Abstract
The proteasome regulatory particle (RP) initiates the degradation of ubiquitin-protein conjugates by unfolding the substrate and translocating it into the proteasome core particle (CP) to be degraded1. Here we show that in yeast three proteins are found associated with RP but not RP-CP holoenzyme: Nas6, Rpn14, and Hsm3. Mutations in these genes confer proteasome loss of function phenotypes, despite their virtual absence from holoenzyme. These effects result from deficient RP assembly. Thus, Nas6, Rpn14, and Hsm3 are RP chaperones. The RP contains six ATPases–the Rpt proteins–and each RP chaperone binds to the C-terminal domain of a specific Rpt. We show in an accompanying study2 that RP assembly is templated through the Rpt C-termini, apparently by their insertion into binding pockets in the CP. Thus, RP chaperones may regulate proteasome assembly by directly restricting the accessibility of Rpt C-termini to the CP. In addition, competition between the CP and RP chaperones for Rpt engagement may explain the release of RP chaperones as proteasomes mature.
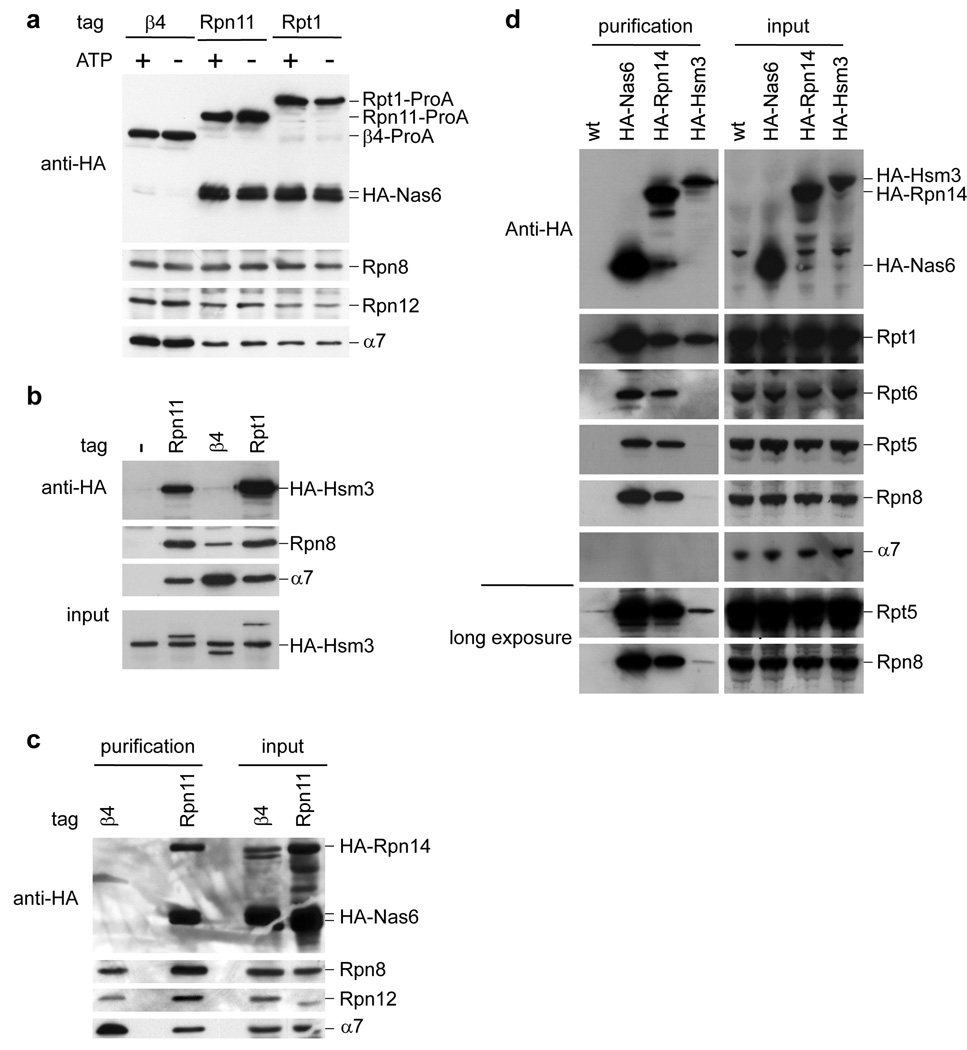
Affinity purification has allowed identification of proteins that associate with proteasomes, but are not true subunits1,3–5. Nas6, one such protein4, is the apparent homolog of gankyrin, a liver oncoprotein. Gankyrin is thought to interact with Rb, Mdm2, CDK4, and the proteasome6. Using recombinant GST-Nas6 we purified all 19 RP subunits from yeast lysates (Supplementary Table 1). However, to our surprise no CP subunit was recovered. Similarly, affinity purified samples from RP-tagged strains contained Nas6 while those from CP-tagged strains did not (Fig. 1a). Thus, Nas6 may bind a subpopulation of RP that is not associated with CP (hereafter ‘free RP’). The specific association of Nas6 with the RP was validated through additional experiments (Supplementary Fig 1).
Figure 1. Nas6, Hsm3 and Rpn14 bind to free RP.
a, Proteasomes were affinity-purified from strains expressing HA-tagged Nas6. Samples were resolved by SDS-PAGE and immunoblotted for Nas6 (anti-HA), RP (anti-Rpn8 and anti-Rpn12) and CP (anti-α7).
b, Proteasomes were affinity-purified from strains expressing HA-tagged Hsm3, eluted, and analyzed for the presence of Hsm3, RP, and CP.
c, Proteasomes were affinity-purified from strains expressing HA-tagged Nas6 and Rpn14. After elution, samples were analyzed as in a.
d, HA-specific immunoprecipitations were tested for RP and CP by immunoblotting.
Mass spectrometric analysis of immunoprecipitates of HA-tagged Nas6 revealed two other proteins that bind specifically to free RP, Hsm3 and Rpn14. Hsm3 was recently identified as a proteasome interacting protein7. Hsm3 co-purified with the RP but not CP (Fig. 1b), confirming that it is a second RP-specific component. Although we cannot exclude the possibility that a fraction of these proteins shows bona fide proteasome holoenzyme association, their previous assignment as proteasome subunits may reflect that proteasome subcomplexes are often found in holoenzyme preparations. The mammalian homolog of Rpn14, PAAF1, was recently suggested to bind free RP8. Likewise, Rpn14 associated specifically with free RP (Fig. 1c). Hsm3 differed from Rpn14 and Nas6 in associating most strongly with only a subset of RP subunits, potentially representing an RP assembly intermediate (Fig. 1d).
The identification of multiple free RP binding proteins suggests a common function distinct from those of known proteasome-associated proteins. Free RP might be actively sequestered from CP by an inhibitor of the proteasome, a function proposed for PAAF1 (refs. 8,9). Alternatively, free RP might provide a non-proteolytic pathway of RP function, suggested for transcriptional control and for PAAF1 in particular9,10. Thirdly, free RP-binding proteins might act as RP assembly factors. The mechanisms involved in RP assembly11–13 are largely unknown. In contrast, CP assembly has been studied intensively and involves specific chaperones14.
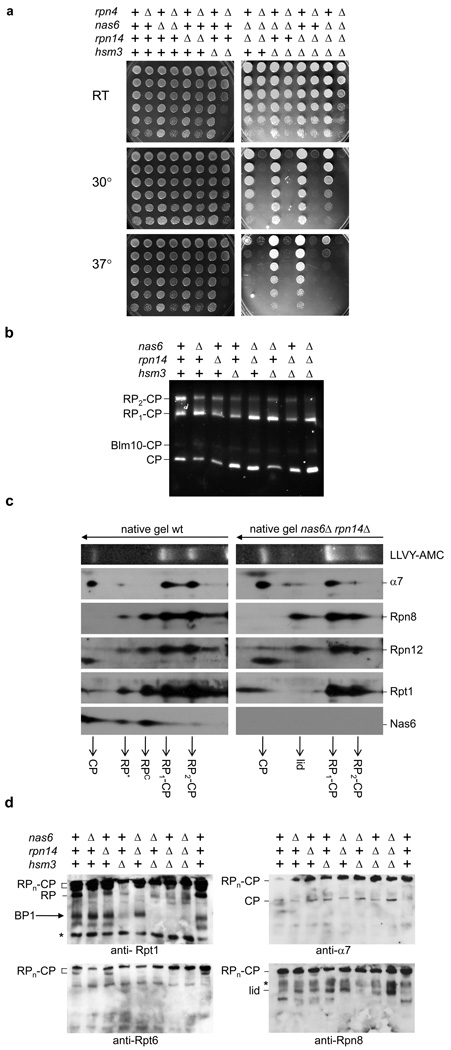
An initial test for a role of RP-binding proteins in the major proposed non-proteolytic function of the RP, transcriptional elongation, proved negative (Supplementary Fig. 2). We therefore tested whether RP-binding proteins play a more general role in proteasome function. Proteasome inhibitor mutants should behave as proteasome hypermorphs, whereas assembly factor mutants should be hypomorphic. Double mutants of the RP-binding factors showed sensitivity to elevated temperature, as typical of proteasome hypomorphs (Fig. 2a). Additional phenotypes are described in Supplementary Fig. 3. To verify that nas6Δ, hsm3Δ, and rpn14Δ mutants are hypomorphic for proteasome function, we deleted the gene for Rpn4, which mediates homeostatic proteasome regulation15. Mutant phenotypes were exacerbated, suggesting that the severity of nas6Δ, hsm3Δ, and rpn14Δ phenotypes is masked through Rpn4-based compensation (Fig. 2a). Accordingly, accumulation of ubiquitin-conjugates was observed in total cell lysates from many of the mutants in the rpn4Δ background (Supplementary Fig. 4).
Figure 2. Phenotypic analysis of nas6Δ, hsm3Δ, and rpn14Δ mutants.
a, Strains with indicated genes deleted were spotted on plates in four-fold dilutions and grown at the indicated temperature.
b, Cell lysates (75µg) were resolved on native gels. Gels were stained for LLVY-AMC hydrolytic activity.
c, Cell lysates of wild-type or nas6Δ rpn14Δ strains were resolved by two-dimensional native-SDS/PAGE gel electrophoresis followed by immunoblotting.
d, Cell lysates were resolved on 5.25% native gels and immunoblotted. BP1 assignment based on ref. 2. *, background band.
The results above indicate that Nas6, Hsm3, and Rpn14 are positive regulators of proteasome function in yeast, despite showing little or no association with proteasome holoenzyme. The function of one RP-binding protein to is not strictly dependent on other proteins in this group, in agreement with the results of Supplementary Figure 5, which show that they can bind RP or RP subassemblies independently of one another.
To analyze proteasomes in the mutants, cell lysates were resolved by native PAGE and active proteasome species visualized. The mutants showed reduced levels of doubly-capped proteasomes, while CP levels were increased (Fig. 2b). Thus, RP-binding proteins are important for maintaining normal proteasome levels, suggesting a role in assembly.
The activity assay only shows CP-containing proteasome species. To follow RP species, we ran lysates on native gels, followed by second dimension SDS-PAGE and immunoblotting. In wild-type lysates we detected RPc, which behaves as a true RP species16, and RP*, which appears to contain a subset of RP subunits (Fig. 2c). Importantly, Nas6 co-migrates with these two species and is virtually absent from holoenzyme. In nas6Δ rpn14Δ mutants, both species are undetectable (Fig. 2c), consistent with defective RP assembly. Instead of RP, we observed a species that migrated in the position of the 9-subunit lid subassembly of the RP16, and tested positive only for lid subunits. The lid mediates substrate deubiquitination1. Size exclusion chromatography of lysates from nas6Δ rpn14Δ hsm3Δ mutants showed results consistent with Fig. 2c (Supplementary Fig. 6). In addition to the lid, the RP contains a 10-subunit assembly known as the base. Free lid complex could indicate a base assembly defect2,12,13, assuming that assembled base consumes free lid as the final step in RP assembly. In summary, the data above suggest that Nas6, Rpn14, and Hsm3 function collectively as RP-chaperones.
To identify base precursors we used native gels with higher acrylamide content and observed a specific species whose presence strongly depended on Hsm3 but was independent of Nas6 and Rpn14 (Fig. 2d). This species, which we termed base precursor 1 (BP1), consists of Rpn1, Rpt1, Rpt2, Hsm3, and Rpt5 (see ref. 2).
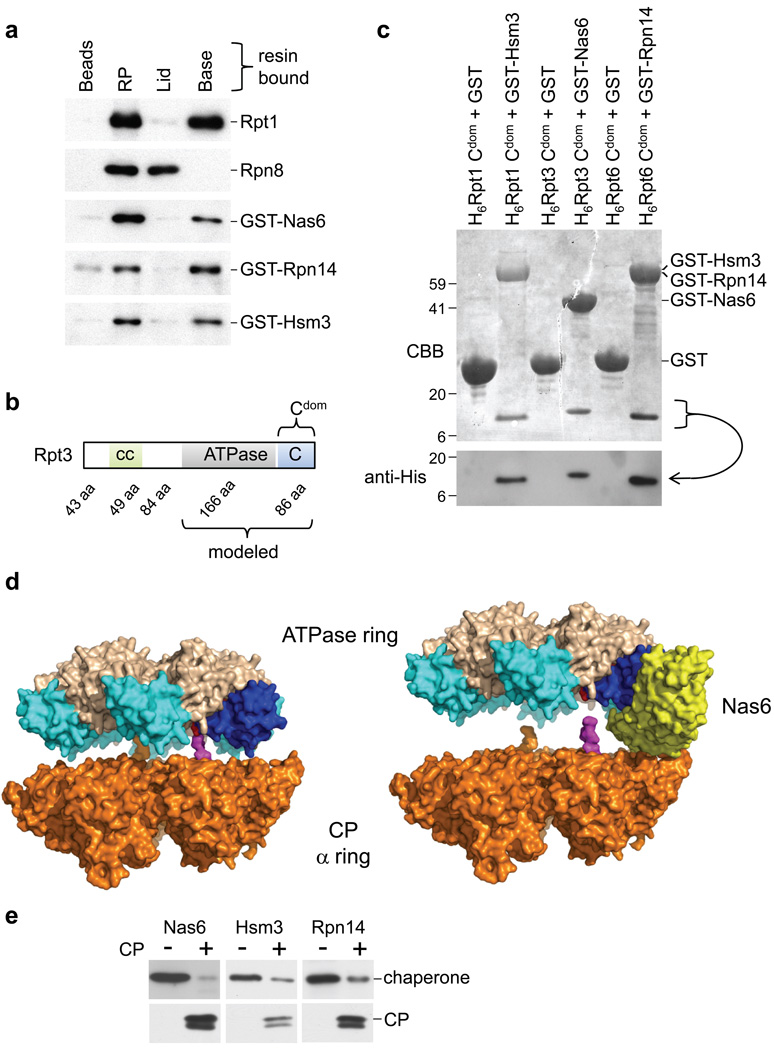
A role for RP-chaperones in base formation was corroborated by the finding that all three RP-chaperones bound specifically to the base (Fig. 3a). Six ATPases of the base, the Rpt proteins, are thought to contact the CP through their C-termini17–19 and to form a heterohexameric ring. Because Nas6 binds the C-terminal domain (Fig. 3b; C-domain20) of Rpt321, we tested the binding of the RP-chaperones to C-domains of all Rpt subunits by pairwise co-expression in E. coli. Though the six Rpt C-domains are closely related21,22, we observed strong discrimination among Rpt proteins by Nas6, Hsm3, and Rpn14: Nas6 bound to Rpt3 uniquely, Hsm3 bound Rpt1, while Rpn14 bound Rpt6 and, less robustly, Rpt4 (Fig. 3c and Supplementary Fig. 7). These results indicate a related mechanism of action of the three chaperones in base assembly.
Figure 3. RP chaperones bind to the C-domains of Rpt proteins in proximity to the CP.
a, RP, lid, base bound to IgG beads or untreated IgG beads were incubated with GST-Nas6, GST-Rpn14, or GST-Hsm3 purified from E. coli. Beads were analyzed for bound proteins.
b, Domain composition of Rpt3; other Rpt proteins have similar architecture. “cc” predicted coiled-coil region; “ATPase” ATPase domain; “C” C-domain20; “aa” amino acids.
c, His-tagged C-domains were co-expressed with GST or GST-tagged Nas6, Rpn14, or Hsm3 in E. coli. Glutathione-Sepharose purified samples were immunoblotted using His-tag antibody followed by Coomassie Blue staining.
d, Modeling of the interaction of Rpt ring with CP in the presence (right) and absence (left) of Nas6: Nas6 binding to the ATPase ring appears to block the C-terminal tail of Rpt3 from docking into the CP. The CP α-ring (beige)24, was combined with the PAN hexameric ATPase23 (ATPase domain grey; C-domains blue). The Nas6-Rpt3 structure21 was mapped onto the ATPase ring by aligning the Rpt3 structure with the PAN C-domain (dark blue; C-terminus red), with R.M.S. 1.1 Å over 283 atoms. The last twelve residues of the PA26 C-terminus17 (magenta) are docked into CP17–19. The length corresponds to that of Rpt3 C-terminal tail that is absent from the Nas6-Rpt3 structure.
e, Affinity-purified, resin-bound base was incubated with indicated RP-chaperone, washed, then loaded with purified CP. Resin-bound proteins were eluted and analyzed by immunoblotting.
The direct interaction of all three RP-chaperones with distinct Rpt proteins is consistent with their ability to function in each other’s absence (Fig. 2a). These results also explain why Hsm3 but not Nas6 or Rpn14 associates with BP1; their binding partners are absent from BP12. In support of the functional significance of the Hsm3-Rpt1 interaction and of BP1, we found that overexpressed Rpt1 rescued phenotypes of rpn4Δ hsm3Δ, nas6Δ hsm3Δ, and rpn14Δ hsm3Δ mutants but not a nas6Δ rpn14Δ mutant (Supplementary Fig. 8). Thus, Rpt1 overexpression relieves the deleterious effects of the absence of Hsm3. Taken together, the data suggest that Hsm3 chaperones Rpt1 or an Rpt1-containing complex.
The Rpt C-termini mediate RP interaction with the CP17–19, and are moreover critical for base assembly2, suggesting that assembly is templated on the CP. In contrast to the RP-CP interaction, binding of RP-chaperones to C-domains of their cognate ATPases does not involve the extreme C-terminal residues (Supplementary Fig. 7). We propose that, despite non-overlapping binding sites, RP-chaperones compete with CP for binding to Rpt proteins, thus controlling the exposure of Rpt C-termini to the CP.
To study the effect of chaperone binding to the Rpt on RP-CP interactions, we modeled the Rpt3-Nas6 structure21,22 into an ATPase ring based on recent structural studies of PAN, a closely related ATPase from the archaeon M. jannaschii23. The ATPase ring was lowered onto the known structure24 of the CP α ring. The modeling predicts that Nas6 physically occludes the formation of proper CP-RP contacts (Fig. 3d), because much of Nas6 projects from the RP towards the CP, so as to prevent close RP-CP apposition.
A prediction of physical occlusion model is that the CP should be able to expel RP-chaperones from the RP. To test this, a base sample loaded with chaperones was immobilized on resin. Application of purified CP to the resin resulted in efficient elution of the RP-chaperones (Fig. 3e). This experiment may mimic key steps in proteasome assembly. Moreover, the direct competition between CP and RP-chaperones shown here appears to explain the absence of RP-chaperones from mature proteasomes. Additional tests of the model are described in the accompanying study2.
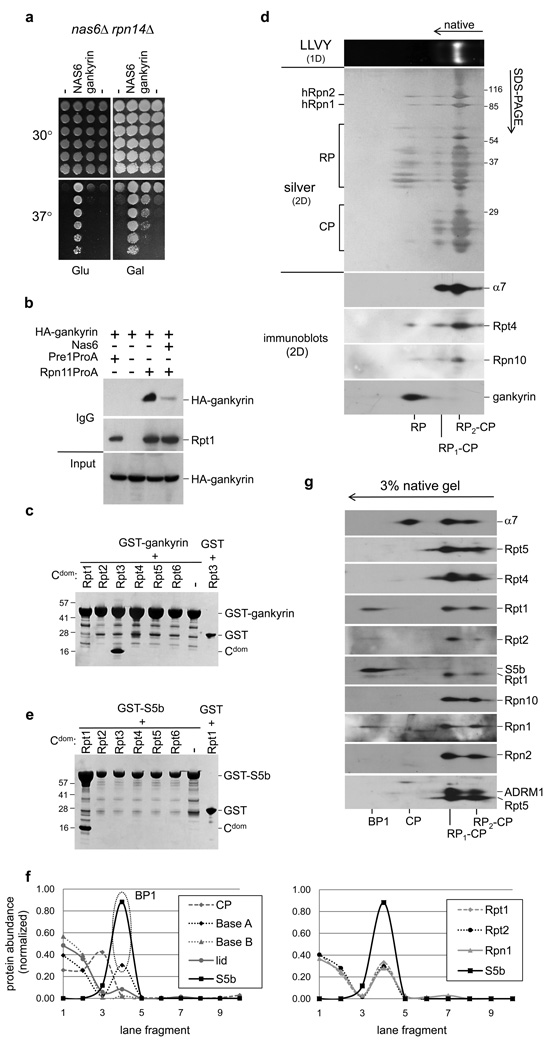
The yeast protein Nas6 and the human oncoprotein gankyrin are closely related by structure, and bind orthologous Rpt proteins21,22. In contrast to Nas6, however, gankyrin is thought to be a major proteasome component6. To test for functional conservation between Nas6 and gankyrin, we expressed gankyrin in a nas6Δ rpn14Δ strain. Gankyrin rescued this strain’s temperature sensitivity, showing strong evolutionary conservation (Fig. 4a). Gankyrin expressed in yeast associated specifically with free RP (Fig. 4b). Binding of gankyrin to RP was reduced when Nas6 was overexpressed, indicating that gankyrin and Nas6 bind the same RP surface in yeast (Fig. 4b). Consistently, gankyrin bound the C-domain of human Rpt3 but not those of other Rpt proteins22 (Fig. 4c). Moreover, preparations of human proteasomes also showed specific association of endogenous Gankyrin with free RP and not holoenzyme (Fig. 4d). Considering the low levels of free RP in the preparation, the specificity of Gankyrin for free RP is appreciable. Taken together, these data indicate that the functions of gankyrin and Nas6 have indeed been conserved in evolution.
Figure 4. Functional conservation between Nas6 and gankyrin.
a, nas6Δ rpn14Δ strains expressing NAS6 or gankyrin from a GAL promoter were spotted onto YPD plates or YEP Raf/Gal plates in four-fold dilutions, and incubated at indicated temperatures.
b, Proteasomes were purified from yeast strains expressing HA-tagged gankyrin and analyzed by immunoblotting.
c, His-tagged C-domains of indicated human Rpt proteins were co-expressed in E. coli with GST-tagged gankyrin. Glutathione-Sepharose purified samples were separated by SDS-PAGE, and stained with Coomassie Blue. Band assignments were confirmed by immunoblotting (data not shown).
d, Human proteasomes purified via HTBH-tagged Rpn11 (Ref. 5) were were resolved by two-dimensional native-SDS/PAGE gel electrophoresis. Proteins were silver stained or analyzed by immunoblotting.
e, as c, only GST-gankyrin was replaced by GST-S5b.
f, HeLa cell lysate was resolved on Blue native gels. Fractions were analyzed by mass spectrometry, and protein abundance estimated using spectral counting. Based on their abundance profiles, most proteasome subunits can be divided into four groups (left graph); CP (α1–7 and β1–7), lid (Rpn3,5,6,7,8,9,10,11), baseB (Rpt3,4,5,6 and Rpn2) and BaseA (Rpt1,2, and Rpn1). Interestingly, S5b is absent from proteasomes (fraction 1 and 2), but abundant in fraction 4. Right panel, S5b and individual profiles of BaseA members.
g, HeLa cell lysates from e were separated by 2D native-SDS-PAGE and analyzed by immunoblotting.
Although BLAST searches did not reveal a human ortholog of Hsm3, the only remaining mammalian proteasome subunit not found in S. cerevisiae, S5b, contains ARM repeats, like Hsm3. S5b can apparently bind to an Rpn1-Rpt1-Rpt2 complex25, further suggesting that these two proteins are orthologous. When tested against a panel of human Rpt proteins, S5b bound specifically to Rpt1, as predicted from the specificity of Hsm3 for yeast Rpt1 (Fig. 4e). To test for BP1 in human cells, we analyzed HeLa cells lysates on native gels, and subjected gel slices to mass spectrometry. The results showed that S5b comigrated with hRpn1, hRpt1, and hRpt2 in a complex that was clearly resolved from proteasomes and RP (Fig. 4f). The existence of BP1 in human cells was confirmed through immunoblotting (Fig. 4g).
Here we identify three factors that assist in assembly of the proteasome base through a common mechanism of action: they associate with C-domains of Rpt proteins, thus negatively regulating interaction of the C-domain with specific pockets within the CP α ring. Modeling indicates that RP-chaperones prevent CP interaction by steric occlusion, and as a result compete with the CP for occupancy of C-domains. This model was tested in three ways: free CP releases RP-chaperones from the base complex (Fig. 3e); single amino acid deletions of Rpt C-termini fail to expel their cognate RP-chaperones2, indicating that proper docking of the C-terminus is needed to expel the chaperone; and insertions of as little as one residue proximal to Rpt C-termini prevent chaperone expulsion, even when the Rpt C-terminus is productively docked in the CP pocket, indicating that RP-chaperone expulsion is precise and requires a close approach of the CP to the RP2. Despite the explanatory potential of this model, we do not exclude the possibilities that the RP-chaperones may promote assembly through additional mechanisms, and perhaps regulate proteasomes in additional ways.
The ability of the all chaperones to bind Rpt C-termini, compete with CP and promote assembly may reflect convergent evolution, considering that the chaperones are unlikely to have diverged from a common ancestor. For example, Nas6 is composed mainly of ankyrin repeats, whereas Hsm3 is rich in ARM repeats and Rpn14 in WD40 repeats. Key features of base assembly, including the BP1 intermediate, are conserved in mammals. The chaperone functions of yeast proteins Rpn14, Hsm3, and Nas6 are apparently paralleled in mammals by PAAF1, and S5b, and most interestingly the oncoprotein gankyrin. It is likely that Gankyrin and S5b are misassigned as proteasome subunits. Thus, conservation of the assembly pathway unexpectedly implies a more general conservation, that the subunit compositions of yeast and mammalian proteasomes are actually identical. While this paper was under review, Le Tallec et al. reported the participation of Hsm3 and S5b in RP assembly, consistent with our findings26.
Nas6, Rpn14, and Hsm3 fit the definition of a molecular chaperone closely27. They assist in assembly of an oligomeric complex, do not act as templates for the complex, and are absent from the mature complex. Chaperones typically function to prevent undesired and nonspecific interactions of their target proteins, but RP-chaperones represent an interesting case in which the interaction suppressed by the chaperone is a proper interaction in the mature complex, and moreover an interaction that is specifically required for assembly itself. These properties of RP-chaperones point to a role in controlling the timing and ordering of maturation events.
How does competition of RP-chaperones with the CP for occupancy of the Rpt C-domains promote proper base or Rpt ring assembly? In the simplest version of templated assembly, the ring would assemble in contact with the CP in a growing arc, subunit by subunit, until, with the inclusion of its last subunit, the ring is closed. In this “one-template model,” the Rpt-CP interaction, in combination with Rpt-Rpt interactions, would guide the proper choice of subunit and stabilize the nascent, open ring. Such a was proposed for assembly of the CP β ring28. Interestingly, β ring assembly is, like Rpt ring assembly, guided by the α ring, albeit the opposite face of the α ring. In the case of the Rpt ring, the one-template model may not apply, based in part on our identification of an apparent precursor to the base2, BP1. This complex contains Rpn1, Rpt1, Rpt2, Rpt5, and Hsm3. Thus, the ring is likely to be assembled in patchwork fashion, partly from preformed ring segments.
The scaffold for BP1 appears to be a non-Rpt subunit, Rpn12. Rpn2 is related to Rpn1, and may play a complementary scaffolding role for most or all base components that are absent from BP1. In this three-scaffold model, the CP α-ring, Rpn1, and Rpn2 each help to template the ring. Both Rpn1 and Rpn2 are toroidal29,30, a property likely to underlie their ability to nucleate ring assembly. A key feature of the three-template model is that the ordering of Rpt addition is achieved by interactions among multisubunit base precursors. An example of how RP-chaperones could order the assembly pathway is that Hsm3 might help BP1 to assemble with other base precursors late in the assembly pathway by suppressing premature interaction of Rpt1 with the CP.
Methods summary
Yeast strains, plasmids and antibodies
See Supplementary Table 2 for genotypes of strains used. Standard methods were used for strain construction and growth. The reported sequence of Hsm3 in the Saccharomyces Genome Database differs from the sequence in our strain background at the C-terminus (see Supplementary Information). Plasmids and antibodies used are described in Supplementary Table 3 and 4 respectively.
Immunoprecipitations and protein purifications
Recombinant proteins were expressed in and purified from Rosetta 2 cells (Novagen) as detailed in Supplementary Methods. For pulldown experiments, yeast cells were lysed by French press or glass beads. Lysate was cleared and incubated with the specific resin. For details see Supplement.
Native gels
For native gels, cell lysates were made in an ATP-containing buffer, cleared by centrifugation, and loaded on a 3.6% or 5.25% polyacrylamide gel. To transfer proteins from native gels to PVDF, the gels were pre-soaked for 10 min in 1X SDS-PAGE running buffer and transferred according to standard protocols. For 2D-Immunoblotting, a lane was excised and loaded on top of an SDS-PAGE gel. Proteins were transferred from the second dimension gel to PVDF and probed with indicated antibodies. Detailed protocols for all experimental procedures can be found in Supplementary Information.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgements
We thank Carl Mann for Cim5/Rpt1 and Cim3/Rpt6 antibodies, Kazuishi Kubota for preparing HeLA lysates and especially Lan Huang for the hRpn11-HTBH expressing cell line. We also thank John Hanna, Marion Schmidt and members of the Finley lab, Suzanne Elsasser especially, for critically reading the manuscript. This work was supported by the NIH (GM043601 to D.F.), a NIH NRSA postdoctoral fellowship (5F32GM75737-2 to SP) and an EMBO long-term fellowship (J.R.).
Footnotes
Author Information Reprints and permissions information is available at npg.nature.com/reprintsandpermissions.
References
- 1.Finley D. Recognition and Processing of Ubiquitin-Protein Conjugates by the Proteasome. Annu. Rev. Biochem. 2009;78 doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park S, et al. Hexameric Assembly of the Proteasomal ATPases is Templated through their C-termini. Nature. doi: 10.1038/nature08065. (Submitted; accompanying paper). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leggett DS, et al. Multiple associated proteins regulate proteasome structure and function. Mol. Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 4.Verma R, et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, et al. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry (Mosc) 2007;46:3553–3565. doi: 10.1021/bi061994u. [DOI] [PubMed] [Google Scholar]
- 6.Dawson S, Higashitsuji H, Wilkinson AJ, Fujita J, Mayer RJ. Gankyrin: a new oncoprotein and regulator of pRb and p53. Trends Cell Biol. 2006;16:229–233. doi: 10.1016/j.tcb.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Guerrero C, Milenkovic T, Przulj N, Kaiser P, Huang L. Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13333–13338. doi: 10.1073/pnas.0801870105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park Y, et al. Proteasomal ATPase-associated factor 1 negatively regulates proteasome activity by interacting with proteasomal ATPases. Mol. Cell. Biol. 2005;25:3842–3853. doi: 10.1128/MCB.25.9.3842-3853.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lassot I, et al. The proteasome regulates HIV-1 transcription by both proteolytic and nonproteolytic mechanisms. Mol. Cell. 2007;25:369–383. doi: 10.1016/j.molcel.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 10.Collins GA, Tansey WP. The proteasome: a utility tool for transcription? Curr. Opin. Genet. Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Imai J, Maruya M, Yashiroda H, Yahara I, Tanaka K. The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J. 2003;22:3557–3567. doi: 10.1093/emboj/cdg349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isono E, et al. The assembly pathway of the 19S regulatory particle of the yeast 26S proteasome. Mol. Biol. Cell. 2007;18:569–580. doi: 10.1091/mbc.E06-07-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kusmierczyk AR, Kunjappu MJ, Funakoshi M, Hochstrasser M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat. Struct. Mol. Biol. 2008;15:237–244. doi: 10.1038/nsmb.1389. [DOI] [PubMed] [Google Scholar]
- 14.Kusmierczyk AR, Hochstrasser M. Some assembly required: dedicated chaperones in eukaryotic proteasome biogenesis. Biol. Chem. 2008 doi: 10.1515/BC.2008.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie Y, Varshavsky A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3056–3061. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleijnen MF, et al. Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat. Struct. Mol. Biol. 2007;14:1180–1188. doi: 10.1038/nsmb1335. [DOI] [PubMed] [Google Scholar]
- 17.Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol. Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 18.Gillette TG, Kumar B, Thompson D, Slaughter CA, Demartino GN. Differential roles of the C-termini of AAA subunits of PA700 (19S regulator) in asymmetric assembly and activation of the 26s proteasome. J. Biol. Chem. 2008 doi: 10.1074/jbc.M805935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith DM, et al. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol. Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ammelburg M, Frickey T, Lupas AN. Classification of AAA+ proteins. J. Struct. Biol. 2006;156:2–11. doi: 10.1016/j.jsb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura Y, et al. Structural basis for the recognition between the regulatory particles Nas6 and Rpt3 of the yeast 26S proteasome. Biochem. Biophys. Res. Commun. 2007;359:503–509. doi: 10.1016/j.bbrc.2007.05.138. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura Y, et al. Structure of the oncoprotein gankyrin in complex with S6 ATPase of the 26S proteasome. Structure. 2007;15:179–189. doi: 10.1016/j.str.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Zhang F, et al. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol. Cell. doi: 10.1016/j.molcel.2009.04.021. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groll M, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 25.Gorbea C, Taillandier D, Rechsteiner M. Mapping subunit contacts in the regulatory complex of the 26 S proteasome. S2 and S5b form a tetramer with ATPase subunits S4 and S7. J. Biol. Chem. 2000;275:875–882. doi: 10.1074/jbc.275.2.875. [DOI] [PubMed] [Google Scholar]
- 26.Le Tallec B, Barrault MB, Guerois R, Carre T, Peyroche A. Hsm3/S5b participates in the assembly pathway of the 19S regulatory particle of the proteasome. Mol. Cell. 2009;33:389–399. doi: 10.1016/j.molcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Ellis RJ. Molecular chaperones: assisting assembly in addition to folding. Trends Biochem. Sci. 2006;31:395–401. doi: 10.1016/j.tibs.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Hirano Y, et al. Dissecting beta-ring assembly pathway of the mammalian 20S proteasome. EMBO J. 2008;27:2204–2213. doi: 10.1038/emboj.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Effantin G, Rosenzweig R, Glickman M, Steven AC. Electron Microscopic Evidence in Support of α-Solenoid Models of Proteasomal Subunits Rpn1 and Rpn2. J. Mol. Biol. 2009;386:1204–1211. doi: 10.1016/j.jmb.2009.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenzweig R, Osmulski PA, Gaczynska M, Glickman MH. The central unit within the 19S regulatory particle of the proteasome. Nat. Struct. Mol. Biol. 2008;15:573–580. doi: 10.1038/nsmb.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.