Summary
Tail-anchored (TA) proteins are characterized by a C-terminal transmembrane region that mediates posttranslational insertion into the membrane of the endoplasmic reticulum. We have investigated the requirements for membrane insertion of three TA proteins, RAMP4, Sec61β and cytochrome b5. We show here that newly synthesized RAMP4 and Sec61β can accumulate in a cytosolic, soluble complex with the ATPase Asna-1/TRC40 before insertion into ER-derived membranes. Membrane insertion of these TA proteins is stimulated by ATP, sensitive to redox conditions and blocked by alkylation of SH groups by N-ethylmaleimide (NEM). In contrast, membrane insertion of cytochrome b5 is not found to be mediated by Asna-1, not stimulated by ATP and not affected by NEM or an oxidative environment. Asna-1 mediated pathway of membrane insertion of RAMP4 and Sec61β may relate to functions of these proteins in the ER stress response.
Keywords: tail-anchored protein, RAMP4, Sec61β, Cytochrome b5, endoplasmic reticulum, Asna-1/TRC40
Introduction
Insertion of proteins into the membrane of the endoplasmic reticulum (ER) can proceed by different types of mechanisms (Rapoport et al., 1996). The best characterised of these is the signal recognition particle (SRP)- dependent pathway in which the nascent chain ribosome complex is targeted to the ER membrane by SRP and insertion into the Sec61 translocation channel is coupled with further chain elongation (Keenan et al., 2001) Proteins inserted by this pathway usually contain an N-terminal signal or signal anchor sequence. In contrast, tail-anchored (TA) proteins represent a class of membrane proteins that lack an N-terminal signal sequence. They are anchored to the membrane by a single stretch of hydrophobic amino acid residues close to the COOH terminus, exposing their larger NH2-terminal, and usually functional portion, to the cytoplasm. Among TA proteins are components of the ER translocation site, Sec61β, Sec61γ and the ribosome associated membrane protein 4 (RAMP-4) also called stress-associated endoplasmic reticulum protein 1 (SERP1) as well as others including synaptobrevin/vesicle-associated membrane protein (VAMP), cytochrome b5 (b5) or Bcl-2 family members (Borgese et al., 2003b; Yamaguchi et al., 1999). The latter ones are not exclusively located at the ER but are also found in the mitochondrial outer membrane (Annis et al., 2004). TA proteins studied so far can insert into membranes post-translationally but requirements for targeting to and insertion into the ER membrane are not well understood and it seems that different pathways of membrane insertion exist (Annis et al., 2004; Borgese et al., 2003a; Borgese et al., 2003b; Janiak et al., 1994; Wattenberg and Lithgow, 2001). The integration of b5 can occur post-translationally and requires low concentrations of ATP (Anderson et al., 1983; Borgese et al., 2003b; Yabal et al., 2003). Membrane insertion of synaptobrevin 2 was found to be dependent on ATP consistent with a role for cytosolic chaperones in maintaining the precursor in an insertion-competent state (Kim et al., 1997; Kutay et al., 1995). On the other hand some TA proteins, namely synaptobrevin 2 and Sec61β were found to associate post-translationally with SRP and require GTP for their membrane targeting/insertion (Abell et al., 2004). Although proteins are implicated in the insertion of TA proteins there are also indications that some TA proteins insert into membranes unassisted (Borgese et al., 2003a; Brambillasca et al., 2006; Brambillasca et al., 2005). Recently Stefanovic and Hegde reported the identification of Asna-1 as a component of a post-translational pathway of TA membrane protein insertion (Stefanovic and Hegde, 2007). Asna-1 was found to interact with the transmembrane domain (TMD) of Sec61β, VAMP2 and a syntaxin prior to their membrane insertion. It was suggested that Asna-1 together with other proteins of the TMD recognition complex (TRC) targets TA proteins for insertion into the ER membrane (Stefanovic and Hegde, 2007). A different conclusion was reached in another study in which Sec61β was reported to interact with the molecular chaperones Hsc70 and Hsp40 and that these components are capable to facilitate the ATP dependent delivery of Sec61β to the ER (Abell et al., 2007).
We have investigated the requirements for membrane insertion of three TA proteins: RAMP4 (Ribosome Associated Membrane Protein 4), also called SERP-1 (Stress-Associated Endoplasmic Reticulum Protein 1), a protein of the ER translocation site that assists membrane insertion of proteins and is involved in the ER quality control and stress response (Hori et al., 2006; Schröder et al., 1999; Yamaguchi et al., 1999), Sec61β, a subunit of the Sec61 translocation complex (Abell et al., 2007) and cytochrome b5 (b5) (Borgese et al., 2003a; Brambillasca et al., 2006; Brambillasca et al., 2005; Yabal et al., 2003). Based on an interaction with Asna-1, ATP requirement and redox or NEM sensitivity membrane insertion of RAMP4 and Sec61β can clearly be distinguished from the NEM insensitive membrane targeting/insertion of b5.
Results
Post-translational insertion of RAMP4op (R4op)
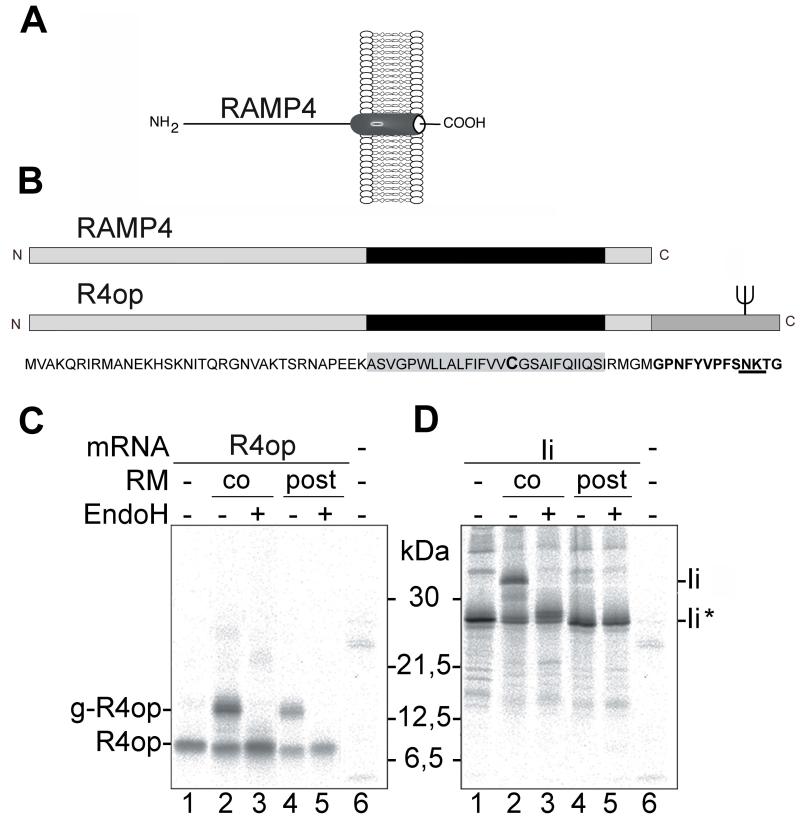
RAMP4 is a small tail-anchored (TA) membrane protein that exposes its N-terminus on the cytoplasmic and its C-terminus on the lumenal side of the ER membrane (Fig. 1A). To study membrane insertion of RAMP4 we have added an opsin tag to its C-terminus and thereby generated RAMP4op (R4op) (Fig. 1B). The opsin tag is recognized by the monoclonal anti-opsin antibody R2-15 (α-op) and contains a N-glycosylation site. As N-glycosylation is a lumenal event this modification indicates that R4op exposes its C-terminus on the lumenal side like authentic RAMP4.
Fig. 1.
Posttranslational membrane insertion of R4op
(A) Topology of RAMP4 in the ER membrane. RAMP4 is a tail-anchored ER membrane protein that exposes its N-terminus on the cytosolic and the C-terminus on the lumenal side of the membrane. (B) Schematic representation of RAMP4 and RAMP4op (R4op). R4op contains at its C-terminus a bovine opsin tag comprising 13 amino acid residues (represented by a dark grey box). The tag provides an N-glycosylation site (fork). The predicted transmembrane domain (TMD) is represented as a black box. A single cysteine residue in the TMD is typed in bold. (C and D) In vitro translation and membrane insertion of R4op and the type II membrane protein Invariant chain (Ii) respectively. Proteins were synthesized in the rabbit reticulocyte lysate (RRL), in the absence (lanes 1, 4, 5 and 6) or presence of rough microsomes (RM co) (lanes 2 and 3). To samples shown in lanes 4 and 5, RM were added after completion of translation (RM post). Where indicated, samples were treated with EndoH to remove N-linked oligosaccharides. Proteins were immunoprecipitated using either anti-opsin (C) or anti-Ii (D) antibodies, separated by SDS-PAGE and visualized by autoradiography. g-R4op: glycosylated R4op; Ii*: non-glycosylated Ii;
To investigate membrane targeting and insertion of R4op we synthesized this protein in a rabbit reticulocyte lysate (RRL) translation system. In order to see whether R4op can be inserted post-translationally, we added rough microsomal membranes (RM) from canine pancreas either at the start of translation or after termination of protein synthesis following the addition of puromycin. As a control for co-translational membrane insertion we used invariant chain (Ii), a doubly glycosylated type II membrane protein. When Ii was synthesized in the RRL in the absence of RM, a protein of about 26 kDa could be detected consistent with the molecular weight of unglycosylated Ii (Fig. 1D, lane 1). This protein was not present in the control reaction lacking mRNA (Fig. 1D, lane 6). When the synthesis of Ii was carried out in the presence of RM an additional, 33 kDa protein was seen (Fig. 1D, lane 2). Removal of N-linked carbohydrates with endogylcosidase H (EndoH), led to the disappearance of the 33 kDa protein and increase in the amount of non-glycosylated Ii (Fig. 1D, lane 3). This indicates that Ii is inserted into RM membranes and becomes glycosylated. When RMs were added after completion of Ii synthesis, no glycosylated Ii was detected (Fig. 1D, lanes 4 and 5), confirming that Ii can only co-translationally be inserted into RM.
When mRNA encoding R4op was translated in the RRL, a protein of about 8 kDa was immunoprecipitated using anti-opsin antibody (Fig. 1C, lane 1). The estimated molecular weight suggests that this is non-glycosylated R4op. When the translation of R4op mRNA was conducted in the presence of RM, a higher molecular weight protein appeared (Fig. 1C, lane 2). Treatment with EndoH reduced the size of this larger protein and lead to an increase in the amount of non-glycosylated R4op (Fig. 1C, lanes 3). When RM were added post-translationally, glycosylated R4op was also seen (Fig. 1C, lanes 4 and 5). Taken together, these results show that R4op synthesized in vitro in the RRL can efficiently be inserted into RM co-as well as post-translationally.
Characterisation of the cytosolic R4op complex
The ability of newly synthesized R4op to become post-translationally inserted into RM suggests that R4op, despite being a membrane protein, can be maintained in an insertion-competent state when exposed to the hydrophillic cytosolic environment. To see how long cytosolic R4op can be maintained in an insertion-competent state, we incubated newly synthesized R4op for different times before adding RM. To see whether membrane insertion of R4op can proceed independently of the presence of ribosomes, we depleted ribosomes by ultracentrifugation. RM were added at 0, 30 and 60 minutes after R4op synthesis. As can be seen in Fig. 2A, efficient membrane insertion and glycosylation can be detected even when the RM were added 60 min. after terminaton of R4op synthesis. Quantification of glycosylated and unglycosylated R4op showed that even after one hour incubation in the cytosol about 60% of R4op was glycosylated and thus membrane inserted (see histogram). We conclude that R4op released from ribosomes can be maintained for an extended time period in a membrane insertion competent state and this does not depend on the presence of ribosomes.
Fig. 2.
Insertion competence and size of the cytosolic R4op complex.
(A) Time dependence of R4op insertion into RM. R4op was synthesized in the RRL, translation was stopped by the addition of puromycin and ribosomes were removed by sedimentation. Reactions were further incubated for the times indicated. RMs were then added and incubation continued for 30 minutes. R4op was immunoprecipitated and characterised by SDS-PAGE and autoradiography. The amounts of immunoprecipitated non-glycosylated R4op (black bars) and glycosylated g-R4op (grey bars) were quantified (see histogram). (B) Sucrose density gradient analysis of cytosolic R4op. R4op was synthesized in the RRL. Aliquots were loaded on top of 10-20% sucrose density gradients containing 2mM ATP (left) or ADP (right). After centrifugation and fractionation, proteins were analyzed by SDS PAGE and autoradiography. Black arrowheads and numbers above the gel indicate migration positions of proteins used as molecular markers and their molecular weights in kDa respectively.
We next asked whether cytosolic R4op accumulates in a distinct complex. Newly synthesized R4op in RRL was separated on 10-20 % sucrose gradients containing either ATP or ADP, to see whether complex formation is dependent on the continued presence of these nucleotides. As can be seen in Fig. 2B, R4op migrates in both sucrose gradients as a distinct complex of about 60 - 70 kDa. In the presence of ATP, R4op migrated with a slightly lower molecular weight than in the presence of ADP. These data suggest that newly synthesized R4op can be maintained in a distinct soluble complex in the absence of RM. Judged by the distance migrated in the gradient we deduce that cytosolic RAMP4 is associated with a cytosolic partner (or partners) of about 50 kDa.
Probing the molecular environment of newly synthesized R4op by chemical crosslinking
In order to directly investigate the molecular surrounding of cytosolic R4op in RRL we applied chemical crosslinking using a crosslinker specific for NH2-groups (DSS) or for SH groups (BMH). Amino acids with NH2 groups are found in the hydrophilic parts of R4op, while a single cysteine residue is found close to the middle of the transmembrane domain (TMD) of R4op (Figure 1B). R4op was synthesized in the RRL and crosslinkers were added in the absence of nucleotides and small molecules or in the presence of ATP or ADP As can be seen in Fig. 3A, both DSS (lane 2 to 4) and BMH (lane 5) induce formation of a crosslinked product of about 46 kDa that is not seen in the absence of crosslinker (compare lane 1 with lanes 2-5). Consistently we found that the amount of crosslinked product was highest when BMH was used. As the 46 kDa crosslinked product was obtained after immunoprecipitation with the anti-opsin antibody it must be an adduct of R4op. Taking into the account the molecular weight of R4op, we estimate the size of its interacting partner to be about 40kDa and accordingly we named it p40. Presence of ATP (lane 3), or ADP (lane 4) did not affect the efficiency of R4op crosslinking to p40. Crosslinking between R4op and p40 does not depend on the presence of the opsin tag as the same crosslinking partner is also seen when authentic RAMP4 is used in the reaction instead of R4op (supplementary Fig. S1).
Fig. 3.
Crosslinking of R4op.
(A) Crosslinking of cytosolic R4op in the presence or absence of nucleotides. After termination of R4op translation in RRL, nucleotides were removed and either no nucleotides or 2mM ATP or ADP added. After the crosslinking by either DSS (D) or BMH (B), R4op was immunoprecipitated and characterised by SDS-PAGE and autoradiography. (B) Crosslinking of R4op in the presence or absence of ribosomes. After translation, ribosomes were removed by ultracentrifugation. A total fraction (total), the resuspended pellet (pellet) and the supernatants (Sn) were crosslinked by BMH or left uncrossliked (-). (C) Crosslinking of R4op in the presence of Triton X-100. After synthesis of R4op in the RRL, the samples were adjusted to the indicated amount of Triton X-100 and then BMH was added. (D) Crosslinking of R4op in the absence or presence of RM. After synthesis of R4op, BMH was added either immediately or after the additional incubation with RM. g-R4op: glycosylated R4op. R4op × p40: crosslinked product of R4op and a 40 kDa protein.
To determine whether p40 is a ribosomal protein, we sedimented ribosomes from the R4op translation mixture prior to crosslinking. As can be seen in Fig. 3B, the crosslinked product between R4op and p40 was detected exclusively in the supernatant fraction (compare lanes 4 and 2) suggesting that p40 is not a ribosomal protein. The efficiency of sedimentation was confirmed by Western blot analysis of an aliquot of the supernatant and pellet fractions with an antibody against the ribosomal protein L23a (supplementary Fig. S2)
Crosslinking of R4op to p40 via the single cysteine residue present within the TMD domain of R4op (Fig. 3A, lane 5), suggests that hydrophobic interactions play a role in the interaction between R4op and p40. To test this possibility we added different amounts of the detergent Triton X-100 before adding BMH. Very low concentrations of Triton X-100 (0.02% v/v) were sufficient to completely abolish crosslinking between R4op and p40 (Fig. 3C). In contrast, ionic interactions do not break the association between these two proteins since crosslinking was observed even when high ionic strength buffer (1M KOAc) was used in the crosslinking reaction (supplementary Fig. S3).
When RMs are added to newly synthesized R4op in the RRL, R4op is inserted into the membranes and becomes glycosylated. To see whether R4op is released from p40 when RMs are added, we applied crosslinking before and after the addition of RM. In the absence of RMs, newly synthesized R4op was found crosslinked to p40 (Fig. 3D, lane 2). When RMs were present in the reaction no crosslinking to R4op was seen anymore. Instead glycosylated R4op appeared (Fig. 3D, lane 3). This indicates that upon RM addition R4op is released from p40 followed by its insertion into RM membranes and its glycosylation.
Requirements for membrane insertion of R4op
ER membrane insertion of some TA proteins can depend on SRP and the SRP receptor (Abell et al., 2004). To test whether the SRP system is involved in the insertion of R4op we treated rough microsomal membranes (RM) with high salt, puromycin (PKRM) and low amounts of trypsin, 1 or 2 μg/ml (PKRM-T1 and PKRM-T2) respectively. Under these conditions the cytoplasmically exposed portion of the SRP receptor is liberated from the membrane rendering RM incompetent for the SRP-dependent translocation (Meyer and Dobberstein, 1980). Recombinant soluble SRP-receptor (SR) can then be added to restore the SRP-dependent translocation (Fulga et al., 2001). We tested such trypsin-treated membranes for their ability to allow insertion of the type II membrane protein invariant chain (Ii) and of R4op. Membrane insertion was again determined by glycosylation of these proteins and a shift to higher molecular forms. Fig. 4 shows that Ii is inserted into PKRM and mock treated PKRM, but not into trypsin treated membranes, PKRM-T1 or -T2 (lanes 1-4 and 5 and 7). When the purified SRP-receptor was added, membrane insertion of Ii was re-established (lanes 6 and 8). This confirms that the trypsin treatment was effective and that the soluble SRP-receptor can restore membrane insertion of Ii. When membrane insertion of R4op was similarly tested the trypsin treated membranes were found to still promote membrane insertion (glycosylation) of RAMP4 (lanes 13 and 15). This demonstrates that membrane insertion of R4op does not depend on the SRP system.
Fig. 4.
Membrane requirements for R4op and Ii insertion into PKRM.
R4op and Ii were synthesized in the RRL. Puromycin/high salt washed membranes (PKRM), mock treated (PKRM mock) or membranes treated with 1 or 2 μg/ml trypsin (PKRM-T 1 and PKRM-T 2) were present during the synthesis of Ii. R4op was incubated with these membranes post-translationally. Where indicated, 100nm of soluble SRP receptor (SR) was added. Proteins were analyzed by SDS-PAGE and autoradiography. Ii*: nonglycosylated Ii; g-R4op: glycosylated R4op.
Immunoaffinity purification of the R4op/p40 complex and identification of p40 as Asna-1
To isolate and identify the interaction partner of newly synthesized R4op, p40, we pursued an immunoaffinity purification strategy using a large volume of RRL in which R4op was synthesized in the absence of membranes. As a control, another in vitro translation reaction was used to which buffer was added instead of R4op mRNA. Both reaction mixtures were batch adsorbed to CNBr-sepharose beads coupled to the anti-opsin antibody. After binding, the beads were extensively washed with a buffer containing a high salt concentration, a treatment that does not interfere with the R4op - p40 association (supplementary Fig. S3). p40 was then released from R4op by using a buffer containing Triton X-100, and remaining proteins were finally eluted from the affinity matrix using an acidic glycine buffer.
As can be seen in Fig. 5A, lane 1, a prominent band of a protein with an apparent MW of about 40 kDa was detected after silver staining of a gel containing the Triton X-100 eluted sample. No such protein was found in the sample obtained from the control reaction lacking newly synthesized R4op (lane 3).
Fig. 5.
Identification of p40
(A) Immunoaffinity purification of p40 associated with R4op. Large volume RRL translation reactions were incubated with (lanes 1 and 2) or without R4op mRNA (lanes 3 and 4). R4op-containing complexes were affinity purified on anti-opsin antibody beads and proteins released from R4op by elution with 0.1% Triton X-100 (TX) (lanes 1 and 3). Remaining bound proteins were eluted from the column using an acidic glycine buffer (gly) (lanes 2 and 4). Eluted proteins were separated by SDS PAGE and silver stained. The protein band of about 40 kDa was cut out, proteins were eluted and peptide sequences determined by mass spectroscopy. Peptide sequences identified p40 as Asna-1. (B) Immunoprecipitation of R4op × p40 crosslinked product. R4op was synthesized in RRL and aliquots of the reaction were either crosslinked with BMH (+) or incubated with DMSO solvent alone (-). Aliquots of both reactions were either directly applied to the gel (lanes 1 and 2) or immunoprecipitated by anti-opsin antibody (lanes 3 and 4), an anti-Asna-1 antibody (lanes 5 and 6) or a pre-immune serum (lanes 7 and 8) and characterised by SDS PAGE and autoradiography.
The 40 kDa protein band was cut out from the gel, eluted and analyzed by ESI-QUAD-TOF mass spectrometry. Fourteen of the detected peptides correspond to sequences of human Asna-1, an arsenite-stimulated ATPase (protein coverage of 41%). To confirm the identity of p40, we performed immunoprecipitation of the R4op-p40 (Asna-1) crosslinked product using an anti-Asna-1 antibody (Fig. 5B, lane 6). This antibody immunoprecipitated exclusively the 46 kDa crosslinked product, but not non-crosslinked R4op. In the reaction where no BMH was used, anti-Asna-1 could not precipitate any radiolabelled protein (lane 5). The antibody directed against the opsin tag immunoprecipitated both, non-crosslinked as well as crosslinked R4op (lanes 3 and 4 respectively)
We conclude that the 40 kDa cytosolic interacting partner of R4op synthesized in the RRL in the absence of RM is the ATPase Asna-1.
Does Asna-1 also interact with other membrane proteins?
To see whether Asna-1 interacts also with other membrane proteins we tested the C-terminally tagged TA proteins Sec61βop (S61βop), cytochrome b5op (b5op) and the type II membrane protein Ii. S61βop and b5op, like R4op are efficientely post-translationally inserted into ER membranes while Ii is not (supplementary Fig. S4). To see whether these proteins associate with Asna-1 we used again the crosslinkers BMH and DSS. As b5op does not contain a cystein residue (Fig. 6A), we used in this case only DSS. After translation in the RRL and crosslinking we immunoprecipitated crosslinked complexes with an anti-opsin, anti-Asna-1 or a non-related anti-Myc antibody. As can be seen in Fig 6, B-E, efficient crosslinking to Asna-1 and Asna-1 containing higher molecular weight complexes (indicated by a rhombus) was only found for R4op and S61βop, but not for b5op (lanes 3 and 4) and Ii (lanes 5 and 6).
Fig 6.
Cytosolic R4op (RAMP4op) and S61βop (Sec61op) but not b5op (cytochrome-b5op) or Ii (invariant chain) can be crosslinked to Asna-1.
A) Outline of the sequences around the TM (black) of R4op, S61βop, b5op and Ii. The sequences are aligned by the relative position of their TM domains. Lysine (K) and cysteine (C) residues that can function in crosslinking with DSS and BMH respectively are indicated. B - E) Crosslinking of the TA proteins and Ii in the RRL: R4op (A), S61βop (B), b5op (C) and Ii (D) were in vitro synthesized in the RRL and small molecules were removed by gel filtration and either DMSO (-) or the crosslinker BMH (B) or DSS (D) were added. The TA proteins were immunoprecipitated with anti-opsin (α-op), anti-Asna-1 (α-Asna-1) or the unrelated anti-Myc (α-Myc) antibodies and characterised by SDS-PAGE and autoradiography. x-Asna-1: crosslinked to Asna-1; The rhombus indicates yet unidentified higher molecular weight complexes.
Nucleotide and redox state dependence of TA protein insertion into the ER membrane
Previous studies by several groups have shown that membrane insertion of TA proteins can depend on the presence of ATP or GTP or require even no nucleotides. Based on these observations different pathways for membrane insertion of TA proteins were suggested (Abell et al., 2007; Borgese et al., 2003b; Brambillasca et al., 2006). To determine the requirements for nucleotides, we synthesized R4op, S61βop and b5op in the RRL and released nucleotides from proteins by chelating Mg2+ with EDTA. We then removed small molecules from the lysates by gel filtration. Mg(OAc)2 was then supplied to all further reactions.
The homologue of Asna-1 in Saccharomyces cerevisiae, Arr4/Get3 may be a redox sensitive protein since copper-mediated redox stress changes the conformation of the protein as detected in non-reducing SDS-PAGE gels (Metz et al., 2006). Furthermore 5 of the 8 cyteines are conserved from yeast to human. We therefore tested, whether the redox state of the cytosol could influence the nucleotide requirement for the membrane targeting / insertion of the TA proteins. We depleted lysates of small molecules and added H2O2 to 2mM (Fig. 7A, lanes 3-6) or DTT to 2mM (Fig. 7A, lanes 7-10) and either no nucleotides or ATP (A), GTP (G) or CTP (C) as indicated in Fig. 7A to lysates depleted of small molecules. After the addition of RM the lysates were incubated and the labeled TA proteins analysed by SDS PAGE and autoradiography. The percent of glycosylated TA proteins was determined densitometrically. Most efficient membrane insertion (glycosylation) of R4op and S61βop is seen in the presence of ATP under all redox conditions tested (Fig. 7A and supplementary Fig. S5 (0,25 - 8 mM H2O2 or DTT) although it is slightly less efficient under oxidizing conditions. Under reducing conditions a lower level of membrane insertion is seen when nucleotides were removed or in the presence of GTP or CTP (Fig. 7A). Membrane insertion of b5op, in contrast, was not affected by the removal of nucleotides or the addition of nucleotides but is slightly more efficient under reducing (DTT) than oxidizing (H2O2) conditions (Fig. 7A).
Fig 7.
Comparison of the requirements for membrane insertion of R4op, S61βop and b5op.
A) Nucleotide and redox state dependence of the post-translational membrane insertion. After the synthesis of the TA proteins in the RRL (lane 1) RM were added either directly (lane 2) or after treatments as indicated (lanes 3-10). To test nucleotide tri-phosphate (NTP) and redox conditions required for membrane insertion of these TA proteins small molecules were removed from the lysates by gel filtration (lanes 3-10) and 2 mM H2O2 (lanes 3-6), 2 mM DTT, 3 mM ATP (A), GTP (G) or CTP (C) added as indicated. After incubation, proteins were separated by SDS-PAGE and visualized by autoradiography. The percent (%) of glycosylated TA protein (glyc) was quantified (right panel).
B) Nucleotide depletion by apyrase and membrane insertion of R4op, S61βop and b5op. The three TA proteins were synthesized in the RRL (lane 1) and RM added either directly (lane 2) or after removal of small molecules by gel filtration and addition of H2O2, DTT, apyrase or ATP as indicated (lanes 3-8).
C) Release of R4op from Asna-1 and membrane insertion. After synthesis of R4op in the RRL, small molecules were removed by gel filtration and lysates were adjusted to either 2 mM H2O2 (lanes 1-8) or 2 mM DTT (lanes 9-16) and 3 mM ATP and then incubated with RM as indicated. After the membrane insertion small molecules were removed by gel filtration and BMH crosslinking induced where indicated. Proteins were immunoprecipitated by anti-opsin antibodies (α-op), separated by SDS-PAGE and visualized by autoradiography.
D) Free sulfhydryl groups on cytosolic proteins are required for the membrane insertion of R4op and S61βop but not of b5op.
After synthesis of the three TA proteins in the RRL (lane 1) RM were added (lane 2). and the insertion reaction incubated for 30 min at 30°C. To test whether free sulfhydryl groups are needed for membrane insertion, lysates were adjusted to 5mM N-ethylmaleimide (NEM) where indicated. After an incubation, small molecules were removed by gel filtration and the lysates adjusted to 2 mM H2O2 (lanes 3-6), 2mM DTT (lanes 7-10) and 3mM ATP as indicated and incubated with RM. Proteins were separated by SDS-PAGE and visualized by autoradiography.
As membrane insertion of b5 is known to require low concentrations of ATP (Yabal et al., 2003) and the gel filtration may not have removed all nucleotides, we used in addition apyrase to deplete nucleotides. As can be seen in Fig. 7B all three TA proteins were not any more glycosylated neither under oxidizing nor reducing conditions (lanes 5 and 8). Complete depletion of nucleotides lead to a reduction in the amounts of S61bop and b5op and the accumulation of smaller molecular weight forms of these proteins suggesting proteolytic processing. Efficient membrane insertion of R4op and S61op required the addition of ATP while efficient membrane insertion of b5op was not increased by the addition of ATP (lanes 3,4 and 6,7).
To see how the redox conditions affect the interaction (crosslinking) between R4op and Asna-1 and the release of Asna-1 upon addition of RM, we crosslinked proteins after the membrane insertion reaction with BMH. R4op × Asna-1 complexes were either immunoprecipitated with the anti-opsin antibody (Fig. 7C) or with anti-Asna-1 antibody (supplementary Fig. S6). We find that release of R4op from Asna-1 and membrane insertion (glycosylation) requires the presence of RM and ATP (cf. lanes 13 and 15) and thus suggests that the ATP-dependent pathway is mediated by Asna-1. The release however is less efficient under oxidizing conditions (cf. lanes 5 and 7). R4op is not released from Asna-1 in the absence of ATP (lanes 5 and 13). A low amount of glycosylated R4op is also seen in the absence of added ATP.
To test whether free sulfhydryl groups on cytosolic proteins are essential for membrane insertion of the three TA proteins, we treated the translation reaction with N-ethylmaleimide (NEM), eliminated excess NEM by gel filtration and added, where indicated, H2O2 or DTT or ATP to the lysates and incubated them with RM (Fig. 7D). We find that NEM treated cytosol prevents membrane insertion (glycosylation) of R4op as well as S61βop, but not that of b5op (Fig. 7D).
Discussion
We have investigated requirements for membrane insertion of R4op, S61βop and b5op. We show here that these three TA proteins can insert post-translationally into membranes of the ER in a cell free system. In the absence of membranes newly synthesized R4op and S61βop but not b5op can be crosslinked to Asna-1, an arsenite-stimulated ATPase that promotes various membrane related functions (Auld et al., 2006; Kao et al., 2007; Shen et al., 2003; Tseng et al., 2007). We identified Asna-1 after immunoisolation of the soluble R4op complex by mass spectrometry and by immunoprecipitation with an anti-Asna-1 antibody. As the release of R4op from Asna-1 requires the presence of RM and ATP we propose that Asna-1 is a functional intermediate in the membrane insertion of R4op and probably also of S61βop. Asna-1 mediated targeting / membrane insertion of R4op and S61βop is sensitive to oxidizing conditions and is blocked when free SH groups are modified by NEM. In contrast, membrane insertion of b5op is not blocked by NEM and low level of nucleotides are sufficient for membrane insertion of this TA protein (Borgese et al., 2003a; Yabal et al., 2003).
While the initial version of this manuscript was prepared Stefanovic and Hegde reported the finding that Asna-1 interacts with transmembrane domains (TMDs) of several TA proteins, Sec61β, VAMP2 and two members of the syntaxin family, Nsyn1 and Stx1 (Stefanovic and Hegde, 2007). Our findings add RAMP4/SERP1 to this list of TA proteins that interact with Asna-1 in a cytosolic complex before membrane insertion. Using sucrose gradient centrifugation we find a size of 60-70 kDa for the soluble R4op complex. However distinct higher molecular weight complexes are identified by crosslinking with R4op as well as with S61βop (Fig. 6 B, C). These may represent crosslinks to dimers of Asna-1(Kurdi-Haidar et al., 1998a). A size of 100 - 500 kDa is found by Stefanovic and Hegde for the soluble Sec61β complex, named TMD recognition complex (TRC) (Stefanovic and Hegde, 2007). This complex is proposed to contain besides Asna-1 (named TRC40) additional proteins that function in the membrane targeting of TA proteins (Stefanovic and Hegde, 2007).
Several lines of evidence show that Asna-1 interacts with the TMD of TA proteins. We show that Asna-1 can be crosslinked to a single SH group (Cys) in the transmembrane region of R4op. Binding to a hydrophobic region can also be deduced from the fact that a mild detergent but not the addition of high salt can break the interaction of Asna-1 with R4op. Stefanovic and Hegde show directly that the TMD of Sec61β is required for an interaction with Asna-1/TRC40. Deletion of the TMD of Sec61β prevents interaction with Asna-1/TRC40 (Stefanovic and Hegde, 2007). However the interaction of Asna-1 with TA proteins is probably not restricted to the TMD as crosslinking to the single cysteine in the N-terminal hydrophilic region of S61βop is also observed (Fig. 6A). Asna-1 does not seem to interact with b5op or Ii as no crosslinking of Asna-1 to these proteins could be observed (Fig. 6 D, E). Consistent with this conclusion is also the fact that membrane insertion of b5op is not stimulated by additional ATP (Fig. 7B). The hydrophobic signal anchor domain of nascent Ii is known to interact with SRP (High et al., 1993). Our data show that Asna-1 is not in proximity to any hydrophobic TMD of newly synthesized membrane proteins that accumulate in the cytosol.
Different targeting pathways for the membrane insertion of R4op and S61βop and b5op
As RAMP4 and Sec61β are ER stress induced proteins (Hori et al., 2006) and one of the functions of Asna-1/Get3 is related to cellular stress (Metz et al., 2006; Shen et al., 2003), we investigated how cytosolic redox conditions and nucleotides affect membrane insertion of these proteins. Our results are schematically summarized in Table 1. We find that the cytosolic requirements for membrane insertion of R4op and S61βop are very similar: free SH groups in a cytosolic protein are required for membrane insertion of these proteins as NEM treated cytosol does not support membrane insertion, neither under oxidizing, nor reducing conditions. Efficient membrane insertion of R4op and S61 op requires ATP and is not supported by GTP or CTP or non-hydrolysable nucleotides (AMPPNP, GMPPNP) (Fig. 7A and data not shown). The reduction of nucleotides by gel filtration revealed a striking difference between the membrane insertion of R4op and S61βop on the one hand and b5op on the other: while low levels of nucleotides fully support the membrane insertion of b5op they support only a basel level insertion of R4op and S61βop. Additional ATP is required for release of R4op from Asna-1 and efficient membrane insertion of R4op and S61βop. In contrast to R4op and S61βop, membrane insertion of b5op is not NEM sensitive and is not affected by oxidative cytosolic conditions. The free SH group(s) affected by NEM may reside in the substrate protein, Asna-1 or another targeting factor. Both, R4op and S61βop contain a cystein in the TMD and the N-terminal domain respectively, while b5op does not contain a cysteine.
Table 1.
Summarized results for the requirements for membrane insertion of R4op & S61bop and b5op. Most efficient glycosylation (membrane insertion): +++, less: ++, drastically reduced: +, not detected: -
| DTT | H2O2 | |||||||
|---|---|---|---|---|---|---|---|---|
| +ATP | low NTP |
-NTP | NEM +ATP |
+ATP | low NTP |
-NTP | NEM +ATP |
|
| RAMP4 & Sec61β | +++ | + | - | - | ++ | - | - | - |
| Cytochrome-b5 | +++ | +++ | - | +++ | ++ | ++ | - | ++ |
Different pathways for the membrane insertion of TA proteins have been proposed previously. They were defined by the requirement for different cytosolic factors and ATP, GTP or no nucleotides (Abell et al., 2003; Abell et al., 2004; Borgese et al., 2003a; Kim et al., 1997; Kutay et al., 1995; Kutay et al., 1993; Rabu and High, 2007; Steel et al., 2002). For Sec61β it was found that it interacts with SRP and the molecular chaperones Hsc70 and Hsp40, and that Hsc70 stimulates membrane insertion in conjunction with Hsp40 (Abell et al., 2007). It is well conceivable that these chaperones in addition to Asna-1 mediate membrane insertion of TA proteins like RAMP4 and Sec61β. The higher molecular weight complexes seen in the R4op an S61βop crosslinks may represent Asna-1 dimers (Kurdi-Haidar et al., 1998a) and / or complexes with such chaperones (Fig. 6, B, C) (Rabu and High, 2007). Another possibility is that the low nucleotide requiring membrane insertion of R4op and S61βop is mediated by the HSP70/HSP40 chaperone system and SRP (Abell et al., 2003; Abell et al., 2004; Abell et al., 2007).
The ATPase Asna-1 is implicated to function in membrane associated processes
Mammalian Asna-1 is a 37 kDa cytoplasmic ATPase with a single ATP-binding cassette (Kurdi-Haidar et al., 1998b). Asna-1 is homologous to bacterial ArsA and to Arr4p/Get3 of Saccharomyces cerevisiae (Bhattacharjee et al., 2001; Shen et al., 2003). Yeast Arr4/Get3 ATPase has been implicated in multiple membrane-dependent pathways including metal ion homeostasis and heat tolerance (Metz et al., 2006; Shen et al., 2003). In C. elegans and in mammals Asna-1 is mainly expressed in cells with high secretory activity, in particular in β-islet cells (Kurdi-Haidar et al., 1996; Kurdi-Haidar et al., 1998c). The data presented here and previously by Stefanovic and Hegde suggest that membrane insertion of a subset of TA proteins is mediated by Asna-1(Stefanovic and Hegde, 2007). Our data furthermore suggest, that the Asna-1 pathway may not be the only pathway by which TA proteins like R4op and S61βop can be inserted into the ER membrane. In the presence of low nucleotides, when Asna-1 is not released from R4op, a proportion of R4op is found inserted into membranes (Fig. 7A and C). This pool is rather sensitive to oxidative conditions (Fig. 7A). Such conditions may arise when for instance metal ions accumulate in the cytosol. Such an assumption is also supported by considering functions observed for the yeast homolog of Asna-1, Arr4/Get3 (Auld et al., 2006; Metz et al., 2006; Schuldiner et al., 2005). Arr4/Get3 has been implicated in many membrane-dependent pathways including ion homeostasis, ER-associated degradation and regulation of membrane transport processes (Auld et al., 2006; Metz et al., 2006; Schuldiner et al., 2005; Shen et al., 2003). Arr4/Get3 and Asna1 may contribute to these functions by mediating efficient membrane insertion of a certain subset of TA proteins (Auld et al., 2006; Metz et al., 2006; Schuldiner et al., 2005). Clearly more work is required to elucidate the role of Asna-1/TRC40 in membrane insertion of RAMP4 and other TA proteins.
Materials and methods
Cloning procedures and plasmids
Constructs used in this study were made by standard methods (Maniatis et al., 1982) and verified by sequencing. Vector pGem4Ii used for the synthesis of invariant chain was described previously (High et al., 1993). A plasmid containing mouse RAMP4 cDNA (MNCb-2442) was obtained from the National Institute of Infectious Diseases, Division of Genetic Resources, Tokyo, Japan. An EcoRI / PstI fragment was ligated into the pGEM4Z plasmid (Promega) to give pGEM4Z-MNCb-RAMP4. A tag containing the N terminal 13 amino acids residues of bovine opsin was added to the C-terminus of mouse RAMP4 encoded by the plasmid pGEM4Z-RAMP4/3′UTR (mouse RAMP4 with authentic 3′ UTR) using PCR-based mutagenesis ExSite kit from Stratagene and the following oligonucleotides: GG_F1 (5′- GGCCCAAACTTCTACGTGCCTTTCTCCAACAAGACGGGCTGAAGTGACTGACCTTGA -3′) pGG_R1 (5′- CATGCCCATCCTGATACTTTGAATAATCTGGAAAATTGCAGAGCCACAGACAA-3′) The plasmids encoding the human cytochrome b5 (b5) and Sec61β cDNAs containing the C-terminal opsin tag, pcDNA5-Cb5OPG and pcDNA5-Sec61βOPG (Abell et al., 2007) respectively were a kind gift from Stephen High.
In vitro transcription/translation, membrane insertion and denaturing immunoprecipitation
Messenger RNA was synthesized from the SP6 or T7 promotor using linearized plasmid DNA and standard methods as described previously (Schröder et al., 1999). Proteins were synthesized in the rabbit reticulocytes lysate (RRL) according to manufacturers instructions (Promega RRL kit for in vitro translation) and using L-[35S] methionine (7.5μCi per 10μl reaction). 150ng of in vitro synthesized mRNA was used per 10μl reaction.
Rough microsomes (RM) and puromycin-high salt washed membranes (PKRM) were prepared as described by Walter and Blobel and resuspended at 50 OD280/ml in RM buffer (50 mM Hepes-KOH pH7.6, 50 mM KOAc, 2 mM Mg(OAc)2, 2mM DTT) (Walter and Blobel, 1983). To prepare trypsin-treated PKRM, the amount of trypsin indicated in the figure legend was added to 1ml of PKRM (Meyer and Dobberstein, 1980). This mixture was incubated for 60 minutes on ice. The reaction was stopped by addition of RM buffer containing 1mM PMSF, 10 μg/ml aprotinin and 0.75M KOAc. Membranes were pelleted by centrifugation and resuspended in 1ml of RM buffer. Mock-treated membranes were prepared in the same way, except water was used instead of trypsin.
Translation reactions were incubated for 30 minutes at 30°C and stopped by addition of puromycin to the final concentration of 2mM. In reactions where proteins were inserted cotranslationally, RM from canine pancreas were present during translation. For post-translational insertion RM were added after termination of translation and incubation continued for 30 minutes at 30°C.
When insertion into trypsin-treated microsomal membranes was reconstituted, soluble recombinant SRP receptor SRhisα/βΔN was added to the final concentration of 100nm (Fulga et al., 2001).
To test redox state and nucleotide requirements for membrane insertion of TA proteins, we synthesized the TA proteins in the RRL and then chelated Mg2+ by adding EDTA to 5 mM. To test the the effect of NEM on membrane insertion of the TA proteins, NEM (Sigma-Aldrich, Steinheim Germany) freshly dissolved in water, was added to the lysates after translation to a final concentration of 5mM. Where indicated in figure legends small molecules were removed by gel filtration using prepacked G-25 MicroSpin columns (GE Healthcare) that were equilibrated in 50 mM Hepes-KOH pH 7.6 and 80mM KOAc. Reactions were then adjusted to 10 mM Mg(OAc)2, 2 mM H2O2 or DTT and 3 mM of the nucleotide indicated in the figures. In some experiments apyrase was added to a final concentration of 0.4 U/μl and the samples incubated for 30 min at 30° C.
At the end of the RM insertion assays, proteins were precipitated with ammonium sulfate and 1/5 of the starting reaction was prepared for SDS PAGE. Where indicated antigens were immunoprecipitated under denaturing conditions using specific antibodies and protein A-coupled sepharose beads (Amersham Pharmacia) as previously described (High et al., 1993; Schröder et al., 1999).
Proteins were separated by SDS PAGE on 15% gels and radiolabelled molecules were visualized by autoradiography.
Chemical crosslinking
Crosslinkers were purchased from Pierce, dissolved in DMSO and stored at -20°C. Before crosslinking, small molecules were removed by gel filtration using Microspin columns pre-packed with Sephadex G-25 (GE Healthcare) that were equilibrated in 20 mM Hepes-KOH pH 7.6, 80 mM KOAc, 0.5 mM Mg(OAc)2. If not specified otherwise, crosslinker were used at a final concentration of 250 μM. Crosslinking was conducted for 15 minutes at room temperature. The reaction was quenched by the addition of glycine and DTT to the final concentration of 10 mM each and incubation on ice for further 5 minutes. Samples were then processed for denaturing immunoprecipitation and analyzed by a 15% or 6-15% gradient SDS gel and autoradiography. As a control, one sample was incubated with DMSO lacking the crosslinker.
Sucrose density gradient analysis
To analyze R4op-containing cytosolic complexes, R4op was synthesized in vitro in 20 μl RRL reaction. After termination of protein synthesis by puromycin, aggregates were removed by centrifugation for 10 min at 13.000 rpm in a tabletop centrifuge. Supernatants and molecular weight marker proteins were loaded onto linear 10-20% sucrose gradients containing 50mM Hepes-KOH pH 7.5, 80mM KOAc, 1mM Mg(OAc)2, 1mM DTT and either 2 mM ATP or 2mM ADP, Gradients were centrifuged in a SW60 rotor at 4°C for 15 hours at 45000 rpm. Fractions were collected from the bottom using an ISCO density gradient fractionator. Fraction 12 contains the resuspended pellet. Proteins in the fractions were precipitated by TCA resusupended in 1% SDS, 10mM Tris-Cl pH 7.5, 80mM KOAc, 1mM Mg(OAc)2 and R4op was immunoprecipitated by the anti-opsin antibody. Proteins were then characterised by SDS PAGE and autoradiography. Marker proteins, run on parallel gradients were: lysozyme (13 kDa), chymotrypsin A (25kDa), albumin (67 kDa).
Affinity purification of R4op and associated proteins
To purify R4op-containing complexes, we have coupled the monoclonal anti-opsin R2-15 antibody to CNBr-Sepharose beads (Amersham Pharmacia). The beads were washed with buffer I (0.5 M KOAc, 10 mM Tric-Cl pH 7.6) and equilibrated in ice cold Buffer II (250 mM KOAc, 10mM Tris-Cl pH 7.6).
R4op or no protein were synthesized in each 5 ml of rabbit reticulocyte lysates obtained from Green Hectares (Wisconsin, USA). After the synthesis aggregates were removed by centrifugation in the Sorvall SS34 rotor for 15 minutes at 10000 rpm. The resulting supernatants were added to 200 μl anti-opsin coupled beads. The mixture was incubated with shaking for 3 hours at room temperature. Beads were then washed 2 times with 4ml of buffer W1 (1M KOAc, 10 mM Tris-Cl pH 7.6), transferred to fresh tubes and washed 8 times with 4ml of W2 buffer (500 mM KOAc, 10 mM Tris-Cl pH 7.6). Proteins were then eluted by incubating the beads with 150 μl of buffer E1 (0.1% Triton X-100, 500 mM KOAc, 10 mM Tris-Cl pH 7.6) for 10 minutes at room temperature. After centrifugation, proteins in the eluate were precipitated with 20% TCA / 80% acetone and resuspended in 30 μl sample buffer for SDS PAGE. Beads were washed once with 4ml of buffer II, and then incubated in 150 μl of 100 mM glycine (pH 2.5) for 10 minutes. After centrifugation proteins in the glycine eluate were precipitated by TCA. Proteins were separated on a 15% SDS gel and visualized by standard silver staining except that the gel was developed in a solution of 0.01% formaldehyde (v/v) and 2% sodium carbonate (w/v) followed by washing with 1% acetic acid. To determine the identity of the ~40 KDa protein, the band was cut from the gel and analyzed by ESI-QUAD-TOF mass spectrometry.
Antibodies
Rabbit antibodies against invariant chain (anti Ii), ribosomal protein L23 (anti L23) and RAMP4 (anti-RAMP4) have been described previously (High et al., 1993; Lipp and Dobberstein, 1986; Pool et al., 2002). Hybridoma cell line secreting mouse monoclonal anti-opsin antibody (R2-15) was generated by Dr. Paul Hargrave (Adamus et al., 1991). Antibodies against Asna-1 were raised against a maltose-binding protein-Asna-1 fusion protein in guinea pig (Peptide Speciality Laboratories, Heidelberg). Secondary antibodies used for western blotting were purchased from Sigma-Aldrich, Steinheim Germany.
Supplementary Material
Acknowledgements
This work was supported by grants from the Deutsche Forschungsgemeinschaft, SFB 638/A2 to BD and SFB638/A4 to BS. We gratefully thank Thomas Ruppert and Armin Bosserhoff for mass spectrometry, Stephen High for providing plasmids and Dr. Paul Hargrave for providing the hybridoma cell line expressing anti-opsin antibody. We are grateful for gifts of reagents and technical support from I. Sinning, T. Fulga, O. Schlenker. We thank Stephen High and members of our groups for stimulating discussions.
References
- Abell BM, Jung M, Oliver JD, Knight BC, Tyedmers J, Zimmermann R, High S. Tail-anchored and signal-anchored proteins utilize overlapping pathways during membrane insertion. J Biol Chem. 2003;278:5669–78. doi: 10.1074/jbc.M209968200. [DOI] [PubMed] [Google Scholar]
- Abell BM, Pool MR, Schlenker O, Sinning I, High S. Signal recognition particle mediates post-translational targeting in eukaryotes. Embo J. 2004;23:2755–64. doi: 10.1038/sj.emboj.7600281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abell BM, Rabu C, Leznicki P, Young JC, High S. Post-translational integration of tail-anchored proteins is facilitated by defined molecular chaperones. J Cell Sci. 2007 doi: 10.1242/jcs.002410. [DOI] [PubMed] [Google Scholar]
- Adamus G, Zam ZS, Arendt A, Palczewski K, McDowell JH, Hargrave PA. Anti-rhodopsin monoclonal antibodies of defined specificity: characterization and application. Vision Res. 1991;31:17–31. doi: 10.1016/0042-6989(91)90069-h. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Mostov KE, Blobel G. Mechanisms of integration of de novo synthesized polypeptides into membranes: signal recognition particle is required for integration into microsomal membranes of calcium ATPase and of lens MP26 but not of cytochrome b5. Proc. Natl. Acad. Sci. USA. 1983;80:7249–7253. doi: 10.1073/pnas.80.23.7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annis MG, Yethon JA, Leber B, Andrews DW. There is more to life and death than mitochondria: Bcl-2 proteins at the endoplasmic reticulum. Biochim Biophys Acta. 2004;1644:115–23. doi: 10.1016/j.bbamcr.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Auld KL, Hitchcock AL, Doherty HK, Frietze S, Huang LS, Silver PA. The conserved ATPase Get3/Arr4 modulates the activity of membrane-associated proteins in Saccharomyces cerevisiae. Genetics. 2006;174:215–27. doi: 10.1534/genetics.106.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee H, Ho YS, Rosen BP. Genomic organization and chromosomal localization of the Asna1 gene, a mouse homologue of a bacterial arsenic-translocating ATPase gene. Gene. 2001;272:291–9. doi: 10.1016/s0378-1119(01)00522-4. [DOI] [PubMed] [Google Scholar]
- Borgese N, Brambillasca S, Soffientini P, Yabal M, Makarow M. Biogenesis of tail-anchored proteins. Biochem Soc Trans. 2003a;31:1238–42. doi: 10.1042/bst0311238. [DOI] [PubMed] [Google Scholar]
- Borgese N, Colombo S, Pedrazzini E. The tale of tail-anchored proteins: coming from the cytosol and looking for a membrane. J Cell Biol. 2003b;161:1013–9. doi: 10.1083/jcb.200303069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambillasca S, Yabal M, Makarow M, Borgese N. Unassisted translocation of large polypeptide domains across phospholipid bilayers. J Cell Biol. 2006;175:767–77. doi: 10.1083/jcb.200608101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambillasca S, Yabal M, Soffientini P, Stefanovic S, Makarow M, Hegde RS, Borgese N. Transmembrane topogenesis of a tail-anchored protein is modulated by membrane lipid composition. Embo J. 2005;24:2533–42. doi: 10.1038/sj.emboj.7600730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulga TA, Sinning I, Dobberstein B, Pool MR. SRbeta coordinates signal sequence release from SRP with ribosome binding to the translocon. Embo J. 2001;20:2338–47. doi: 10.1093/emboj/20.9.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High S, Martoglio B, Görlich D, Andersen SSL, Ashford AJ, Giner A, Hartmann E, Prehn S, Rapoport TA, Dobberstein B, et al. Site-specific photo-crosslinking reveals that Sec61p and TRAM contact different regions of a membrane inserted signal sequence. J. Biol. Chem. 1993;268:26745–26751. [PubMed] [Google Scholar]
- Hori O, Miyazaki M, Tamatani T, Ozawa K, Takano K, Okabe M, Ikawa M, Hartmann E, Mai P, Stern DM, et al. Deletion of SERP1/RAMP4, a component of the endoplasmic reticulum (ER) translocation sites, leads to ER stress. Mol Cell Biol. 2006;26:4257–67. doi: 10.1128/MCB.02055-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiak F, Glover JR, Leber B, Rachubinski RA, Andrews DW. Targeting of passenger protein domains to multiple intracellular membranes. Biochem J. 1994;300:191–9. doi: 10.1042/bj3000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao G, Nordenson C, Still M, Ronnlund A, Tuck S, Naredi P. ASNA-1 Positively Regulates Insulin Secretion in C. elegans and Mammalian Cells. Cell. 2007;128:577–87. doi: 10.1016/j.cell.2006.12.031. [DOI] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–75. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- Kim PK, Janiak-Spens F, Trimble WS, Leber B, Andrews DW. Evidence for multiple mechanisms for membrane binding and integration via carboxyl-terminal insertion sequences. Biochemistry. 1997;36:8873–82. doi: 10.1021/bi970090t. [DOI] [PubMed] [Google Scholar]
- Kurdi-Haidar B, Aebi S, Heath D, Enns RE, Naredi P, Hom DK, Howell SB. Isolation of the ATP-binding human homolog of the arsA component of the bacterial arsenite transporter. Genomics. 1996;36:486–91. doi: 10.1006/geno.1996.0494. [DOI] [PubMed] [Google Scholar]
- Kurdi-Haidar B, Heath D, Aebi S, Howell SB. Biochemical characterization of the human arsenite-stimulated ATPase (hASNA-I) J Biol Chem. 1998a;273:22173–6. doi: 10.1074/jbc.273.35.22173. [DOI] [PubMed] [Google Scholar]
- Kurdi-Haidar B, Heath D, Lennon G, Howell SB. Chromosomal localization and genomic structure of the human arsenite-stimulated ATPase (hASNA-I) Somat Cell Mol Genet. 1998b;24:307–11. doi: 10.1023/b:scam.0000007134.16744.8b. [DOI] [PubMed] [Google Scholar]
- Kurdi-Haidar B, Heath D, Naredi P, Varki N, Howell SB. Immunohistochemical analysis of the distribution of the human ATPase (hASNA-I) in normal tissues and its overexpression in breast adenomas and carcinomas. J Histochem Cytochem. 1998c;46:1243–8. doi: 10.1177/002215549804601104. [DOI] [PubMed] [Google Scholar]
- Kutay U, Ahnert-Hilger G, Hartmann E, Wiedenmann B, Rapoport TA. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. Embo J. 1995;14:217–23. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Hartmann E, Rapoport TA. A class of membrane proteins with a C-terminal anchor. Trends in Cell Biol. 1993;3:72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- Lipp J, Dobberstein B. Signal recognition particle-dependent membrane insertion of mouse invariant chain: a membrane-spanning protein with a cytoplasmically exposed amino terminus. J Cell Biol. 1986;102:2169–75. doi: 10.1083/jcb.102.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular cloning: A Laboratory Manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, New York: 1982. [Google Scholar]
- Metz J, Wachter A, Schmidt B, Bujnicki JM, Schwappach B. The yeast Arr4p ATPase binds the chloride transporter Gef1p when copper is available in the cytosol. J Biol Chem. 2006;281:410–7. doi: 10.1074/jbc.M507481200. [DOI] [PubMed] [Google Scholar]
- Meyer DI, Dobberstein B. Identification and characterization of a membrane component essential for the translocation of nascent proteins across the membrane of the endoplasmic reticulum. J Cell Biol. 1980;87:503–8. doi: 10.1083/jcb.87.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool MR, Stumm J, Fulga TA, Sinning I, Dobberstein B. Distinct modes of signal recognition particle interaction with the ribosome. Science. 2002;297:1345–8. doi: 10.1126/science.1072366. [DOI] [PubMed] [Google Scholar]
- Rabu C, High S. Membrane protein chaperones: a new twist in the tail? Curr Biol. 2007;17:R472–4. doi: 10.1016/j.cub.2007.04.031. [DOI] [PubMed] [Google Scholar]
- Rapoport TA, Jungnickel B, Kutay U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- Schröder K, Martoglio B, Hofmann M, Hölscher C, Hartmann E, Prehn S, Rapoport TA, Dobberstein B. Control of glycosylation of MHC class II-associated invariant chain by translocon-associated RAMP4. EMBO J. 1999;18:4804–4815. doi: 10.1093/emboj/18.17.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–19. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Shen J, Hsu CM, Kang BK, Rosen BP, Bhattacharjee H. The Saccharomyces cerevisiae Arr4p is involved in metal and heat tolerance. Biometals. 2003;16:369–78. doi: 10.1023/a:1022504311669. [DOI] [PubMed] [Google Scholar]
- Steel GJ, Brownsword J, Stirling CJ. Tail-anchored protein insertion into yeast ER requires a novel posttranslational mechanism which is independent of the SEC machinery. Biochemistry. 2002;41:11914–20. doi: 10.1021/bi026105r. [DOI] [PubMed] [Google Scholar]
- Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–59. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- Tseng YY, Yu CW, Liao VH. Caenorhabditis elegans expresses a functional ArsA. Febs J. 2007;274:2566–72. doi: 10.1111/j.1742-4658.2007.05791.x. [DOI] [PubMed] [Google Scholar]
- Walter P, Blobel G. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 1983;96:84–93. doi: 10.1016/s0076-6879(83)96010-x. [DOI] [PubMed] [Google Scholar]
- Wattenberg B, Lithgow T. Targeting of C-terminal (tail)-anchored proteins: understanding how cytoplasmic activities are anchored to intracellular membranes. Traffic. 2001;2:66–71. doi: 10.1034/j.1600-0854.2001.20108.x. [DOI] [PubMed] [Google Scholar]
- Yabal M, Brambillasca S, Soffientini P, Pedrazzini E, Borgese N, Makarow M. Translocation of the C terminus of a tail-anchored protein across the endoplasmic reticulum membrane in yeast mutants defective in signal peptide-driven translocation. J Biol Chem. 2003;278:3489–96. doi: 10.1074/jbc.M210253200. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Hori O, Stern DM, Hartmann E, Ogawa S, Tohyama M. Stress-associated endoplasmic reticulum protein 1 (SERP1)/ribosome-associated membrane protein 4 (RAMP4) stabilizes membrane proteins during stress and facilitates subsequent glycosylation. J. Cell Biol. 1999;147:1195–1204. doi: 10.1083/jcb.147.6.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.