Abstract
The increasing number of publicly available biological databases reflects the evolving need for managing and evaluating abundant and complex data in biological, clinical and computational research. Currently there are over 1000 biologically relevant databases in the public domain with varied content and diverse approaches to capturing and presenting data. This review summarizes the comparatively small niche of sophisticated databases and other resources that aim to enhance understanding of chemicals and their biological actions. The databases reviewed include one that emphasizes environmental chemicals and 9 that emphasize drugs and small molecules. These databases and their associated resources are incrementally strengthening the expanding field of toxicogenomics-based research by providing centralized sources of manually and computationally curated datasets and highly sophisticated tools for the meta-analysis of continually increasing environmental chemical, drug and small molecule datasets.
Keywords: Chemical, Drug, Database, Toxicogenomics
Background
Toxicology is the study of adverse effects of chemicals on living organisms. Understanding the molecular actions of chemicals will be key to improving: a) understanding about the complex relationships between environmental chemicals and human disease; b) therapeutic drug design; and c) the basis of variable susceptibility. The combination of cross-species genomic sequencing initiatives, advances in high-throughput experimental technologies and developments in information technology are changing the course and potential for toxicology research.
Databases serve an essential role in biological and clinical research. This role is evidenced by the increasing number of databases introduced each year (Galperin, 2008). Among the more than 1000 databases in the public domain, only a few are dedicated to chemicals (environmental chemicals or drugs) and their mechanisms of action (Table I). This review describes and distinguishes these resources.
Table I.
Publicly available databases with an emphasis on environmental chemicals or drugs and small molecules
| Database | URL | Recent Citation |
|---|---|---|
| Environmental Chemicals | ||
| Comparative Toxicogenomics Database (CTD) | http://ctd.mdibl.org | (Mattingly et al., 2006) |
|
| ||
| Drugs and Small Molecules | ||
| ChemBank | http://chembank.broad.harvard.edu/ | (Seiler et al., 2008) |
| Chemical Effects in Biological Systems (CEBS) | http://www.niehs.nih.gov/cebs-df/ | (Waters et al., 2008) |
| DrugBank | http://www.drugbank.ca | (Wishart et al., 2008) |
| Kyoto Encyclopedia of Genes and Genomes (KEGG) | http://www.genome.jp/kegg/ | (Kanehisa et al., 2008) |
| Pharmacogenetic Effect Database (PharmGED) | http://bidd.cz3.nus.edu.sg/phg/ | (Zheng et al., 2007) |
| Pharmacogenetics and Pharmacogenomics Knowledge Base (PharmGKB) | http://www.pharmgkb.org | (Hernandez-Boussard et al., 2008) |
| Potential Drug Target Database (PDTD) | http://www.dddc.ac.cn/pdtd/ | (Gao et al., 2008) |
| PubChem | http://pubchem.ncbi.nlm.nih.gov/ | (Wheeler et al., 2008) |
| Search Tool for Interactions of Chemicals (STITCH) | http://stitch.embl.de | (Kuhn et al., 2008) |
Databases possessing a combination of the following characteristics were selected for review: inclusion of chemical-gene/protein relationships; inclusion of chemical-disease relationships; molecular pathway representation; cross-species data; external database cross-links; use of manual curation; breadth of data; user-friendly interface; data analysis capabilities; and public availability (Table I). Databases were described broadly by their content, data statistics (where available), query capabilities, results and analysis capabilities. They are clustered into two categories – those that emphasize environmental chemicals and those that emphasize drugs and small molecules (Table II). This review is not intended to be comprehensive but instead samples databases that provide a range of content and functionality while aiming to enhance understanding about chemical actions.
Table II. Feature summary of public databases with an emphasis on environmental chemicals or drugs and small molecules.
A check indicates the presence of a feature, however the extent of these features (e.g., data content, functionality, curation) varies widely among the databases. A dash indicates the absence of a feature
| Features | Environ- mental |
Drugs and Small Molecules | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CTD | ChemBank | CEBS | DrugBank | KEGG | PharmGED | PharmGK B |
PDTD | PubChem | STITCH | |
| Chemicals | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Genes/proteins | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | — | ✓ |
| Diseases/phenotypes | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | — | — |
| Chemical- gene/protein associations | ✓ | ✓ | ✓ | ✓ | — | ✓ | ✓ | ✓ | — | ✓ |
| Chemical-disease relationships | ✓ | ✓ | ✓ | ✓ | — | ✓ | ✓ | ✓ | — | — |
| Molecular pathway data | ✓ | — | — | ✓ | ✓ | — | ✓ | ✓ | — | ✓ |
| Cross-species data | ✓ | ✓ | ✓ | ✓ | ✓ | — | — | — | — | ✓ |
| Diverse external database links | ✓ | ✓ | ✓ | ✓ | ✓ | — | ✓ | — | ✓ | — |
| Manual literature curation | ✓ | ✓ | — | ✓ | ✓ | ✓ | ✓ | ✓ | — | — |
| Data analysis capabilities | ✓ | ✓ | ✓ | ✓ | — | — | ✓ | — | — | ✓ |
| Publicly available | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
Databases
Environmental chemicals
The Comparative Toxicogenomics Database (CTD) is a publicly available database and research tool for building hypotheses about the effects of environmental chemicals on human disease (Mattingly et al., 2006). It is the only highly curated database that focuses on the actions of environmental chemicals in invertebrates and vertebrates. CTD provides a unique set of curated data describing specific molecular interactions between chemicals, genes and proteins as well as chemical- and gene-disease relationships (Figure 1). Currently, CTD contains 121.166 molecular interactions involving 4.043 chemicals and 13,553 genes; 6,248 gene-disease relationships; and 3,067 chemical-disease relationships. Integration of these three types of binary relationships allows users to make novel connections between chemicals, genes/proteins and human diseases (Figure 1). For example, CTD now contains over 381,000 and 84,000 inferred gene-disease and chemical-disease relationships, respectively. All data in CTD are curated using controlled vocabularies that ensure consistency of curation and data retrieval in addition to providing hierarchical searching options. For example, users may search for chemicals or diseases by specific terms (e.g., mercury or breast cancer, respectively) or broad categories (e.g., heavy metals or cancer, respectively). Users may query data from a range of perspectives including chemicals, genes/proteins, curated interactions, diseases or references. In addition to manually curated data, CTD integrates data for over 124,000 chemicals, 2.6 million sequences and their GO and KEGG pathway annotations, 128,000 taxonomic terms and 6,300 human diseases. Batch query and data download options facilitate meta-analyses of datasets in CTD.
Figure 1. Curated data relationships in the Comparative Toxicogenomics Database (CTD).
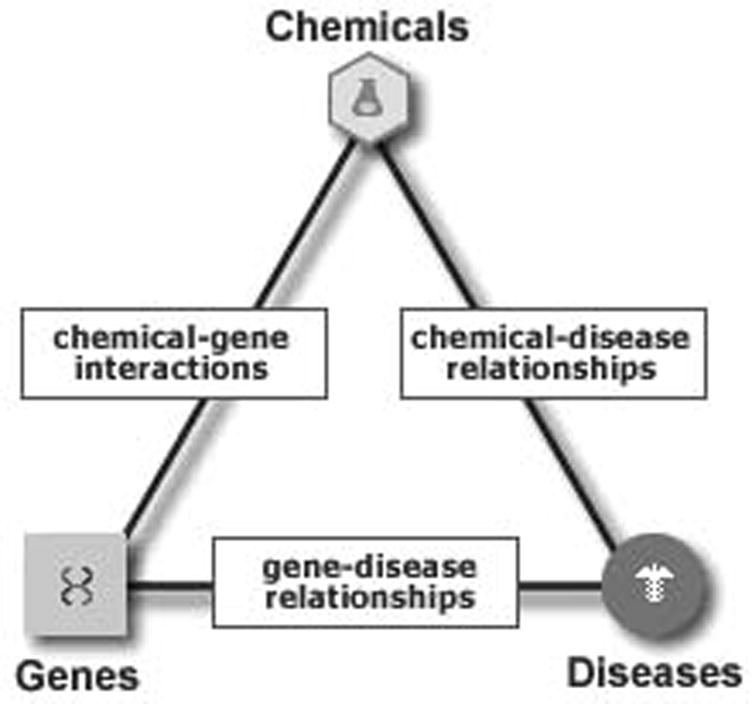
CTD manually curates chemical-gene/protein interactions and chemical- and gene-disease relationships from the peer-reviewed published literature. By integrating these relationships and interactions, CTD facilitates development of hypotheses about the etiologies of environmentally influenced diseases. Other databases described in this review address various aspects of these essential relationships between drugs and small molecules, genes/proteins and diseases/phenotypes.
Drugs and small molecules
ChemBank provides data derived from small molecules and small-molecule screens, and aims to guide the synthesis of novel compounds, identification of small molecules that perturb biological pathways, and the drug discovery process (Seiler et al., 2008). ChemBank provides information on hundreds of thousands of small molecules and hundreds of biomedically relevant assays performed at the Broad Institute in collaboration with the international research community. Users may search the data by assay, protein or small molecules by name, substructure, similarity or function. All proteins are associated with a small molecule in some way, but some associations are not necessarily apparent on the pages at this time. Representation of these associations will be made more complete in the future (personal communication). Manual curation uses controlled vocabularies and is focused on capturing information about the biological activity of small molecules and the biological process tested in small-molecule assays. ChemBank provides analysis tools to interrogate the data provided. Query mechanisms and analysis tools are not necessarily intuitive but there are substantial help pages and tutorials to assist new users.
The Chemical Effects in Biological Systems (CEBS) is a public repository and tool for toxicogenomics research that includes microarray, proteomics, clinical chemistry, hematology and histopathology data (Waters et al., 2008). Users may access data through a number of “browse and search” mechanisms. Users may search proteomics data to identify proteins with differential responses to treatments. Multiple microarray studies can be selected and used to conduct a range of comparative analyses such as identifying differentially expressed genes or results based on platform type. Unique to microarray databases, CEBS users may also search individual subjects based on response. For example, microarray data may be identified for all subjects with elevated alanine aminotransferase, which is a measure of liver toxicity. Data are integrated with study design information (e.g., timeline, subject description, chemical exposures), histopathology data and information from external data sources such as KEGG (Kanehisa et al., 2008). CEBS aims to be a cross-species database and currently contains data from human, mouse, rat, yeast and zebrafish. A new release of CEBS is planned that will substantially expand the current data mining and analysis capabilities.
DrugBank integrates drug data with information about drug targets including sequences, structures and pathways (Wishart et al., 2008). The database contains approximately 4,800 drug entries including over 1,480 FDA-approved small molecule drugs, 128 FDA-approved biotech (protein/peptide) drugs, 71 nutraceuticals and over 3,200 experimental drugs. Additionally, more than 2,500 non-redundant drug target sequences are linked to these FDA approved drug entries. Users may browse data by chemical categories (e.g., approved drugs, nutraceuticals) or search for particular compounds of interest. Results are summarized by chemical with associated formulas, structures, CAS numbers, therapeutic category and indication. Each chemical is linked to a “drugcard,” that summarizes information largely integrated from other resources about the drug and its targets/proteins. Chemical-drug relationships are established by a combination of text mining and expert review.
The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a collection of 19 databases containing information about genes and proteins (GENES), chemical substances (LIGAND), molecular diagrams of interaction networks (PATHWAY), hierarchies and relationships of biological objects (BRITE), structure information about all approved drugs in Japan and the United States (DRUGS) and simple relationships between disease genes, pathways, drugs and diagnostic markers (DISEASE) (Kanehisa et al., 2008). Users may search the databases by gene, pathway, chemical or disease. Currently, KEGG contains 371 reference pathways, over 15,000 compounds, 7,000 drugs, 10,000 glycans and 7,600 reactions. PATHWAY, BRITE, LIGAND and DRUG data are manually curated whereas much of the GENE database information is integrated from other sources or computationally derived. Data are highly integrated, allowing navigation between different datasets. Interactions and relationships between entities are provided in pathways and on detail pages; however, specifics about these interactions/relationships are not described and their precise source references are not provided clearly.
The Pharmacogenetic Effect Database (PharmGED) integrates information about the effects of protein polymorphisms, non-coding region mutations, splicing alterations or expression variations on drug response (Zheng et al., 2007). It currently contains 1,825 entries covering 266 distinct proteins, 693 polymorphisms, and 414 drugs/ligands cited in 856 references. Users may search by names of proteins, drugs/ligands, and diseases or by drug class. Query results are returned in table format anchored by the query type: proteins and their associated gene symbols, polymorphism rules (which are undefined but presumably refer to known polymorphisms), drug/ligand names and/or drug classifications. Querying by protein name retrieves additional information including descriptions of protein function, and pharmacogenetic effects. These data appear to be constructed using free text, rather than controlled vocabularies. The process by which associations are made between proteins, diseases and drugs is not described.
The Pharmacogenetics and Pharmacogenomics Knowledge Base (PharmGKB) aims to facilitate understanding how genetic variation contributes to drug response (Hernandez-Boussard et al., 2008). PharmGKB is part of the NIH Pharmacogenetics Research Network, a national collaborative research consortium. It provides manually curated relationships between drugs, diseases/phenotypes and genes including their variations and gene products. Currently, PharmGKB contains data for 608 genes with variants, 386 variants of interest, 546 drugs, 53 pathways, 542 diseases, 36 Very Important Pharmacogenes (an initiative to provide annotated information about genes, alleles, haplotypes and splice variants of particular relevance for pharmacogenetics and pharmacogenomics) and 2 SNP arrays. PharmGKB also permits registered users to submit primary data, including genotype, phenotype, pathway, SNP array, microarray and other data. Users may search data by gene, drug, disease, pathways, submissions and variants. Data in PharmGKB is highly integrated allowing users to navigate among related data.
The Potential Drug Target Database (PDTD) provides information about structurally solved proteins that are potential targets of drugs (Gao et al., 2008). The database is integrated with a molecular docking tool, TarFis Dock that allows users to query potential protein targets using the molecular structure of a drug, drug candidate or other compound. PDTD contains information for over 1,200 entries with 834 known and potential drug targets. Users may search data by a Protein Data Bank (PDB) ID, target name or disease. Query results are returned in table format anchored by the resulting PDB IDs and their associated biochemical type (e.g., receptor), therapeutic area (e.g., neoplastic diseases), target name (i.e., protein name) and related disease. Protein-disease relationships are identified from the literature and databases such as the Therapeutic Target Database (TTD), Thomson Pharma and DrugBank. The process and extent of manual curation and the use of controlled vocabularies are not described. External links to other databases with associated data are provided.
PubChem is part of the NIH Molecular Library Roadmap Initiative and provides users with information on the biological activities of small molecules (Wheeler et al., 2008). It comprises three primary databases, PCSubstance, PCCompound, and PCBioAssay that are integrated with each other and other NCBI resources. PCSubstance records contain substance information electronically submitted by identified depositors. PCCompound contains a non-redundant set of chemical structures and calculated properties such as molecular weight. PubMed queries can be executed using chemical terms from records in the Substance and Compound databases. These linked queries leverage PubMed MeSH annotation for chemicals but no other filters such as associated diseases or molecular interactions. PCBioAssay provides information about bioactivity screens of chemicals. Users may search by chemical terms, accession IDs or structure. The latter enables chemical similarity searches.
The Search Tool for Interactions of Chemicals (STITCH) aims to help researchers explore known and predicted interactions between chemicals and proteins (Kuhn et al., 2008). STITCH contains information on the interactions between more than 68,000 chemicals, including 2,200 drugs, and 1.5 million proteins across 373 species. The data set is large, however, coverage among different species is variable and most relationships are computationally predicted. Specifically, protein-protein interactions are computationally derived and incorporated from the STRING database using information from genomic context, gene fusion and co-expression (von Mering et al., 2007). Chemical-protein relationships are predicted by text-mining the literature. Users may search STITCH data by chemical and protein names and accession IDs. Results include a predicted summary network of associated chemicals and proteins and experiments, databases and text mining results from which relationships were derived. Tools for manipulating networks are provided.
Other resources for chemicals, drugs and small molecules
In addition to the databases described above, there are other resources that facilitate incorporation, integration and curation of chemically based data. Two commonly used sources for controlled vocabularies include the National Library of Medicine’s Medical Subject Headings (MeSH; http://www.nlm.nih.gov/mesh/MBrowser.html) and Chemical Entities of Biological Interest (ChEBI; http://www.ebi.ac.uk/chebi/) (Nelson et al., 2002; Degtyarenko et al., 2008). MeSH is a hierarchical controlled vocabulary that covers diverse concepts from organisms to the humanities (Nelson et al., 2002). The component dedicated to “Chemicals and Drugs” consists of more general descriptors as well as supplementary concepts, or specific chemicals and their synonyms. Records include descriptive notes, annotations and chemical registry numbers. ChEBI is an ontology of primarily small chemical compounds defined as constitutionally or isotopically distinct atom, molecule, ion, ion pair, radical, radical ion, complex, conformer, etc. (Degtyarenko et al., 2008). Relationships between the compounds, such as “is a conjugate base of,” are specified. ChEBI uses IUPAC and IUBMB-approved nomenclature. Records include different types of chemical structures, chemical properties and registry numbers. Other repositories of chemical and small molecule information include ChemID (http://chem.sis.nlm.nih.gov/chemidplus/), Superdrug (http://bioinf.charite.de/superdrug/) and ChemDB (http://cdb.ics.uci.edu/CHEM/Web/) (Goede et al., 2005; Chen et al., 2007).
Conclusion
Recent years have ushered an exponential growth of information from diverse areas of research bound by the common goal of understanding how environmental chemicals, drugs and small molecules interact with a cell’s molecular machinery to cause disease or to remedy its emergence. These vast datasets have necessitated the development of creative database solutions to aid in their archival, but more importantly, to provide investigators with a set of unique tools with which they can generate hypotheses, establish connections between otherwise disconnected datasets and create opportunities for new fields of inquiry within the area of toxicogenomics. This article has attempted to summarize the currently available chemical databases for toxicogenomics research, pointing out their strengths and complementarities and aiming to provide the reader with a broad sense of their power and utility.
Acknowledgments
CTD is supported by NIH grants from the National Institute of Environmental Health Sciences (ES014065) and the INBRE program of the National Center for Research Resources (RR016463). The author thanks the CTD team for their hard work and dedication and Dr. AJ Planchart for critically reading this manuscript.
Footnotes
Conflict of Interest Statement
The author declares she has no financial conflicts of interest. Dr. Mattingly and the CTD group have worked collaboratively with developers of the CEBS, DrugBank and PubChem databases to establish reciprocal links between their resources.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chen JH, Linstead E, Swamidass SJ, Wang D, Baldi P. ChemDB update--full-text search and virtual chemical space. Bioinformatics. 2007;23:2348–2351. doi: 10.1093/bioinformatics/btm341. NLM Medical Subject Headings; http://www.nlm.nih.gov/mesh. [DOI] [PubMed]
- Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcantara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. The Molecular Biology Database Collection: 2008 update. Nucleic Acids Res. 2008;36:D2–4. doi: 10.1093/nar/gkm1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Li H, Zhang H, Liu X, Kang L, Luo X, Zhu W, Chen K, Wang X, Jiang H. PDTD: a web-accessible protein database for drug target identification. BMC bioinformatics. 2008;9:104. doi: 10.1186/1471-2105-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goede A, Dunkel M, Mester N, Frommel C, Preissner R. SuperDrug: a conformational drug database. Bioinformatics. 2005;21:1751–1753. doi: 10.1093/bioinformatics/bti295. [DOI] [PubMed] [Google Scholar]
- Hernandez-Boussard T, Whirl-Carrillo M, Hebert JM, Gong L, Owen R, Gong M, Gor W, Liu F, Truong C, Whaley R, Woon M, Zhou T, Altman RB, Klein TE. The pharmacogenetics and pharmacogenomics knowledge base: accentuating the knowledge. Nucleic Acids Res. 2008;36:D913–918. doi: 10.1093/nar/gkm1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M, von Mering C, Campillos M, Jensen LJ, Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008;36:D684–688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly CJ, Rosenstein MC, Davis AP, Colby GT, Forrest JN, Jr, Boyer JL. The comparative toxicogenomics database: a cross-species resource for building chemical-gene interaction networks. Toxicol Sci. 2006;92:587–595. doi: 10.1093/toxsci/kfl008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SJ, Powell T, Humphreys BL. The Unified Medical Language Sstem (UMLS) Project. In: Kent A, Hall CM, editors. Encyclopedia of Library and Information Science. Marcel Dekker, Inc; New York: 2002. pp. 369–378. [Google Scholar]
- Seiler KP, George GA, Happ MP, Bodycombe NE, Carrinski HA, Norton S, Brudz S, Sullivan JP, Muhlich J, Serrano M, Ferraiolo P, Tolliday NJ, Schreiber SL, Clemons PA. ChemBank: a small-molecule screening and cheminformatics resource database. Nucleic Acids Res. 2008;36:D351–359. doi: 10.1093/nar/gkm843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Kruger B, Snel B, Bork P. STRING 7--recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M, Stasiewicz S, Merrick BA, Tomer K, Bushel P, Paules R, Stegman N, Nehls G, Yost KJ, Johnson CH, Gustafson SF, Xirasagar S, Xiao N, Huang CC, Boyer P, Chan DD, Pan Q, Gong H, Taylor J, Choi D, Rashid A, Ahmed A, Howle R, Selkirk J, Tennant R, Fostel J. CEBS--Chemical Effects in Biological Systems: a public data repository integrating study design and toxicity data with microarray and proteomics data. Nucleic Acids Res. 2008;36:D892–900. doi: 10.1093/nar/gkm755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Ostell J, Pruitt KD, Schuler GD, Shumway M, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Tatusov RL, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng CJ, Han LY, Xie B, Liew CY, Ong S, Cui J, Zhang HL, Tang ZQ, Gan SH, Jiang L, Chen YZ. PharmGED: Pharmacogenetic Effect Database. Nucleic Acids Res. 2007;35:D794–799. doi: 10.1093/nar/gkl853. [DOI] [PMC free article] [PubMed] [Google Scholar]


