Abstract
In two decades the study of circadian rhythms in cyanobacteria has gone from observations of phenomena in intractable species to the development of a model organism for mechanistic study, atomic-resolution structures of components, and reconstitution of a circadian biochemical oscillation in vitro. With sophisticated biochemical, biophysical, genetic, and genomic tools in place, the circadian clock of the unicellular cyanobacterium Synechococcus elongatus is poised to be the first for which a systems-level understanding can be achieved.
Introduction
Initial observations of what are now accepted as clock-controlled processes in cyanobacteria were reported in the mid-1980s by scientists working with natural isolates of diverse species (Ditty et al. 2003). Diurnal rhythms in nitrogen fixation, photosynthesis, amino acid uptake, and cell division were not immediately interpreted as the consequence of an endogenous biological clock. At least two groups recognized that these rhythms were reminiscent of eukaryotic circadian cycles (Sweeney and Borgese 1989; Huang et al. 1990), and the group of T.-C. Huang performed experiments to demonstrate that rhythms of amino acid uptake in Synechococcus (now Cyanothece) RF-1 persist in constant conditions, reset the timing of peak activity in response to an environmental signal, and are temperature compensated (Huang et al. 1990). With these three criteria fulfilled, a cyanobacterial rhythm was demonstrated to share the hallmarks of eukaryotic circadian rhythms (Dunlap et al. 2004).
Emergence of Synechococcus elongatus PCC 7942 as a prokaryotic model organism for circadian research
None of the strains in which rhythmic behavior were first identified are amenable to genetic manipulation, which left the investigation of the cyanobacterial circadian mechanism at a standstill until a genetic model system could be developed. The cyanobacterium Synechococcus elongatus had not been reported to exhibit any rhythmic behaviors, but S. elongatus PCC 7942 was then and remains among the best-developed cyanobacterial genetic systems, and our laboratory was at the forefront of developing molecular tools and protocols for the species (Golden et al. 1987; Golden 1988). M.R. Schaefer in the S. Golden laboratory had noted rhythmic reporter expression in S. elongatus during growth in continuous culture conditions over several days (unpublished data). Technician C.A. Strayer had already created marine bacterial luciferase gene fusions as a tool for studying light-regulated photosynthesis genes, based on work in Streptomyces by A. Schauer (Schauer et al. 1988). Moreover, we were aware of the reports of circadian rhythmicity in Synechococcus (Cyanothece) RF-1 through interactions with our colleague V. Cassone. Circadian biologists C.H. Johnson (Vanderbilt University) and T. Kondo (then at the National Institute for Basic Biology in Okazaki, Japan) were in search of a tractable microbial circadian system, and they contacted our lab with the premise that perhaps all cyanobacteria have a clock, and that reporter genes might turn a genetically tractable strain into a circadian model organism, as had been done by the S.A. Kay laboratory for the plant Arabidopsis thaliana (Millar et al. 1992). A cyanobacterial genetic model was born in 1992, when expression of luciferase, driven by the promoter for the photosynthesis gene psbAI, proved to be robustly rhythmic (Kondo et al. 1993).
During the next few years, an international team established baseline data and sophisticated manipulation and analysis tools for circadian rhythms in S. elongatus. T. Kondo and colleague M. Ishiura in Okazaki were joined by students S. Aoki and S. Kutsuna; C.H. Johnson at Vanderbilt by student Y. Liu; and our lab at Texas A&M by postdoctoral scientists N.F. Tsinoremas and C.R. Andersson, and student N.V. Lebedeva. Together, these researchers quickly showed that luciferase accurately reports gene expression in S. elongatus (Liu et al. 1995a), that essentially all loci in the organism are expressed rhythmically (Liu et al. 1995b), and that circadian rhythms of gene expression continue even when cells are dividing faster than once per day (Kondo et al. 1997). A critical advance was the design by T. Kondo of an instrument and software that could acquire bioluminescence data from thousands of colonies over the course of several days, allowing the detection of mutants defective in specific circadian properties (Kondo et al. 1994). This custom instrument, affectionately referred to in the Golden and Johnson labs as the ‘Kondotron’ (and more modestly as ‘the turntable’ in Japan) was used extensively in the Kondo-Ishiura laboratory. Large-scale screening capabilities were extended to the Johnson and Golden labs using, respectively, Kondotron technology and the Packard TopCount luminometer which had been developed for circadian use by the S.A. Kay laboratory (Strayer et al. 2000).
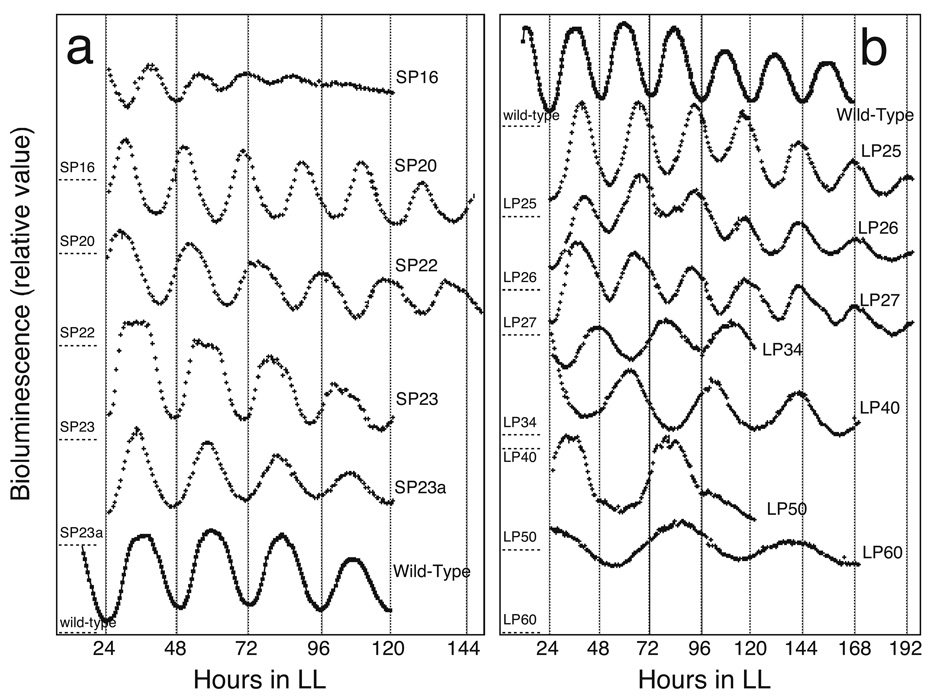
The first mutants to emerge from large-scale mutagenesis procedures turned out to be the most central (Kondo et al. 1994); however, they were not the first whose loci were identified (Tsinoremas et al. 1996; Kutsuna et al. 1998), owing to the greater ease of retrieving physically-tagged loci than chemically-induced mutant alleles. Kondotron screening yielded a plethora of mutants with altered circadian periods or arrhythmic reporter gene expression after chemical mutagenesis (Kondo et al. 1994) (Fig. 1). Strikingly, all mutant phenotypes were complemented or otherwise altered by the introduction of a single segment of S. elongatus DNA from a library (Ishiura et al. 1998). The overlapping segment among several positive library clones carried three open reading frames that did not match any characterized genes in sequence databases and bore no resemblance to cloned clock genes from eukaryotic organisms. They were named kaiA, kaiB, and kaiC for the Japanese kanji Kai, which implies a cycle. Subsequent work in the Kondo lab would show conclusively that their products comprise a circadian oscillator (Nakajima et al. 2005).
Fig. 1.
Bioluminescence rhythms of clock mutants that display altered periods. Free-running bioluminescence rhythms of period mutants together with that of wild type under the standard assay conditions (36 min white light at 46 µE m-2s-1 followed by 9 min of darkness, 30°C) are shown. A, short period mutants and wild type; B, long period mutants and wild type. For all experiments, colonies were grown in LL, given a 12-hour dark pulse, then returned to LL for the measurement of rhythmicity depicted here. Vertical scales for bioluminescence intensity are approximately equivalent. The baseline (dark) levels for the traces are shown by dashed lines on the left vertical axes that are labeled with the name of the mutant. Figure and legend reprinted with permission from (Kondo et al. 1994).
The Kai oscillator of S. elongatus
The kaiABC locus is arranged as three adjacent genes, with a promoter preceding kaiA and kaiB (Ishiura et al. 1998). The kaiB and kaiC genes are expressed as a dicistronic operon. Because overexpression of kaiA stimulates activity of the kaiBC promoter and overexpression of kaiC represses it, parallels were drawn to the transcription-translation feedback loops of eukaryotic circadian clock models (Ishiura et al. 1998). From the beginning, however, some of the data did not fit this scheme. For example, the influence of kaiC on kaiBC expression is not solely negative; KaiC is necessary for full expression of the kaiBC promoter. Subsequent work would show that the KaiABC oscillator uses a fundamentally post-translational mechanism of action (Xu et al. 2003; Nakahira et al. 2004; Ditty et al. 2005; Tomita et al. 2005).
The kaiA, kaiB, and kaiC nucleotide sequences yielded little predictive information regarding their functions (Ishiura et al. 1998). Their only homologs at the time of their initial description in 1997 were unannotated open reading frames in the first entirely sequenced cyanobacterial genome, that of Synechocystis PCC 6803 (http://bacteria.kazusa.or.jp/cyanobase/). Among dozens of finished cyanobacterial genome sequences available today (http://www.genomesonline.org/gold.cgi), the kaiB and kaiC genes are present and adjacently arranged in all but one, Gloeobacter violaceus PCC 7421, which is also the only known strain to lack thylakoid membranes to house the photosynthetic machinery (http://bacteria.kazusa.or.jp/cyanobase/). Potential homologs of kaiB and kaiC are also present in some green and purple photosynthetic bacteria and some Archaea, but kaiA appears to be exclusively cyanobacterial (Dvornyk et al. 2003). Some cyanobacteria, notably the abundant Prochlorococcus species of the open ocean, also lack kaiA.
Only KaiC had some predictable activities. Its Walker box-P loop motifs suggested nucleotide binding (Ishiura et al. 1998), the similarity of its N- and C-terminal halves suggested an iteration in the structure, and general resemblance to the RecA/DNAB superfamily (Leipe et al. 2000; Vakonakis and LiWang 2004) hinted at a multimeric ring structure. Indeed, KaiC binds ATP, autophosphorylates on serine and threonine residues, and forms hexameric rings (Mori et al. 2002; Hayashi et al. 2003; Nishiwaki et al. 2004; Pattanayek et al. 2004; Vakonakis et al. 2004b). The featureless sequences of KaiA and KaiB required that their functions be inferred indirectly by their influence on KaiC phosphorylation.
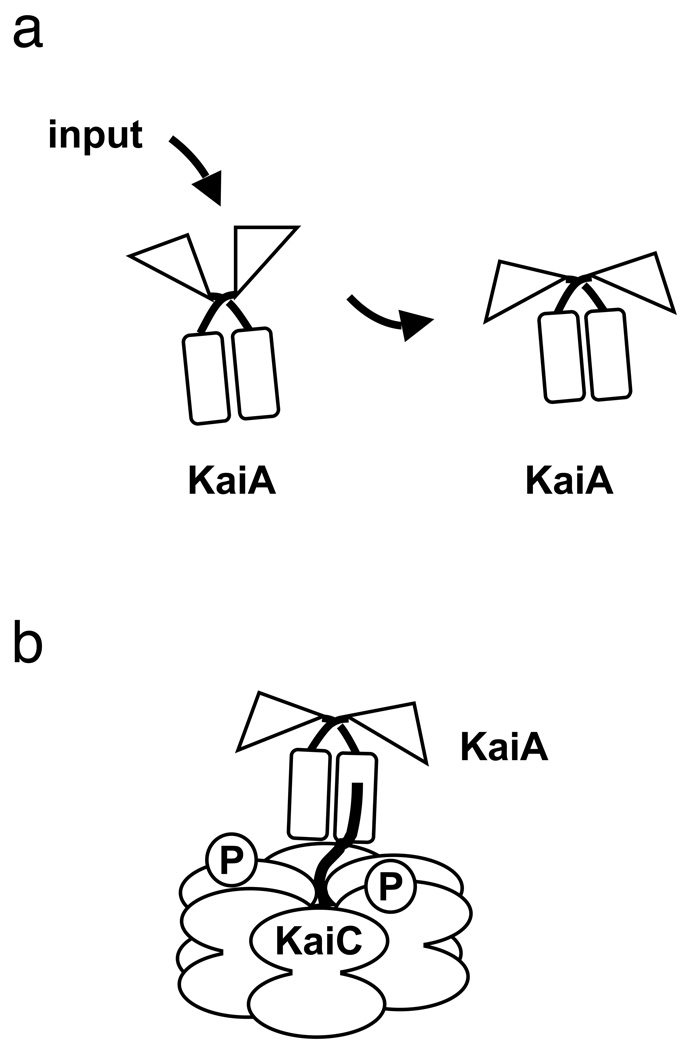
Structural analysis revealed that KaiA has two independently folded domains, of which the C-terminal portion (C-KaiA) is dimerized and shares with the full-length KaiA protein the activity of stimulating KaiC phosphorylation (Williams et al. 2002; Uzumaki et al. 2004; Vakonakis et al. 2004b). The lack of bioinformatics data for this domain reflected genuine novelty, as C-KaiA established a new self-contained, X-class, four-helix bundle fold (Vakonakis et al. 2004b). The N-terminal domain (N-KaiA), not involved in direct interaction with KaiC and not universally present among diverse cyanobacterial species, shares conservation in structure, although not in primary sequence, with signal transduction receiver domains (Williams et al. 2002). The biochemical function of N-KaiA is different from that of bona fide receiver domains as it lacks the active site residues needed for phosphoryl transfer, which is the established activity of receivers. More likely, associations at the interface of N- and C-terminal domains modify interaction of the C-terminal dimer in its interaction with KaiC. Such a mechanism provides an entry point via N-KaiA for external stimuli to impinge on the Kai oscillator (Fig. 2a).
Fig. 2.
(a) Environmental input is predicted to be interpreted (indirectly) by the N-terminal pseudo-receiver domain of KaiA (N-KaiA, triangular domains), likely through its interactions with other input pathway components. This stimulation of N-KaiA is predicted to trigger a conformational change at its C-terminal KaiC-binding domain (C-KaiA, rectangular domains). (b) Interaction of C-KaiA with KaiC stimulates the autokinase activity of KaiC to result in a hyperphosphorylated KaiC protein. The cartoon is not intended to imply specific structural details of protein-protein interactions or phosphorylation sites. Figure and legend adapted with permission from (Mackey and Golden. 2007).
The interaction of KaiA with KaiC occurs specifically with a peptide at the extreme C-terminus of KaiC (Vakonakis and LiWang 2004). The recent structural determination of KaiC reveals that part of this peptide is tucked into the main hexameric ring as a loop, with an extension protruding from the doughnut (Pattanayek et al. 2004). The straightened conformation of this KaiC segment in complex with C-KaiA (Vakonakis and LiWang 2004), the receiver-fold identity of N-KaiA, and the observation that N-KaiA affects KaiC phosphorylation (Williams et al. 2002) (apparently without direct interactions) all come together in the following model, developed in collaboration with A. LiWang, that connects the input pathways of the clock with the oscillator in S. elongatus. We propose that an input pathway impinges on the N-KaiA, altering the interface between the KaiA N- and C-terminal domains and thereby modifying the dimeric structure of C-KaiA (Fig. 2a). This conformational change affects the affinity of C-KaiA for the C-terminal extension on KaiC. In this model, the function of KaiA is to grab the protruding peptide from KaiC and pull the loop out of the folded domain, thereby stimulating the autokinase activity of KaiC (Fig. 2b). In this way, oscillator activity could be hastened or slowed by the input pathways and their influence over KaiA-KaiC binding activity.
KaiB remains the least understood of the Kai oscillator proteins. It is multimeric in structure, having been reported by different groups to be a dimer (Garces et al. 2004) or a tetramer (Hitomi et al. 2005; Iwase et al. 2005); this lack of agreement may reflect a difference among species, genuine alternative quaternary structures of the protein, or simply differences in interpretation of structural data. The known function of KaiB is to block KaiA-induced autophosphorylation of hyper- but not hypophosphorylated KaiC (Williams et al. 2002; Kitayama et al. 2003; Xu et al. 2003; Mori et al. 2007). The combined effects of KaiA and KaiB on KaiC phosphorylation, and the significance of these activities for circadian oscillation, were shown most dramatically by the T. Kondo lab, who reconstituted in vitro a temperature-compensated oscillation of KaiC phosphorylation from a simple mixture of the Kai proteins and ATP (Nakajima et al. 2005). The cycling appearance of a slower-migrating phosphorylated KaiC band recurs with a cycle time of 22–24 hours. C.H. Johnson and colleagues have visualized protein complexes over a time course of this in vitro reaction and confirmed that KaiB comes into play after KaiA is already associated with KaiC (Mori et al. 2007). This finding was predicted by the fact that KaiB does not seem to stimulate dephosphorylation of KaiC in the absence of KaiA (Williams et al. 2002; Kitayama et al. 2003; Xu et al. 2003).
Connecting the oscillator with the outside world
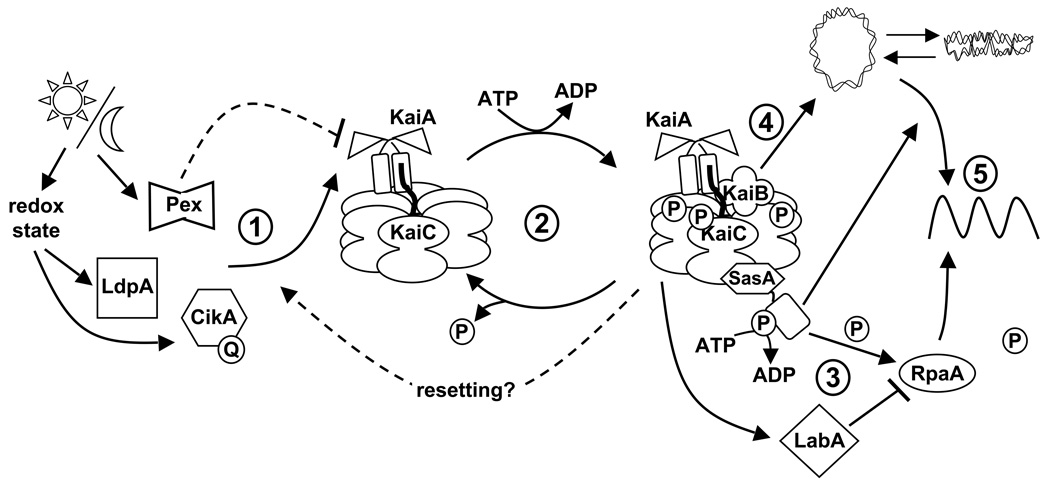
No data have, as yet, confirmed that input pathways converge on N-KaiA. However, some input pathway components have been identified (Fig. 3), and they copurify from cellular extracts with KaiA (Ivleva et al. 2005; Ivleva et al. 2006). The first is LdpA, for light-dependent period, which is an iron-sulfur cluster-containing protein (Katayama et al. 2003). Consistent with its cofactors, LdpA undergoes reduction-oxidation transitions (Ivleva et al. 2005). The protein is named for the phenotype of a null mutant, which has a fixed circadian period regardless of ambient light intensity. Wild-type S. elongatus follows Aschoff’s Rule, such that the period is shorter under high light conditions and longer under lower light, with a range of about 24 to 25 h. The ldpA mutant is locked at the short end of the range (Katayama et al. 2003). Over-expression of LdpA lengthens circadian period by about an hour (Ivleva et al. 2005).
Fig. 3.
Schematic representation of the internal timing mechanism in S. elongatus. (1) The Kai-based oscillator is synchronized with the external environment through input pathways. Daily cues, such as fluctuations in light, are interpreted through the input pathway components Pex (described in (Kutsuna et al. 1998)), LdpA, and CikA. LdpA and CikA likely sense changes in light quantity via modifications in metabolism that effect changes in the cellular redox state. LdpA is a redox-active Fe-S protein, and CikA directly binds a quinone molecule (Q), which alters CikA abundance in the cell. Pex protein binds to, and represses expression from, the promoter of kaiA (Kutsuna et al. 1998). The combined information from these proteins is predicted to feed into and slow down the chemical oscillation of the Kai proteins because the absence of any one of the input pathway components results in a shorter circadian rhythm, reflective of a faster clock. (2) The oscillation in KaiC phosphorylation results from the opposing actions of KaiA, which stimulates KaiC autokinase activity, and KaiB, which abolishes the positive effect of KaiA. Increasing evidence supports the hypothesis that the phosphorylation state of KaiC affects the ability of the clock to reset properly. (3) The SasA protein interacts with KaiC through its N-terminus (hexagonal domain), which is similar in sequence, but not structure, to KaiB. Temporal information is transduced from the Kai oscillator to SasA through the stimulation of SasA autophosphorylation by KaiC. RpaA is activated by SasA via transfer of a phosphoryl group. LabA acts as a mediator of negative regulation from KaiC, and is predicted to repress RpaA function (described in (Takai et al. 2006)). (4) Additionally, the Kai oscillator regulates the compaction rhythm of the cyanobacterial nucleoid (double-stranded loop), which would control accessibility of transcriptional machinery to promoter regions. (5) SasA is not required for rhythmic chromosome compaction, but it is necessary (as is RpaA) for overt rhythmicity of gene expression. Arrows and perpendicular lines indicate positive and negative regulation, respectively. The cartoon is not intended to imply specific structural details of protein-protein interactions or phosphorylation sites. Figure and legend adapted with permission from (Mackey and Golden. 2007).
Another factor that is needed to transmit environmental cues to the circadian clock is CikA (circadian input kinase), a histidine protein kinase whose activity is required for resetting the phase of circadian rhythms (Schmitz et al. 2000). A cikA null mutant is ‘blind’ to a 5 h dark pulse administered during free-running conditions in constant light; in contrast the wild type will reset the timing of the next peak by up to 8 h, depending on the circadian time at which the dark pulse is administered. When either LdpA or CikA is expressed with a 6-histidine affinity tag and retrieved from cyanobacterial cells, both KaiA and KaiC copurify with them (Ivleva et al. 2005; Ivleva et al. 2006). Likewise, His-tagged KaiC retrieves CikA (Ivleva et al. 2006). These data provide biochemical evidence that the input and oscillator divisions of the clock are physically associated in S. elongatus.
CikA includes a GAF, a histidine protein kinase (HPK), and a receiver-like domain (Mutsuda et al. 2003). Although GAF is a chromophore-binding domain in phytochrome-type photoreceptors, it does not seem to serve that function in CikA: the cysteine residue necessary for bilin attachment is absent, and CikA purified from S. elongatus does not carry a covalently bound cofactor. The HPK domain conforms both in sequence and biochemical activity with well-characterized members of this family from diverse bacteria. The receiver-like domain is recognizable in sequence as a member of that class, but lacks the crucial aspartate residue needed to carry out receiver phosphoryl transfer activity. Thus, like N-KaiA, the C-terminal domain of CikA is a pseudo-receiver (PsR) (Mutsuda et al. 2003; Gao et al. 2007).
The PsR domain of CikA serves several important functions in the protein (Ivleva et al. 2006; Zhang et al. 2006). It represses kinase activity, likely by associating with the HPK domain as would a bona fide receiver and thereby occluding the active site histidine (Zhang et al. 2006; Gao et al. 2007). Removal of PsR markedly increases autophosphorylation activity of the HPK domain (Zhang et al. 2006). Fusion of a fluorescent protein, ZsGreen, to the N terminus of CikA produced a fully functional protein that localizes to the pole of the cell. Removal of the PsR domain blocked localization and left the fluorescent fusion distributed throughout the cell. These results are consistent with genetic evidence that the PsR domain is used to interact with one or more partners, and that those components are related to the clock. Whereas a cikA null mutant still exhibits circadian rhythms, albeit with a reduced amplitude and shortened circadian period, a strain in which cikA is over-expressed is arrhythmic (Mutsuda et al. 2003; Zhang et al. 2006). This cessation of rhythmicity is dependent upon the presence of the PsR domain in the over-expressed protein, suggesting that this domain titrates out components that are essential for the circadian cycle (Zhang et al. 2006). Most recently, the PsR domain was shown to bind a quinone cofactor, implicating CikA as a direct redox sensor (Ivleva et al. 2006).
Despite several forward genetic screens aimed at identification of clock-related photoreceptors and the mutation of candidate genes of this class, there are no data to suggest that light information is transmitted directly to the clock in S. elongatus. The known input components implicate cellular redox state, which varies as a function of photosynthetic electron transport, as a proxy for light in setting the clock (Ivleva et al. 2005; Ivleva et al. 2006). The extensive photosynthetic apparatus, so central to this obligate photoautotroph, may fulfill the role of photoreceptor for the circadian system.
Work is in progress to identify the links between the known input components LdpA and CikA and the circadian oscillator. Candidate pathway constituents are under investigation to determine whether there is a specific flow of environmental information from these factors to the N-terminal domain of KaiA as proposed in the model (S.R. Mackey and S.S. Golden, unpublished data). We are developing fusions of oscillator proteins to the yellow fluorescent protein, YFP, in order to determine cellular localization of oscillator proteins during the circadian cycle (G. Dong and S.S. Golden, unpublished data).
Connecting the oscillator with the functions it controls
In S. elongatus, the best-documented clock-controlled process is the global expression of genes throughout the genome (Liu et al. 1995b). Two cellular mechanisms are thought to contribute to rhythmic gene expression (Fig. 3). A signal transduction pathway comprised of an HPK protein called SasA (Synechococcus adaptive sensor) and its probable response regulator partner RpaA (regulator of phycobilisome-associated) provide a direct route for information to flow from the oscillator to gene expression (Iwasaki et al. 2000; Takai et al. 2006). SasA, whose presumptive sensor domain resembles KaiB at the primary sequence level but not in structure (Iwasaki et al. 2000; Garces et al. 2004; Vakonakis et al. 2004a; Hitomi et al. 2005), interacts physically with KaiC (Iwasaki et al. 2000). This interaction greatly stimulates the autokinase activity of SasA (Iwasaki et al. 2000), and the phosphorylation of RpaA (Takai et al. 2006). The latter protein carries motifs characteristic of DNA-binding proteins. However, a specific binding site for RpaA has not been determined. Mutants defective for either SasA or RpaA have very attenuated rhythmicity of gene expression, although oscillation of expression from the kaiBC promoter, with a shortened period and reduced amplitude, is evident (Iwasaki et al. 2000; Takai et al. 2006). SasA does not affect the phosphorylation of KaiC in vitro, indicating that the flow of information is from the oscillator to SasA (Smith and Williams 2006).
Visualization of the cyanobacterial nucleoid during the circadian cycle has revealed that the chromosome compacts and de-compacts rhythmically (Smith and Williams 2006). Such a comprehensive change in genome topology could explain the universal rhythmicity of promoter activity in this organism and the residual rhythmicity observed in sasA and rpaA mutants. The chromosome compaction cycle is dependent upon the kai genes, but does not require SasA (Smith and Williams 2006). Thus, the clock appears to include output pathways from the oscillator to gene expression using both direct transcription modulation, via SasA and RpaA, and indirectly through wholesale remodeling of the chromosome. The targets of the former pathway, and the mediators of the latter, have yet to be determined.
Discovery of new connections between the clock and cellular activities
Our understanding of the circadian oscillator and the greater clock in S. elongatus has come a long way in a relatively short period of time, but we still don’t know what the oscillator really does, other than carry out rhythmic phosphorylation of KaiC. Although many large-scale mutant hunts have been performed in S. elongatus and many genes related to the clock have been identified, more participants are predicted to exist in forging connection of the circadian oscillator to the cell it serves. The diminutive size of the S. elongatus genome renders feasible a comprehensive examination of each locus for its potential effect on circadian timing. A functional genomics project is under way in which we have mutagenized segments of the genome with transposons to achieve saturation of mutant alleles (Holtman et al. 2005). Individual genes that carry insertion mutations are introduced into bioluminescent reporter strains where they recombine with native chromosomal alleles; each mutant is then screened for alterations in circadian gene expression. Thus far, 95% of loci have been mutagenized, and mutants defective for 700 different loci, approximately one quarter of the genome, have been screened for circadian defects (C.K. Holtman, Y. Chen, and S.S. Golden, unpublished). From the screened pool, 71 genes not previously associated with the clock have been identified as affecting circadian gene expression; most of these cause long or short circadian periods when mutated, or change the phase of peak reporter gene expression. None that cause complete arrhythmia have been identified, other than the known kaiA, kaiB, and kaiC genes. The newly identified genes fall into diverse categories, including many loci of unknown function, some of which are conserved among diverse cyanobacteria and some that are apparently unique to S. elongatus. Some loci of particular interest are predicted to encode redox active proteins, supporting an emerging picture of redox, rather than light itself, as the chief environment input currency in the cell.
Many of the mutants that show circadian phenotypes carry insertion alleles that cannot be segregated entirely from their wild-type counterparts, indicating that the mutated locus is essential for viability (Holtman et al. 2005). In these situations, the phenotype is observed in merodiploid strains that carry both alleles, and presumably have decreased expression of the target gene. Because S. elongatus carries multiple copies of its chromosome, a strain with a mixed population of mutant and wild-type chromosomes may carry a range of ratios of alleles, which is influenced by selective pressure for an antibiotic resistance gene on the transposon.
The first clock-affecting gene found in the genomics screen reinforces a common theme found in eukaryotic circadian systems: proteolysis is an important factor for maintaining normal circadian rhythms in vivo. The clpP2 and clpX genes form an operon, in which the upstream gene encodes the ClpP2 protease and the downstream gene the ClpX chaperone that delivers substrates to the enzyme. Neither can be inactivated completely in S. elongatus, but merodiploids affected in either locus have long-period phenotypes (Holtman et al. 2005). Conditional expression of an antisense RNA that is complementary to and spans the intergenic region of the discistronic RNA phenocopies the (merodiploid) mutant phenotype. Because bacteria do not possess the dicer pathway that is responsible for gene silencing, the gene suppression is temporary and presumably results from direct interference with transcription or translation, or stimulates a pathway that non-specifically recognizes double-stranded RNA. The ability to conditionally suppress genes that are essential for viability provides a valuable tool for assessing the contributions to circadian timing of genes that tie the clock to critical cellular processes.
Although we don’t yet know how the circadian clock orchestrates the life of the cell, we do know that its activity is important for the organism. The wealth of circadian period mutants generated in the early 1990s provided the means to test the extent to which a circadian clock confers an adaptive advantage. Mutant and wild-type strains that have different free-running periods grow equally well when grown in different T cycles (11 h light: 11 h dark; 15 h light: 15 h dark). However, big differences in survival emerge when these strains are mixed in equal numbers and challenged to compete in days of different lengths (Ouyang et al. 1998). Under the pressure of competition, the strain whose free-running period is closest to that of the T cycle always emerges as the predominant strain. Together, these findings suggest that cyanobacteria actively influence the growth of others in the population, and that resonance of circadian period with the prevailing daily cycle confers a fitness advantage.
How representative is S. elongatus?
S. elongatus is one representative among a handful of cyanobacterial strains that are amenable to genetic manipulation (Koksharova and Wolk 2002). Of these, it has the smallest genome, the most limited metabolic range, and no developmental programs. It is a freshwater mesophile that has been isolated from environments in Texas and California (PCC 6301 and PCC 7942, Pasteur Culture Collection, http://www.pasteur.fr/recherche/banques/PCC/index.html). The cyanobacterial clade encompasses very diverse species, including multicellular organisms that differentiate specialized cell types, species that help to form desert crusts, denizens of mats in alkaline hot springs, abundant phytoplankton in the open ocean, and strains that thrive in Antarctica.
The age of genomics has particularly favored the cyanobacterial clade, with more than two dozen genomes from diverse species available. Sequenced genomes vary in size from 1.7–12 Mbp (http://www.genomesonline.org/gold.cgi), and genome size correlates roughly with the range of metabolic and developmental capabilities of a particular species (Rippka et al. 1979; Herdman et al. 2001). The diversity of lifestyles and genetic makeup suggests that cyanobacteria will show variations in circadian machinery. For example, the presence of multiple copies of kaiB and kaiC in some species, which does not correlate with genome size, suggests the action of multiple oscillators, or at minimum a more complicated oscillatory cycle than has been proposed in S. elongatus. Some input pathway features are certain to vary among species. KaiA in Nostoc punctiforme and Anabaena sp. PCC 7120 lacks the N-terminal domain of the S. elongatus protein (Williams et al. 2002), and the oceanic Prochlorococcus species do not encode a KaiA protein at all (Dvornyk et al. 2003). The CikA kinase, which is essential for circadian resetting in S. elongatus, has no clear homologs in other cyanobacteria. Variations among species in the mechanism for the transfer of environmental light information to the clock is not surprising, because their light-harvesting antenna structures and pigments for photosynthesis are also diverse (Glazer 1989; Ting et al. 2002). Thus, although the findings in S. elongatus have been enlightening and instructive, it should not be assumed that all cyanobacterial circadian clocks will operate just as the S. elongatus clock. Variations in circadian mechanism at least as notable as those that distinguish the clocks of fruit flies and mice should be expected.
Potential homologs of the KaiB and KaiC proteins of the S. elongatus circadian oscillator are encoded in the genomes of some other bacteria and some Archaea (Dvornyk et al. 2003). A question still unanswered is whether these proteins serve circadian functions in their hosts, or perhaps carry out ancestral, non-clock activities. Moreover, many clades in the bacterial domain do not have kai homologs, but might have undiscovered, non-homologous clocks, just as plants and fungi have circadian oscillators all their own. Only concerted research efforts in other prokaryotic models will reveal the extent of circadian biological timing in the Tree of Life.
Conclusions
The choice of S. elongatus as the cyanobacterial circadian model was, in retrospect, quite fortuitous. Robust and high amplitude rhythms of bioluminescence were observed from the first luciferase reporter gene that was tested (psbAI promoter), and these clear rhythms facilitated circadian screening. Another popular genetically tractable cyanobacterium, Synechocystis sp. strain PCC 6803, was found to exhibit lower amplitude rhythms in similar assays (Aoki et al. 1995; Aoki et al. 2002). Moreover, now that the oscillator genes have been identified, the presence of additional copies of kaiB and kaiC in the Synechocystis genome (Dvornyk et al. 2003; Chen 2007) suggests that redundancy might have complicated forward genetics had that organism been chosen initially.
The efforts of many talented students and senior scientists have contributed to the richly developed clock model system we enjoy today with S. elongatus. Among those, the creative technological innovations of T. Kondo, which drove the development of phenotypic screening (Kondo et al. 1994) and the in vitro oscillator system, merit special notice (Nakajima et al. 2005). Clock analysis in this organism is now at a very sophisticated stage, with structural detail (Garces et al. 2004; Pattanayek et al. 2004; Ye et al. 2004; Hitomi et al. 2005), mathematical models (Mori et al. 2007), global genomics tools (Holtman et al. 2005), and cellular imaging probes (Zhang et al. 2006). Fifteen years of progress in describing the parts that comprise the oscillator have led us to the threshold of understanding how the cell harnesses this ticking mechanism to build a clock that tunes the life of a cyanobacterium to thrive in the world it inhabits.
Acknowledgments
I am deeply indebted to C.H. Johnson and T. Kondo for the invitation to join the world of chronobiologists, and for their longtime collaboration and friendship. I also gratefully acknowledge the extensive role of M. Ishiura, who contributed richly to the progress described here. They and their students have shared hopes, ideas, and reagents with my group for over a decade. My colleague A. LiWang is the very personification of an effective and enthusiastic collaborator, and the work in my lab owes an ever-increasing debt to him and his students as our ideas and expertise continue to mesh. The members of the Center for Research on Biological Clocks at Texas A&M provide guidance, inspiration, and collegiality on a daily basis. My own students have given tirelessly and enthusiastically of themselves in investigating the clock while retaining “a feeling for the organism” (Keller 1983). I thank them all, with special nods to C.R. Andersson, N.F. Tsinoremas, S.B. Williams, and S.R. (Mini-Me) Mackey. Work in my lab is supported by grants from the National Institutes of Health (P01 NS039546 and R01 GM062419) and the Department of Energy (DE-FG02-04ER15558).
References
- Aoki S, Kondo T, Ishiura M. Circadian expression of the dnaK gene in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 1995;177(19):5606–5611. doi: 10.1128/jb.177.19.5606-5611.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki S, Kondo T, Ishiura M. A promoter-trap vector for clock-controlled genes in the cyanobacterium Synechocystis sp. PCC 6803. Journal of microbiological methods. 2002;49(3):265–274. doi: 10.1016/s0167-7012(01)00376-1. [DOI] [PubMed] [Google Scholar]
- Chen Y. Biology. College Station, TX: Texas A&M University; 2007. Functional genomics of the unicellular cyanobacterium Synechococcus elongatus PCC 7942; p. 188. [Google Scholar]
- Ditty JL, Canales SR, Anderson BE, Williams SB, Golden SS. Stability of the Synechococcus elongatus PCC 7942 circadian clock under directed anti-phase expression of the kai genes. Microbiology. 2005;151(Pt 8):2605–2613. doi: 10.1099/mic.0.28030-0. [DOI] [PubMed] [Google Scholar]
- Ditty JL, Williams SB, Golden SS. A cyanobacterial circadian timing mechanism. Annu Rev Genet. 2003;37:513–543. doi: 10.1146/annurev.genet.37.110801.142716. [DOI] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ, DeCoursey PJ, editors. Chronobiology: Biological Timekeeping. Sunderland. MA: Sinauer Associates, Inc.; 2004. [Google Scholar]
- Dvornyk V, Vinogradova O, Nevo E. Origin and evolution of circadian clock genes in prokaryotes. Proc Natl Acad Sci U S A. 2003;100(5):2495–2500. doi: 10.1073/pnas.0130099100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T, Zhang X, Ivleva NB, Golden SS, LiWang A. NMR structure of the pseudo-receiver domain of CikA. Protein Sci. 2007;16(3):465–475. doi: 10.1110/ps.062532007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces RG, Wu N, Gillon W, Pai EF. Anabaena circadian clock proteins KaiA and KaiB reveal a potential common binding site to their partner KaiC. Embo J. 2004;23(8):1688–1698. doi: 10.1038/sj.emboj.7600190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer AN. Light guides. Directional energy transfer in a photosynthetic antenna. J Biol Chem. 1989;264(1):1–4. [PubMed] [Google Scholar]
- Golden SS. Mutagenesis of cyanobacteria by classical and gene-transfer-based methods. Methods Enzymol. 1988;167:714–727. doi: 10.1016/0076-6879(88)67083-2. [DOI] [PubMed] [Google Scholar]
- Golden SS, Brusslan J, Haselkorn R. Genetic engineering of the cyanobacterial chromosome. Methods Enzymol. 1987;153:215–231. doi: 10.1016/0076-6879(87)53055-5. [DOI] [PubMed] [Google Scholar]
- Hayashi F, Suzuki H, Iwase R, Uzumaki T, Miyake A, Shen JR, Imada K, Furukawa Y, Yonekura K, Namba K, Ishiura M. ATP-induced hexameric ring structure of the cyanobacterial circadian clock protein KaiC. Genes Cells. 2003;8(3):287–296. doi: 10.1046/j.1365-2443.2003.00633.x. [DOI] [PubMed] [Google Scholar]
- Subsection I: (Formerly Chroococcales Wettstein 1924, emend. Rippka, Deruelles, Waterbury, Herdman and Stanier 1979) In: Herdman M, Castenholz RW, Iteman I, Waterbury JB, Rippka R, editors; Boone DR, Castenholz RW, Garrity GM, editors. Bergey's Manual of Systematic Bacteriology. New York: Springler-Verlag; 2001. p. 721. [Google Scholar]
- Hitomi K, Oyama T, Han S, Arvai AS, Getzoff ED. Tetrameric architecture of the circadian clock protein KaiB. A novel interface for intermolecular interactions and its impact on the circadian rhythm. J Biol Chem. 2005;280(19):19127–19135. doi: 10.1074/jbc.M411284200. [DOI] [PubMed] [Google Scholar]
- Holtman CK, Chen Y, Sandoval P, Gonzales A, Nalty MS, Thomas TL, Youderian P, Golden SS. High-throughput functional analysis of the Synechococcus elongatus PCC 7942 genome. DNA Research. 2005;12:103–115. doi: 10.1093/dnares/12.2.103. [DOI] [PubMed] [Google Scholar]
- Huang T-C, Tu J, Chow TJ, Chen T-H. Circadian rhythm of the prokaryote Synechococcus sp. RF-1. Plant Physiol. 1990;92:531–533. doi: 10.1104/pp.92.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281(5382):1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- Ivleva NB, Bramlett MR, Lindahl PA, Golden SS. LdpA: a component of the circadian clock senses redox state of the cell. EMBO J. 2005;24(6):1202–1210. doi: 10.1038/sj.emboj.7600606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivleva NB, Gao T, LiWang AC, Golden SS. Quinone sensing by the circadian input kinase of the cyanobacterial circadian clock. Proc Natl Acad Sci U S A. 2006;103(46):17468–17473. doi: 10.1073/pnas.0606639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Williams SB, Kitayama Y, Ishiura M, Golden SS, Kondo T. A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell. 2000;101(2):223–233. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- Iwase R, Imada K, Hayashi F, Uzumaki T, Morishita M, Onai K, Furukawa Y, Namba K, Ishiura M. Functionally important substructures of circadian clock protein KaiB in a unique tetramer complex. J Biol Chem. 2005;280(52):43141–43149. doi: 10.1074/jbc.M503360200. [DOI] [PubMed] [Google Scholar]
- Katayama M, Kondo T, Xiong J, Golden SS. ldpA encodes an iron-sulfur protein involved in light-dependent modulation of the circadian period in the cyanobacterium Synechococcus elongatus PCC 7942. J Bacteriol. 2003;185(4):1415–1422. doi: 10.1128/JB.185.4.1415-1422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller EF. A Feeling for the Organism: the life and work of Barbara McClintock. Hillsdale, NY: Adonis Press; 1983. [Google Scholar]
- Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. Embo J. 2003;22(9):2127–2134. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koksharova O, Wolk C. Genetic tools for cyanobacteria. App Microbiol Biotech. 2002;58(2):123–137. doi: 10.1007/s00253-001-0864-9. [DOI] [PubMed] [Google Scholar]
- Kondo T, Mori T, Lebedeva NV, Aoki S, Ishiura M, Golden SS. Circadian rhythms in rapidly dividing cyanobacteria. Science. 1997;275(5297):224–227. doi: 10.1126/science.275.5297.224. [DOI] [PubMed] [Google Scholar]
- Kondo T, Strayer CA, Kulkarni RD, Taylor W, Ishiura M, Golden SS, Johnson CH. Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 1993;90(12):5672–5676. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Tsinoremas NF, Golden SS, Johnson CH, Kutsuna S, Ishiura M. Circadian clock mutants of cyanobacteria. Science. 1994;266(5188):1233–1236. doi: 10.1126/science.7973706. [DOI] [PubMed] [Google Scholar]
- Kutsuna S, Kondo T, Aoki S, Ishiura M. A period-extender gene, pex, that extends the period of the circadian clock in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1998;180(8):2167–2174. doi: 10.1128/jb.180.8.2167-2174.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Aravind L, Grishin NV, Koonin EV. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome research. 2000;10(1):5–16. [PubMed] [Google Scholar]
- Liu Y, Golden SS, Kondo T, Ishiura M, Johnson CH. Bacterial luciferase as a reporter of circadian gene expression in cyanobacteria. J Bacteriol. 1995a;177(8):2080–2086. doi: 10.1128/jb.177.8.2080-2086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Tsinoremas NF, Johnson CH, Lebedeva NV, Golden SS, Ishiura M, Kondo T. Circadian orchestration of gene expression in cyanobacteria. Genes Devel. 1995b;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- Mackey SR, Golden SS. Winding up the cyanobacterial circadian clock. Trends in microbiology; 2007. in press. [DOI] [PubMed] [Google Scholar]
- Millar AJ, Short SR, Chua NH, Kay SA. A novel circadian phenotype based on firefly luciferase expression in transgenic plants. The Plant cell. 1992;4(9):1075–1087. doi: 10.1105/tpc.4.9.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Saveliev SV, Xu Y, Stafford WF, Cox MM, Inman RB, Johnson CH. Circadian clock protein KaiC forms ATP-dependent hexameric rings and binds DNA. Proc Natl Acad Sci U S A. 2002;99(26):17203–17208. doi: 10.1073/pnas.262578499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Williams DR, Byrne MO, Qin X, Egli M, McHaourab HS, Stewart PL, Johnson CH. Elucidating the ticking of an in vitro circadian clockwork. PLoS Biol. 2007;5(4):e93. doi: 10.1371/journal.pbio.0050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutsuda M, Michel KP, Zhang X, Montgomery BL, Golden SS. Biochemical properties of CikA, an unusual phytochrome-like histidine protein kinase that resets the circadian clock in Synechococcus elongatus PCC 7942. J Biol Chem. 2003;278(21):19102–19110. doi: 10.1074/jbc.M213255200. [DOI] [PubMed] [Google Scholar]
- Nakahira Y, Katayama M, Miyashita H, Kutsuna S, Iwasaki H, Oyama T, Kondo T. Global gene repression by KaiC as a master process of prokaryotic circadian system. Proc Natl Acad Sci U S A. 2004;101(3):881–885. doi: 10.1073/pnas.0307411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308(5720):414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- Nishiwaki T, Satomi Y, Nakajima M, Lee C, Kiyohara R, Kageyama H, Kitayama Y, Temamoto M, Yamaguchi A, Hijikata A, Go M, Iwasaki H, Takao T, Kondo T. Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc Natl Acad Sci U S A. 2004;101(38):13927–13932. doi: 10.1073/pnas.0403906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci U S A. 1998;95(15):8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayek R, Wang J, Mori T, Xu Y, Johnson CH, Egli M. Visualizing a circadian clock protein: crystal structure of KaiC and functional insights. Molecular cell. 2004;15(3):375–388. doi: 10.1016/j.molcel.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- Schauer A, Ranes M, Santamaria R, Guijarro J, Lawlor E, Mendez C, Chater K, Losick R. Visualizing gene expression in time and space in the filamentous bacterium Streptomyces coelicolor. Science. 1988;240(4853):768–772. doi: 10.1126/science.3363358. [DOI] [PubMed] [Google Scholar]
- Schmitz O, Katayama M, Williams SB, Kondo T, Golden SS. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science. 2000;289(5480):765–768. doi: 10.1126/science.289.5480.765. [DOI] [PubMed] [Google Scholar]
- Smith RM, Williams SB. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci U S A. 2006;103(22):8564–8569. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289(5480):768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- Sweeney BM, Borgese MB. A circadian rhythm in cell division in a prokaryote, the cyanobacterium Synechococcus WH7803. J Phycol. 1989;25:183–186. [Google Scholar]
- Takai N, Nakajima M, Oyama T, Kito R, Sugita C, Sugita M, Kondo T, Iwasaki H. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc Natl Acad Sci U S A. 2006;103(32):12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CS, Rocap G, King J, Chisholm SW. Cyanobacterial photosynthesis in the oceans: the origins and significance of divergent light-harvesting strategies. Trends in microbiology. 2002;10(3):134–142. doi: 10.1016/s0966-842x(02)02319-3. [DOI] [PubMed] [Google Scholar]
- Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307(5707):251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- Tsinoremas NF, Ishiura M, Kondo T, Andersson CR, Tanaka K, Takahashi H, Johnson CH, Golden SS. A sigma factor that modifies the circadian expression of a subset of genes in cyanobacteria. EMBO J. 1996;15(10):2488–2495. [PMC free article] [PubMed] [Google Scholar]
- Uzumaki T, Fujita M, Nakatsu T, Hayashi F, Shibata H, Itoh N, Kato H, Ishiura M. Crystal structure of the C-terminal clock-oscillator domain of the cyanobacterial KaiA protein. Nat Struct Mol Biol. 2004 doi: 10.1038/nsmb781. [DOI] [PubMed] [Google Scholar]
- Vakonakis I, Klewer DA, Williams SB, Golden SS, LiWang AC. Structure of the N-terminal domain of the circadian clock-associated histidine kinase SasA. J Mol Biol. 2004a;342(1):9–17. doi: 10.1016/j.jmb.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Vakonakis I, LiWang AC. Structure of the C-terminal domain of the clock protein KaiA in complex with a KaiC-derived peptide: implications for KaiC regulation. Proc Natl Acad Sci USA. 2004 doi: 10.1073/pnas.0403037101. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakonakis I, Sun J, Wu T, Holzenburg A, Golden SS, LiWang AC. NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: implications for KaiA-KaiC interaction. Proc Natl Acad Sci U S A. 2004b;101(6):1479–1484. doi: 10.1073/pnas.0305516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SB, Vakonakis I, Golden SS, LiWang AC. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: a potential clock input mechanism. Proc Natl Acad Sci U S A. 2002;99(24):15357–15362. doi: 10.1073/pnas.232517099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Mori T, Johnson CH. Cyanobacterial circadian clockwork: roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. Embo J. 2003;22(9):2117–2126. doi: 10.1093/emboj/cdg168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Vakonakis I, Ioerger TR, LiWang AC, Sacchettini JC. Crystal structure of circadian clock protein KaiA from Synechococcus elongatus. J Biol Chem. 2004;279(19):20511–20518. doi: 10.1074/jbc.M400077200. [DOI] [PubMed] [Google Scholar]
- Zhang X, Dong G, Golden SS. The pseudo-receiver domain of CikA regulates the cyanobacterial circadian input pathway. Molecular microbiology. 2006;60(3):658–668. doi: 10.1111/j.1365-2958.2006.05138.x. [DOI] [PubMed] [Google Scholar]