expression in response to endogenous and exogenous stimuli. Using a suite of sample preparation, fractionation, separation and analysis tools a modern proteomics facility can perform quantitative ‘differential display’ experiments comparing cohorts of control and disease samples, highlighting molecules directly related to and/or indicative of disease processes (Figure 1). Whilst similar data can be gained from gene expression studies, mRNA levels do not always reflect protein expression and cellular phenotype.2 Thus, proteomics provides data not easily obtained from other post-genomic technologies (Figure 2).3 Consequently, proteomic techniques have been applied to wide variety of organisms including plants, bacteria, fungi, and metazoa. In a medical context, proteomic investigations have the potential to uncover proteins of therapeutic and diagnostic potential. However, despite ongoing endeavors, proteomic science has yet to provide a significant breakthrough in this area, with many putative markers failing during validation. In this review we outline proteomic techniques and discuss current technical challenges and likely future developments.
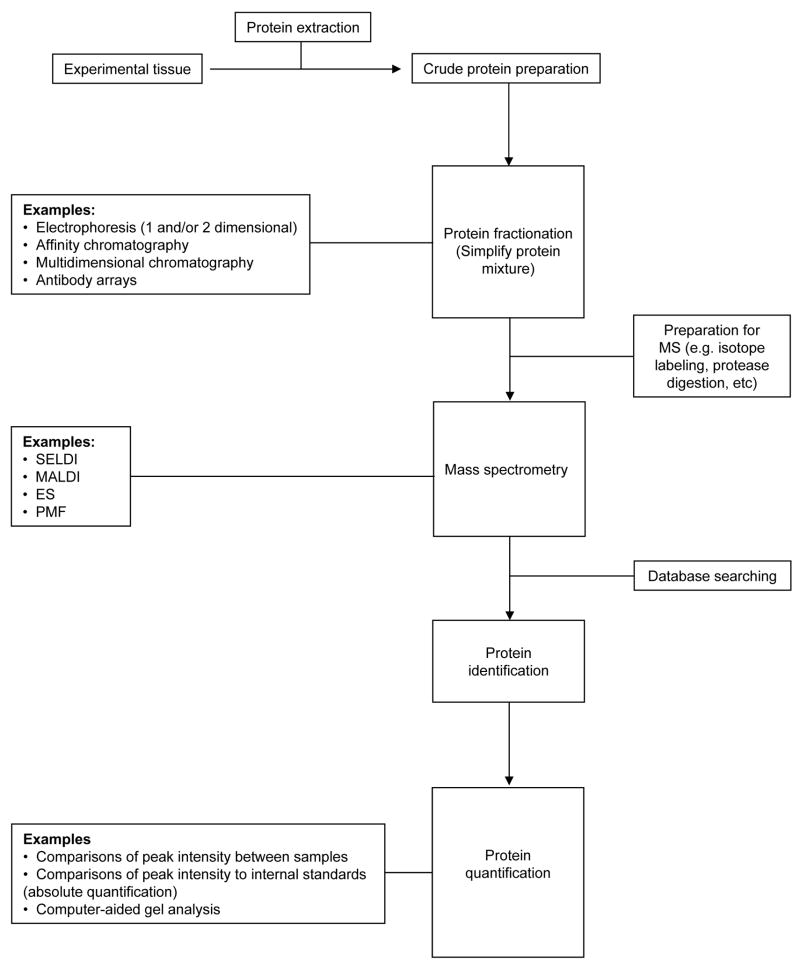
Figure 1.
Standardised proteomics workflow. In a typical proteomics experiment, crude protein mixtures are fractionated to simplify downstream analyses. Proteins of interest are commonly investigated using mass spectrometry and identified following computer-aided database searches. Once identified, protein known proteins can be quantified by comparing relative expression between treatments (relative quantification), or compared to an internal standard of known quantity (absolute quantification). Examples of the techniques available to perform each step are provided. Definitions of acronyms are provided in Table 1.
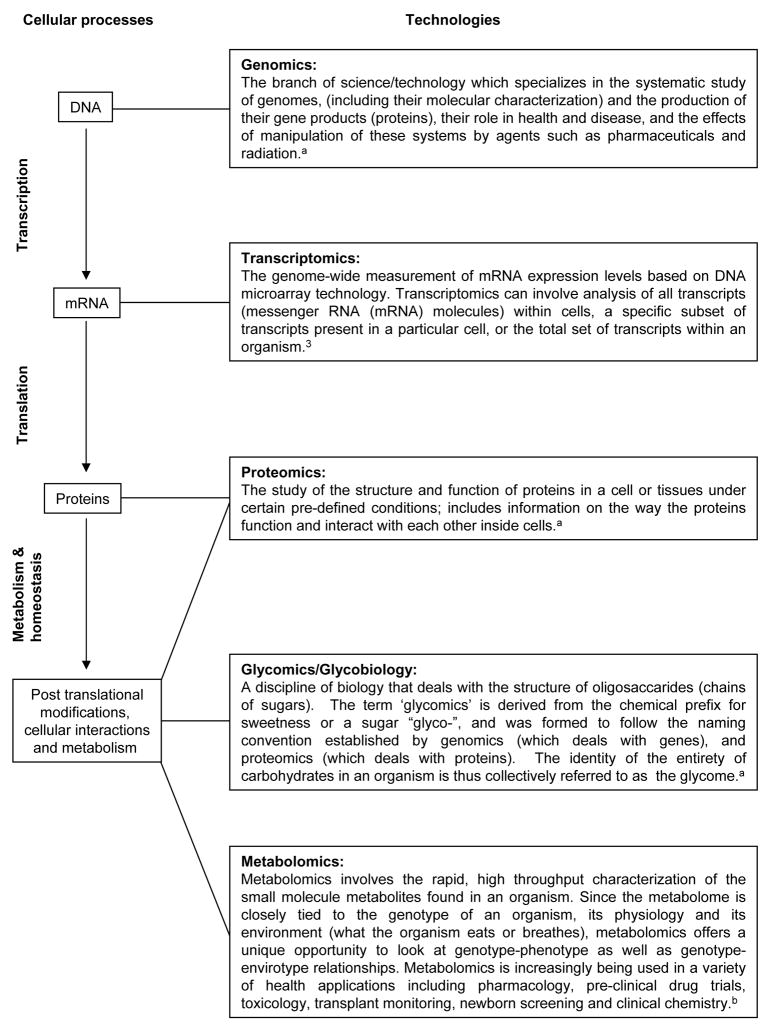
Figure 2.
Overview of cellular processes, and modern technologies available for their study. Definitions were sourced from the Human Proteome Organisation (http://www.hupo.org/overview/glossary/),a the Human Metabolome Project (http://www.metabolomics.ca/),b and a published article by Gomase and Tagore,3 (detailed in references).
Proteomic techniques
The Human Proteome Organisation (HUPO) defines a protein as a complex molecule made up of one or more peptide chains (a peptide consisting of two or more chemically-linked amino acids), which perform a wide variety of functions and are essential to the life of the cell (http://www.hupo.org/overview/glossary/). Further definitions of some common terms used in proteomics are provided in Table 1.
Table 1.
Glossary of common proteomics terms
| Term | Definition |
|---|---|
| ESI | ElectroSpray Ionisation. A method of ionization whereby proteins or peptides are carried by a stream of volatile liquid to an emitter carrying a high voltage. The liquid is sprayed from the emitter and the liquid evaporated, leaving the charged peptides or proteins to be drawn into the mass spectrometer for analysis. Electrospray is advantageous in the analysis of peptides and proteins as multiply charged ions are created which can be discriminated from singly charged ions which are often contaminants |
| High throughput proteomics | The analysis of many protein samples in a short space of time. |
| MALDI | Matrix Assisted Laser Desorption Ionisation. A method of ionizing proteins or peptides whereby proteins are mixed with a chemical matrix and targeted by a laser. The matrix absorbs energy from the laser and is ionized, carrying singly-charged peptides into the gas phase for MS analysis. |
| Mass spectrometry (MS) | The analysis of gas-phase ions (often peptides in the case of proteomics), to determine mass and charge of the intact molecule, or component parts |
| PMF | Peptide Mass Fingerprint. The profile of spectral peaks generated following mass spectrometry which documents the mass of ions within a sample. |
| Reverse-phase chromatography | The process whereby molecules in the liquid phase are separated by their hydrophobic properties. |
| Secretome | A sub-set of proteins secreted from a cell, tissue or organism |
| SELDI | Surface Enhanced Laser Desorption Ionisation. A method of ionising proteins or peptides whereby peptides are prefractionated by reverse phase chromatography and bind to a specialist chip. Bound proteins are exposed to a laser and are ionized into the gas phase for MS analysis. |
| Shotgun proteomics | The process of identifying molecules through MS analysis of a crude protein/peptide mixture prefractionated in one or two-dimensions by chromatographic techniques. |
| TOF | Time Of Flight. A method of measuring the mass of an ion in mass spectrometry, determined by the time taken for an ion to travel a vacuum tube, and reach a detector. |
Proteins are encoded by the genome, with each open reading frame giving rise to an average of 6–7 protein products. Thus, it is estimated that the human genome comprising 23,000–40,000 genes may give rise to up to one million proteins, in addition to approximately 600,000 serum immunoglobulins with slight variations in epitope binding regions.4–6 However, protein structure is not limited to genetic coding as post-translational modifications (PTMs) such as oxidation, phosphorylation and glycosylation are performed throughout cellular metabolism, and can not be predicted by gene analysis. PTMs alter protein function, and consequently have pathological significance. For example the, matrix gamma-carboxyglutamate protein (MGP) is a known inhibitor of cardiovascular calcification, and the uncarboxylated form (uMGP) has been shown to accumulate within calcified tissue. Consequently, circulating uMGP is a proposed biomarker for vascular calcification, with low titres a risk factor for vascular calcification.7 Accordingly, investigations into the expression of proteins and PTMs in disease-states have potential to generate data of clinical significance.
Gel-based proteomics
Two-dimensional polyacrylamide gel electrophoresis (2D-PAGE), separating proteins according to charge (first dimension), and molecular weight (second dimension), can profile thousands of polypeptides within a sample, enabling non-hypothesis led investigations. Changes in protein expression, evidenced through alterations in spot density, are determined through computer-aided comparisons of 2D profiles produced from diseased and control tissues and highlight proteins specifically expressed in response to disease. Spots of interest can be directly excised from gels for identification through mass spectrometry (MS). Many of the published vascular proteomics investigations have been conducted using a gel-based format (Table 2).8–13 For example, gel-based investigations performed by Urbonavicius et al. (2008), confirm the role of oxidative stress in aneurysm rupture, confirmed by the upregulation of stress-response proteins when compared to non-ruptured controls.14
Table 2.
Examples of Gel-based vascular proteomic investigations
| Protein source | Example of findings |
|---|---|
| Serum |
Serum analysis to determine effect of dietary fish oil on circulating biomarkers of inflammation Sera treated to remove abundant proteins then analysed via 2D-PAGE, software, peptide mass fingerprinting and peptide sequencing. Fish oil supplementation caused a reduction in acute phase proteins compared to sunflower oil controls, suggesting a reduction in inflammation. Serum levels of apolipoprotein A1 were also down regulated following fish oil supplementation.8 |
| Tissue |
Comparative analysis of proteins expressed by stable/unstable atherosclerotic plaques Stability of carotid artery plaques assessed histochemically and soluble proteins extracted. Analysis via 2D-PAGE, software and peptide mass fingerprinting. Unstable plaques expressed higher concentrations of ferritin (light subunit), superoxide dismutase 2 and fibrinogen D, and lower concentrations of GST, Rho GDP-Dissociation inhibitor 1, annexin, HSP20 and HSP27.9 |
| Cells |
Identification of proteomic signatures of platelet activation Analysis of low molecular weight soluble cellular proteins using surface-enhanced laser desorption ionisation time of flight (SELDI-TOF) MS. Spectral profiles from resting platelets revealed a characteristic peak pattern when compared to cells activated through shear stress, or exposure to adenosine diphosphate (ADP)- or thrombin receptor activation peptide. Analysis of ADP-stimulated platelets isolated from patients receiving clopidogrel revealed a similar spectral profile to resting cells, demonstrating drug efficacy.10 |
| Cultured cells |
Analysis of cultured neonatal rat VSMCs following homocysteine induced proliferation Analysis of soluble proteins via 2D-PAGE, software and peptide mass fingerprinting. Homocysteine treated cells expressed higher concentrations of WDR1 protein, calreticulin, pyruvate kinase M2, vimentin, aldose reductase and triose phosphate isomerise. Down regulation of lamin C and LIM and SH3 protein 1.11 Analysis of protein expression by human umbilical vein endothelial cells (HUVEC) following digoxin-induced toxicity Cultured HUVEC proteins extracted and analysed via 2D-PAGE, software and peptide sequencing. Pro-apoptotic digoxin treatment results in differential expression of 9 proteins (up regulation of ATP synthetase, Lamin A, Cystatin A, Succinyl Co-A, nuclear riboprotein H3, Proteasome subunit 5; down regulation of HSP60, electron transfer flavoprotein, profilin-1). Induced overexpression of HSP60 reduced digoxin induced apoptosis suggesting that HSP60 is important in cellular protection.12 Analysis of protein expression by HUVECs during replicative senescence to investigate cellular ageing Soluble proteins expressed by cultured HUVECs extracted and analysed via DiGE, software, peptide sequencing and western blot. Senescence seen to be accompanied by expression of proteins involved in cellular stress mechanisms and protein biosynthesis. Reductions in the expression of DNA repair/maintenance proteins is hypothesised to underpin observed nuclear degradation and cytoskeleton disruptions.13 |
Abbreviations: ADP, Adenosine diphosphate; GST, Glutathione transferase; HSP, Heat shock protein; HUVEC, Human umbilical vein endothelial cells; VSMC, Vascular smooth muscle cell
Criticisms of 2D-PAGE include gel-to-gel variability, inability to analyse highly basic and hydrophobic proteins, poor visualization of very high- or very low molecular weight proteins, under-representation of weakly-expressed proteins, and a lack of easy automation.15–19 Some of these criticisms relate to the difficulty of good sample preparation, consequently poor samples are the cause of most poor 2D gels. However, 2D-PAGE is the only technique capable of simultaneously resolving thousands of post-translationally modified proteins due to the, influence such modifications exert on the migration of proteins during electrophoresis which is easily visualized following 2D-PAGE. For example, oxidized 1-Cys peroxiredoxin expressed within the aortas of apolipoprotein E (Apo E−/−) deficient mice has a more acidic charge than the form expressed by Apo E+/+ controls, and migrates to a different position following 2D-PAGE.20 Thus, gel-based investigations generate data difficult to obtain from other proteomic techniques, supporting their continued use.
The introduction of increasingly sensitive protein stains has enhanced the analytical power of 2D-PAGE. The development of the difference gel electrophoresis (DiGE) technique,21 in which fluorescent cyanine dyes are covalently bound to lysine residues within sample proteins, enables analysis of multiple samples within the same gel. Furthermore, DiGE protein expression is compared to a known internal standard, reducing gel-to-gel variation. Currently, the simultaneous analysis of 2 samples (2-plex) is possible, and is particularly useful when comparing protein expression in the same individual at different time points or following interventions, such as a drug treatment. However, the application of DiGE to vascular investigations has been limited, possibly due to the cost of compatible reagents and specialized equipment.13, 22
MS-based proteomics
MS is the central protein identification technique in almost all proteomic investigations, with the exception of chip array experiments. A mass spectrometer measures the mass of ions within a sample, and comprises an ion source, a mass analyzer which measures the mass-to-charge ratio (m/z) of the ions, and a detector to record the frequency of ions at each m/z value.23 In proteomic studies MS analysis is commonly conducted on ionised peptides yielded from enzymatic digestion of sample proteins. MS analysis enables quantitative investigation into complex protein samples regardless of the hydrophobicity, charge and size of constituent components,24 without the need for as large a quantity of sample proteins needed for gel-based approaches.
With this in mind, MS-based proteomic approaches are useful when working with scarce or protein-poor samples. The commercialization of reagents for MS-led investigations has improved access to these approaches. For example, protein labeling techniques using stable ‘heavy’ isotopes (e.g. 15N, 2H, 18O and 13C), generate a detectable shift from native molecular mass, to distinguish between experimental treatments.25–30 These techniques have increased the multiplex capabilities of MS, enabling the simultaneous analysis of several samples. Stable isotopes can also be used to supplement cell culture media, or animal chow to enable MS-based investigations of protein expression in actively metabolising cells and tissues. 31–34 However, MS-based methods do not readily provide information about PTMs which are plainly visible on 2D gels. An online supplement discussing the principles of MS is available.
Protein micro- and macro-array
Protein arrays are equivalent to gene arrays, and assess relative protein expression within sample tissue. Such assays usually consist of specific antibodies immobilized on a chip or membrane which are incubated with proteins extracted from diseased or control samples, enabling antigen/antibody complexes to form. This approach can be used as a means to fractionate crude samples for further analysis (for example in SELDI-MS), or protein binding can be quantified, reflecting antigen levels within patient samples permitting direct investigation into protein expression. The investigative potential of this approach is limited to commercially available antibody panels, thus protein microarrays are more suited to hypothesis-driven research than novel protein discovery per se. However, the two approaches are compatible as proteins discovered through non-biased investigations can be validated in large cohorts using the microarray format, providing that appropriate antibodies are available. It is expected that future developments of this technique will allow more comprehensive protein assessment; however, the range of protein isotypes, fragments and modifications expressed in vivo present considerable challenges for true global analysis using protein arrays.
Challenges of clinical proteomics
Sample complexity
Examination of whole tissue samples is complicated by the presence of abundant proteins which mask more conservatively expressed molecules. This is particularly true of serum or plasma, in which albumin accounts for 55% total protein (normal concentration range 35–50 mg/ml). In contrast other proteins are present in much lower concentrations (e.g. interleukin 6, normal concentration range 0–5 pg/ml).35–38 Thus, plasma protein expression is estimated to span 10 orders of magnitude, often referred to as the ‘dynamic range’ of the sample. Such variation cannot be visualised as current techniques capture a dynamic range spanning 4–5 orders of magnitude and only abundant molecules will be profiled from crude samples.35, 39 Similarly, analysis of vascular tissue is complicated by cellular heterogeneity, favouring proteins expressed by dominant cell types, where the dynamic range spans at least 5–6 orders of magnitude.17 Weakly expressed molecules can be enriched by depleting dominant proteins. However, this can introduce experimental error by removing protein complexes, or misrepresenting genuine variations in the expression of abundant proteins.36, 40
Sample complexity can be reduced by studying a sub-group of proteins expressed within tissues using physical extraction techniques such as laser microdissection to isolate specific cells from tissue preparations,25, 41 biochemical methods,22, 42 or cellular approaches to identify proteins secreted from cell lines or tissue explants (the secretome). Secretome analysis is attractive since soluble secreted proteins are more amenable to proteomic profiling than their hydrophobic counterparts. Furthermore, such studies highlight proteins which may enter the bloodstream, revealing a panel of putative circulating biomarkers.
Secretome analysis is commonly conducted through in vitro incubation of tissue explants,43–45 or cell lines,46, 47 and 2D-PAGE/MS analysis of culture fluid. However, in vitro culture inevitably results in some degree of sample deterioration which releases cellular proteins into the culture media. Under typical conditions cellular proteins cannot be differentiated from secreted molecules at the gel level, potentially resulting in the discovery of false biomarkers. Truly secreted proteins can be putatively revealed by analyzing proteins to detect amino acid motifs involved in protein secretion following MS identification.48 Such analyses generate more robust datasets, although extensive MS investigations are necessary before the truly secreted proteins can be differentiated. Modifications to cell culture and proteomic techniques facilitate identification of secreted proteins, offering an alternative to exhaustive MS/bioinformatic analysis. Radiolabelled amino acids (e.g. 35S-methionine), can be used as metabolic tags are detectable at the gel level; non-labelled proteins can be considered degradation products and discounted from further analysis.49 Potential drawbacks such as the need for specialist radioactive facilities, and radiation-induced alterations in protein expression can be alleviated by using stable isotopes, and SILAC has been used to characterise biomarkers secreted from a variety of tissues.50–52
Experimental complexity
Experimental design
There are currently no formalized guidelines for the design of proteomic experiments and data presentation, and early proteomic investigations often documented two-dimensional protein profiles or mass spectra. Such analyses are no longer considered suitable for publication, and protein identification is essential. The recent publication of the minimum information about a proteomics experiment (MIAPE) document by HUPO’s protein standards initiative (HUPO PSI), has provided more stringent guidelines for reporting proteomic data.57 Whilst these suggestions are not concrete, many of the leading journals in this field employ MIAPE criteria, insisting on the provision of metadata to increase confidence in the proteomic experiment and subsequent analyses (e.g. MS and statistics). Increased visibility of metadata has stimulated the development of open-access online repositories such as the proteomics identifications database (PRIDE),53, 54 which capture MS identifications and annotations from proteomics investigations in a standard format. It is envisaged that such repositories will act as a valuable research tool, whilst upholding HUPO PSI standards and vocabulary.
As with other scientific methodologies, the MIAPE document emphasises the need for replication to generate reliable data. Calculation of suitable sample sizes can be problematic, and depends on the complexity of the proteomes to be analysed and the level of variation anticipated between treatments. Calibration experiments using standard protein mixtures of similar complexity to tissues of interest may help to ensure suitable replication.55, 56 Alternatively, the use of power calculations can help determine suitable sample sizes.57 In the case of gel-based investigations, a minimum of 5 replicates per treatment has been recommended, with modifications to established statistical analyses to limit the number of false protein spots observed.58 Whilst this improves confidence in experimental data, replication in medical experiments may be problematic due to the paucity of suitable samples. Similarly, the availability of control tissue can limit potential investigations, especially in vascular experiments where healthy vasculature is not routinely sampled. Samples recovered at autopsy may increase experimental variation through physiological changes post-mortem and differences in tissue collection protocols. Furthermore, post-mortem samples are of limited use when investigating chronic diseases such as atherosclerosis, where variables including age and prolonged exposure to therapeutic compounds may complicate case/control matching. Small quantities of tissue such as aneurysm neck which can be removed from patients during surgical repair will match experimental samples and may be used as controls, but are unlikely to represent truly healthy vasculature despite being visibly free of complications.
Reproducibility
Biological and technical variations can affect the reproducibility of proteomic investigations although commercialisation of common proteomic consumables has contributed towards increased reproducibility and experimental robustness. The development and standardization of sample preparation protocols appropriate to the experimental tissues represents the greatest challenge to effective and reproducible proteomic profiling. Successful proteomic investigations rely on understanding the sample that is to be analysed and knowledge of how to extract and solubilise the desired proteins whilst eliminating contaminants. Sample preparation can appear daunting primarily because diversity within even the simplest proteome cannot be captured by any single extraction and separation. Over the past decade, fractionation has been widely adopted to reduce sample complexity and enable analysis of weakly expressed proteins. Diversity in sample preparation is reflected by scientific literature.59, 60 Despite this, inter-laboratory variation remains an issue, as discussed by Callesen et al. (2008), who report that only 25% of reported biomarker peaks for breast cancer could be detected by other research groups following SELDI-MS.61
Protein identification
Molecular identification is commonly achieved after MS analysis by searching electronic databases containing protein and/or translated gene sequences. Database searches generate putative identifications based on homology between experimental peptide sequences or fragmentation patterns, to those of known molecules. Putative identifications are quantified by assigning statistical scores (such as statistical error) to reflect the surety of each search; identifications are only accepted when the calculated error falls below accepted thresholds.17 This approach is only possible when databases contain data relating to the experimental proteins and relies on comprehensive datasets. In the case of medical investigations, MS-based protein identification is facilitated by the completion of the human genome project, and increased availability of genomic data for common animal models. However, identification of weakly expressed polypeptides remains challenging in the absence of a PCR equivalent for proteins.
Whilst desireable for publication, protein identification may not always be needed for clinical applications, especially where disease-state protein profiles are distinctly different to healthy controls. For example, MS analysis of low-molecular weight proteins expressed by platelets reveals distinctive changes in spectral peaks which discriminate resting platelets from those aggregated by ADP or thrombin-activator peptide (sensitivity 65%, specificity 89%). Platelets isolated from patients receiving clopidogrel therapy show a 70% reduction in ADP-induced aggregation, this is mirrored by a similar MS profile to resting platelets.10 Such analyses may be of particular benefit when assessing patient response to medication, highlighting appropriate therapeutic regimes. Similarly, case-control comparisons of urine protein profiles in coronary artery disease (CAD) demonstrated increased expression of a panel of 17 polypeptides in CAD patients. Using this pattern as a diagnostic tool, von Zur Muhlen et al. (2009) reported 81% sensitivity and 92% specificity using samples from 26 CAD patients and 12 controls in a blinded study. However, later analyses revealed a similar protein profile in patients with chronic renal failure. suggesting that renal damage may disturb the urine proteome and complicate proteomic analysis.62
Future directions of proteomics
Proteomic techniques document the expression of molecules which directly influence cell phenotype, and therefore provide data of clinical relevance. These techniques can be applied to a wide variety of organisms and biological samples, and can be applied to investigate protein expression in vitro or in vivo. Proteomic tools permit analysis of thousands of proteins and their PTMs and enable non-biased appraisals of the molecular biology of disease states, highlighting molecules which may be overlooked in hypothesis-driven scenarios. As described above, proteomic science is a relatively recent development. Currently proteomics receives many criticisms, notably low reproducibility evidenced by gel-to-gel and inter-laboratory variation. Despite this, it is likely that these issues will diminish as protocols and reporting requirements become standardized and are universally applied. Identification of weakly expressed molecules via MS remains problematic, although techniques such as western blotting may be used to overcome this challenge.
The application of proteomics to vascular disease is at an early stage, and without subsequent downstream analyses, proteomic experiments merely provide lists of protein data with little practical value. Proteomic investigations generate large datasets which require expertise for meaningful analysis, especially in the case of large or complex experiments. The continued use of proteomics has provided the stimulus for improvements in data visualization, contributing to the establishment of bioinformatics as a specialist field. Currently, it is becoming common for proteomic data to be displayed as interaction networks, highlighting associations (e.g. synchronous up/down-regulation) between identified molecules.63, 64 This is of particular benefit when investigating disease mechanisms, or therapeutic pathways. The most routine application of proteomic techniques in vascular medicine is to discover biomarkers of diagnostic, therapeutic and prognostic potential. However, protein identifications generated by proteomic techniques remain putative, requiring validation through more established approaches including ELISA, western blotting and immunohistochemistry, using reliable antibodies. Furthermore, in order to assess biomarker performance molecules of interest must be valididated in large patient populations using high-throughput techniques which is time consuming and limited by access to large patient cohorts.
The investigative power of proteomics is greatly magnified when combined with other post-genomic techniques. Combined proteomic and metabolomic analyses provide direct evidence of the effect of protein expression on cellular processes. Using this approach, Perlman et al. (2009) investigated the cardioprotective mechanisms following nitrate exposure. Nitrate administration resulted in a short-lived increase in cardiac nitrate levels, but substantial elevations in cardiac ascorbate oxidation. This was accompanied by significant improvements in cardiac contractile recovery following ischemia-reperfusion after preconditioning with low (0.1 mg/kg) or high (10 mg/kg) nitrate doses. Proteomic analysis of cardiac mitochondria revealed dose-dependant PTM of isoforms of 3 proteins involved in serine/threonine kinase signaling, anti-oxidant protection and cell metabolism. This led the authors to suggest that a similar mechanism may underpin the cardioprotective value of physical exercise and a diet containing nitrite/nitrate-rich foods.65 Such systemic strategies have been successfully employed in other biological situations, although relatively few vascular investigations have embraced this multi-omics approach. Thus it seems likely that vascular medicine will greatly benefit from the future application of proteomics in a ‘multi-omics’ context.
Acknowledgments
JVM and JG thank Dr Cathy Rush for proof reading and helpful discussion. JG is supported by grants from the NHMRC, Australia (project grants 540403, 540404, 540405) and NIH, USA (RO1 HL080010), and a Practitioner Fellowship from the NHMRC, Australia (431503).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. Journal of Biological Chemistry. 1975;250(10):4007–21. [PMC free article] [PubMed] [Google Scholar]
- 2.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19(3):1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gomase VS, Tagore S. Transcriptomics. Curr Drug Metab. 2008;9(3):245–9. doi: 10.2174/138920008783884759. [DOI] [PubMed] [Google Scholar]
- 4.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, et al. The sequence of the human Genome. Science. 2001;291(5507):1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 5.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.Humphery-Smith I. A human proteome project with a beginning and an end. Proteomics. 2006;4:2519–2521. doi: 10.1002/pmic.200400866. [DOI] [PubMed] [Google Scholar]
- 7.Craenburg EC, Brandenburg VM, Vermeer C, Stenger M, Muhlenbruch G, et al. Uncarboxylated matrix Gla protein (ucMGP) is associated with coronary artery calcification in haemodialysis patients. Thromb Haemostat. 2009;101(2):359–66. [PubMed] [Google Scholar]
- 8.de Roos B, Geelen A, Ross K, Rucklidge G, Reid M, et al. Identification of potential serum biomarkers of inflammation and lipid modulation that are altered by fish oil supplementation in healthy volunteers. Proteomics. 2008;8(10):1965–74. doi: 10.1002/pmic.200700457. [DOI] [PubMed] [Google Scholar]
- 9.Lepedda AJ, Cigliano A, Cherchi GM, Spirito R, Maggioni M, et al. A proteomic approach to differentiate histologically classified stable and unstable plaques from human carotid arteries. Atherosclerosis. 2008:2008. doi: 10.1016/j.atherosclerosis.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin W, Czuchlewski MD, Peerschke EI. Development of proteomic signatures of platelet activation using surface-enhanced laser desorption/ionization technology in a clinical setting. Am J Clin Pathol. 2008;129:862–869. doi: 10.1309/BC0RXR50KKKWCVHM. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, Shen J, Zhan R, Wang X, Wang X, et al. Proteomic analysis of homocysteine induced proliferation of cultured neonatal rat vascular smooth muscle cells. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbapap.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Qiu J, Gao HQ, Liang Y, Yu H, Zhou RH. Comparative proteomics analysis reveals role of heat shock protein 60 in digoxin-induced toxicity in human endothelial cells. Biochim Biophys Acta. 2008;1784(11):1857–64. doi: 10.1016/j.bbapap.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 13.Eman MR, Regan-Klapisz E, Pinkse MW, Koop IM, Haverkamp J, et al. Protein expression dynamics during replicative senescence of endothelial cells studied by 2-D difference in-gel electrophoresis. Electrophoresis. 2006;27(8):1669–82. doi: 10.1002/elps.200500746. [DOI] [PubMed] [Google Scholar]
- 14.Urbonavicius S, Lindholt JS, Vorum H, Urbonaviciene G, Henneberg EW, et al. Proteomic identification of differentially expressed proteins in aortic wall of patients with ruptured and nonruptured abdominal aortic aneurysms. J Vasc Surg. 2008:2008. doi: 10.1016/j.jvs.2008.08.097. [DOI] [PubMed] [Google Scholar]
- 15.Wilkins MR, Gasteiger E, Sanchez JC, Bairoch A, Hochstrasser DF. Two-dimensional gel electrophoresis for proteome projects: the effects of protein hydrophobicity and copy number. Electrophoresis. 1998;19(8–9):1501–5. doi: 10.1002/elps.1150190847. [DOI] [PubMed] [Google Scholar]
- 16.Gygi SP, Corthals GL, Zhang Y, Rochon Y, Aebersold R. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc Natl Acad Sci U S A. 2000;97(17):9390–5. doi: 10.1073/pnas.160270797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett J, Brophy PM, Hamilton JV. Analysing proteomic data. Int J Parasitol. 2005;35(5):543–553. doi: 10.1016/j.ijpara.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 18.Molloy MP, Phadke ND, Chen H, Tyldesley R, Garfin DE, et al. Profiling the alkaline membrane proteome of Caulobacter crescentus with two-dimensional electrophoresis and mass spectrometry. Proteomics. 2002;2(7):899–910. doi: 10.1002/1615-9861(200207)2:7<899::AID-PROT899>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 19.Patton WF, Schulenberg B, Steinberg TH. Two-dimensional gel electrophoresis; better than a poke in the ICAT? Curr Opin Biotechnol. 2002;13(4):321–8. doi: 10.1016/s0958-1669(02)00333-6. [DOI] [PubMed] [Google Scholar]
- 20.Mayr M, Chung YL, Mayr U, Yin X, Ly L, et al. Proteomic and metabolomic analyses of atherosclerotic vessels from apolipoprotein E-deficient mice reveal alterations in inflammation, oxidative stress, and energy metabolism. Arterioscler Thromb Vasc Biol. 2005;25(10):2135–42. doi: 10.1161/01.ATV.0000183928.25844.f6. [DOI] [PubMed] [Google Scholar]
- 21.Unlu M, Morgan ME, Minden JS. Difference gel electrophoresis: A single method for detecting changes in protein extracts. Electrophoresis. 1997;18(11):2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- 22.Sidibe A, Yin X, Tarelli E, Xiao Q, Zampetaki A, et al. Integrated membrane protein analysis of mature and embryonic stem cell-derived smooth muscle cells using a novel combination of CyDye/biotin labeling. Mol Cell Proteomics. 2007;6(10):1788–97. doi: 10.1074/mcp.M600433-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 24.Iliuk A, Galan J, Tao WA. Playing tag with quantitative proteomics. Anal Bioanal Chem. 2008 doi: 10.1007/s00216-008-2386-0. [DOI] [PubMed] [Google Scholar]
- 25.Haqqani AS, Nesic M, Preston E, Baumann E, Kelly J, et al. Characterization of vascular protein expression patterns in cerebral ischemia/reperfusion using laser capture microdissection and ICAT-nanoLC-MS/MS. FASEB J. 2005;19(13):1809–21. doi: 10.1096/fj.05-3793com. [DOI] [PubMed] [Google Scholar]
- 26.Haqqani AS, Kelly J, Baumann E, Haseloff RF, Blasig IE, et al. Protein markers of ischemic insult in brain endothelial cells identified using 2D gel electrophoresis and ICAT-based quantitative proteomics. J Proteome Res. 2007;6(1):226–39. doi: 10.1021/pr0603811. [DOI] [PubMed] [Google Scholar]
- 27.Sethuraman M, McComb ME, Huang H, Heibeck T, Costello CE, et al. Isotope-coded affinity tag (ICAT) approach to redox proteomics: identification and quantitation of oxidant-sensitive cysteine thiols in complex protein mixtures. J Proteome Res. 2004;3(6):1228–33. doi: 10.1021/pr049887e. [DOI] [PubMed] [Google Scholar]
- 28.Sui J, Tan TL, Zhang J, Ching CB, Chen WN. iTRAQ-coupled 2D LC-MS/MS analysis on protein profile in vascular smooth muscle cells incubated with S- and R-enantiomers of propranolol: possible role of metabolic enzymes involved in cellular anabolism and antioxidant activity. J Proteome Res. 2007;6(5):1643–51. doi: 10.1021/pr0605926. [DOI] [PubMed] [Google Scholar]
- 29.Sui J, Zhang J, Tabn TL, Ching CB, Chen WN. Comparative proteomics analysis of vascular smooth muscle cells incubated with S- and R-enantiomers of atenolol using iTRAQ-coupled two-dimensional LC-MS/MS. Mol Cell Proteomics. 2008;7(6):1007–18. doi: 10.1074/mcp.M700485-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Guo Y, Singleton PA, Rowshan A, Gucek M, Cole RN, et al. Quantitative proteomics analysis of human endothelial cell membrane rafts: evidence of MARCKS and MRP regulation in the sphingosine 1-phosphate-induced barrier enhancement. Mol Cell Proteomics. 2007;6(4):689–96. doi: 10.1074/mcp.M600398-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu CC, MacCoss MJ, Howell KE, Matthews DE, Yates JR., 3rd Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal Chem. 2004;76:4951–4959. doi: 10.1021/ac049208j. [DOI] [PubMed] [Google Scholar]
- 32.McClatchy DB, Dong MQ, Wu C, Venable JD, Yates JR., III 15N metabolic Labelling of mammalian tissue with slow protein turnover. J Proteome Res. 2007;6(5):2005–2010. doi: 10.1021/pr060599n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat Protoc. 2006;1(6):2650–60. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 34.Kruger M, Moser M, Ussar S, Thievessen I, Lubar CA, et al. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134(2):353–64. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 35.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–67. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 36.Mayr M, Madhu B, Xu Q. Proteomics and metabolomics combined in cardiovascular research. Trends Cardiovasc Med. 2007;17(2):43–8. doi: 10.1016/j.tcm.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 37.McFarlane I, Bomford A, Sherwood R. Liver disease and laboratory medicine. London: ACB Venture Publications; 2000. [Google Scholar]
- 38.Sacher RA, McPherson RA. Widmann’s clinical interpretation of laboratory tests. Philadelphia: F. A. Davis Co; 2000. [Google Scholar]
- 39.Alvarez-Llamas G, de la Cuesta F, Barderas MG, Darde V, Padial LR, et al. Recent advances in atherosclerosis-based proteomics: new biomarkers and a future perspective. Expert Rev Proteomics. 2008;5(5):679–91. doi: 10.1586/14789450.5.5.679. [DOI] [PubMed] [Google Scholar]
- 40.Pula G, Perera S, Prokopi M, Sidibe A, Boulanger CM, et al. Proteomic analysis of secretory proteins and vesicles in vascular research. Proteomics Clinical Applications. 2008;2:882–891. doi: 10.1002/prca.200800040. [DOI] [PubMed] [Google Scholar]
- 41.Lu Q, Murugesan N, Macdonald JA, Wu SL, Pachter JS, et al. Analysis of mouse brain microvascular endothelium using immuno-laser capture microdissection coupled to a hybrid linear ion trap with Fourier transform-mass spectrometry proteomics platform. Electrophoresis. 2008;29(12):2689–95. doi: 10.1002/elps.200700936. [DOI] [PubMed] [Google Scholar]
- 42.Wu J, Liu W, Sousa E, Qiu Y, Pittman DD, et al. Proteomic identification of endothelial proteins isolated in situ from atherosclerotic aorta via systemic perfusion. J Proteome Res. 2007;6(12):4728–36. doi: 10.1021/pr070537c. [DOI] [PubMed] [Google Scholar]
- 43.Duran MC, Martin-Ventura JL, Mas S, Barderas MG, Darde VM, et al. Characterization of the human atheroma plaque secretome by proteomic analysis. Methods Mol Biol. 2007;357:141–50. doi: 10.1385/1-59745-214-9:141. [DOI] [PubMed] [Google Scholar]
- 44.Duran MC, Mas S, Martin-Ventura JL, Meilhac O, Michel JB, et al. Proteomic analysis of human vessels: application to atherosclerotic plaques. Proteomics. 2003;3(6):973–8. doi: 10.1002/pmic.200300389. [DOI] [PubMed] [Google Scholar]
- 45.Pellitteri-Hahn MC, Warren MC, Didier DN, Winkler EL, Mirza SP, et al. Improved mass spectrometric proteomic profiling of the secretome of rat vascular endothelial cells. J Proteome Res. 2006;5(10):2861–4. doi: 10.1021/pr060287k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peterson DB, Sander T, Kaul S, Wakim BT, Halligan B, et al. Comparative proteomic analysis of PAI-1 and TNF-alpha-derived endothelial microparticles. Proteomics. 2008;8(12):2430–46. doi: 10.1002/pmic.200701029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dupont A, Corseaux D, Dekeyzer O, Drobecq H, Guihot AL, et al. The proteome and secretome of human arterial smooth muscle cells. Proteomics. 2005;5(2):585–596. doi: 10.1002/pmic.200400965. [DOI] [PubMed] [Google Scholar]
- 48.Gao BB, Stuart L, Feener EP. Label-free quantitative analysis of 1D-PAGE LC/MS/MS proteome: Application on angiotensin II stimulated smooth muscle cells secretome. Mol Cell Proteomics. 2008;7(12):2399–409. doi: 10.1074/mcp.M800104-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zwickl H, Traxler E, Staettmer S, Parzefall W, Grasl-Kraupp B, et al. A novel technique to specifically analyze the secretome of cells and tissues. Electrophoresis. 2005;26(14):2779–85. doi: 10.1002/elps.200410387. [DOI] [PubMed] [Google Scholar]
- 50.Gronborg M, Kristiansen TZ, Iwahori A, Chang R, Reddy R, et al. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteomics. 2006;5(1):157–71. doi: 10.1074/mcp.M500178-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.An E, Lu X, Flippin J, Devaney JM, Halligan B, et al. Secreted proteome profiling in human RPE cell cultures derived from donors with age related macular degeneration and age matched healthy donors. J Proteome Res. 2006;5(10):2599–610. doi: 10.1021/pr060121j. [DOI] [PubMed] [Google Scholar]
- 52.Alvarez-Llamas G, Szalowska E, de Vries MP, Weening D, Landman K, et al. Characterization of the human visceral adipose tissue secretome. Mol Cell Proteomics. 2007;6(4):589–600. doi: 10.1074/mcp.M600265-MCP200. [DOI] [PubMed] [Google Scholar]
- 53.Martens L, Hermjakob H, Jones P, Adamski M, Taylor C, et al. PRIDE: the proteomics identifactions database. Proteomics. 2005;5:3537–3545. doi: 10.1002/pmic.200401303. [DOI] [PubMed] [Google Scholar]
- 54.Jones P, Cote RG, Martens L, Quinn AF, Taylor CF, et al. PRIDE: a public repository of protein and peptide identifications for the proteomics community. Nucleic Acids Res. 2006;34(Database issue):D659–D663. doi: 10.1093/nar/gkj138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor CF, Hermjakob H, Jr, Julian RK, Garavelli JS, Aebersold R, et al. The work of the human proteome organisation’s proteomics standards initiative (HUPO PSI) Omics. 2006;10(2):145–151. doi: 10.1089/omi.2006.10.145. [DOI] [PubMed] [Google Scholar]
- 56.Hogan JM, Higdon R, Kolker E. Experimental standards for high-throughput proteomics. Omics. 2006;10(2):152–157. doi: 10.1089/omi.2006.10.152. [DOI] [PubMed] [Google Scholar]
- 57.Lenth RV. Some practical guidelines for effective sample size determination. The American Statistician. 2001;55(3):187–193. [Google Scholar]
- 58.Biron DG, Brun C, Lefevre T, Lebarbenchon C, Loxdale HD, et al. The pitfalls of proteomics experiments without the correct use of bioinformatics tools. Proteomics. 2006;6:5577–5596. doi: 10.1002/pmic.200600223. [DOI] [PubMed] [Google Scholar]
- 59.Dreisbach A, Otto A, Becher D, Hammer E, Teumer A, et al. Monitoring of changes in the membrane proteome during stationary phase adaptation of Bacillus subtilis using in vivo labeling techniques. Proteomics. 2008;8(10):2062–76. doi: 10.1002/pmic.200701081. [DOI] [PubMed] [Google Scholar]
- 60.Graumann J, Hubner NC, Kim JB, Ko K, Moser M, et al. Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol Cell Proteomics. 2008;7(4):672–83. [Google Scholar]
- 61.Callesen AK, Vach W, Jorgensen PE, Cold S, Mogensen O, et al. Reproducibility of mass spectrometry based protein profiles for diagnosis of breast cancer across clinical studies: A systematic review. J Proteome Res. 2008;7(4):1395–1402. doi: 10.1021/pr800115f. [DOI] [PubMed] [Google Scholar]
- 62.von Zur Muhlen C, Schiffer E, Zuerbig P, Kellmann M, Brasse M, et al. Evaluation of urine proteome pattern analysis for its potential to reflect coronary artery atherosclerosis in symptomatic patients. J Proteome Res. 2009;8(1):335–45. doi: 10.1021/pr800615t. [DOI] [PubMed] [Google Scholar]
- 63.Krycer JR, Pang CN, Wilkins MR. High throughput protein-protein interaction data: clues for the architecture of protein complexes. Proteome Science. 2008;26(6):32. doi: 10.1186/1477-5956-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang KY, Filarsky M, Padula MP, Raftery MJ, Herbert BR, et al. Micropreparative fractionation of the complexome by blue native continuous elution electrophoresis. Proteomics. 2009;9(9):2494–502. doi: 10.1002/pmic.200800525. [DOI] [PubMed] [Google Scholar]
- 65.Perlman DH, Bauer SM, Houman A, Bryan NS, Garcia-Saura MF, et al. Mechanistic insights into nitrite-induced cardioprotection using and integrated metabonomic/proteomic approach. Circ Res. 2009;104(6):796–804. doi: 10.1161/CIRCRESAHA.108.187005. [DOI] [PMC free article] [PubMed] [Google Scholar]