Abstract
Automated and accurate segmentation of the aorta in 4D (3D+time) cardiovascular magnetic resonance (MR) image data is important for early detection of congenital aortic disease leading to aortic aneurysms and dissections. A computer-aided diagnosis method is reported that allows one to objectively identify subjects with connective tissue disorders from sixteen-phase 4D aortic MR images. Starting with a step of multi-view image registration, our automated segmentation method combines level-set and optimal surface segmentation algorithms in a single optimization process so that the final aortic surfaces in all 16 cardiac phases are determined. The resulting aortic lumen surface is registered with an aortic model followed by calculation of modal indices of aortic shape and motion. The modal indices reflect the differences of any individual aortic shape and motion from an average aortic behavior. A Support Vector Machine (SVM) classifier is used for the discrimination between normal and connective tissue disorder subjects.
4D MR image data sets acquired from 104 normal and connective tissue disorder MR datasets were used for development and performance evaluation of our method. The automated 4D segmentation resulted in accurate aortic surfaces in all 16 cardiac phases, covering the aorta from the aortic annulus to the diaphragm, yielding subvoxel accuracy with signed surface positioning errors of −0.07 ± 1.16 voxel (−0.10 ± 2.05 mm). The computer aided diagnosis method distinguished between normal and connective tissue disorder subjects with a classification correctness of 90.4%.
Keywords: Aortic MR Images, congenital heart disease, aortic aneurysm, computer-aided diagnosis (CAD), level-set, optimal surface segmentation, principal component analysis (PCA), Support Vector Machine (SVM)
1 Introduction
Aortic aneurysms and dissections are the 18th leading cause of death in the US, representing 0.6% of all deaths in 2006 [1]. Persons with certain congenital connective tissue disorders, such as Marfan Syndrome and Familial Thoracic Aortic Aneurysm Syndrome are at increased risk of developing aortic aneurysm and/or dissection. Therefore, early diagnosis of connective tissue disorders is of a great importance.
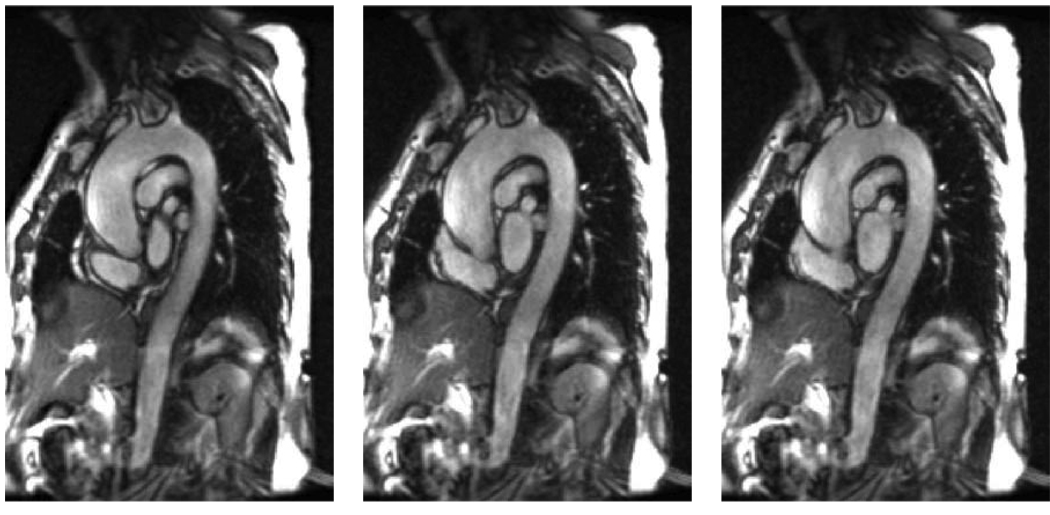
During the last decade, cardiovascular magnetic resonance (MR) imaging contributed substantially to cardiovascular disease diagnosis. A 4D cardiovascular MR image, which consists of 3D images along the cardiac cycle, supplies important functional information about aortic motion that is missing in single-phase 3D images. While there are only limited clinical studies to support the usefulness of motion data, early observations in patients with Marfan syndrome already suggested the importance of examining motion information [2]. The motion information holds promise of improving the diagnosis of the aortic disease. However, 4D MR image analysis is a nontrivial task. When attempting this task manually, obtaining tracings of the aorta in an n-phase 4D image requires expert knowledge and is tedious, time-consuming, and suffers from inter- and intra-observer variability. Fig. 1 shows typical aortic MR data from a connective tissue disorder subject enrolled in this study. Even more importantly, objectively analyzing the small motion and shape differences of the individual aortas throughout the cardiac cycle is difficult if not impossible for a human operator relying solely on qualitative visual means. Therefore, an automated computer-aided diagnosis (CAD) system allowing one to distinguish between normal and abnormal (connective tissue disorder) subjects needs to be developed.
Fig. 1.
Typical aortic MR data from a connective disorder subject enrolled in this study, three cardiac phases shown. Notice that the aorta does not exhibit any aneurysmal malformations.
Many aortic segmentation techniques were developed in 3D using computed tomography (CT) or MR images. Rueckert [3] used Geometric Deformable Models (GDM) to track the ascending and descending aorta. Behrens [4] obtained a coarse segmentation using Randomized Hough Transform (RHT). Bruijne [5] introduced an Adapting Active Shape Model (AASM) for tubular structure segmentation. Subasic [6] utilized a level-set algorithm for segmentation of abdominal aortic aneurysms. Considerable work has been done to establish the abnormalities in vessel mechanics that exist in patients with connective tissue disorders [7–9]. Included among the parameters that have proven useful in assessing the risk of aortic disease progression in Marfan patients is the determination of aortic pulse wave velocity [10].
In a closely related research direction, several authors have proposed techniques for tracking the cardiac movement in 4D cardiac images. Bardinet [11] presented an algorithm for tracking surfaces in 4D cardiac images based on a parametric model. Chandrashekara [12] built a statistical model derived from the motion fields in the hearts of several healthy volunteers to track the movement of the myocardium. McInerney [13] built a dynamic finite element surface model for segmentation. Montagnat [14] presented a 4D cardiac segmentation by introducing time-dependent constraints to the deformable surface framework.
To our knowledge, 4D segmentation algorithms for automated tracking of the aortic movement in 4D have not been presented. In this study, we report a computer-aided diagnosis method for aortic lumen segmentation and subsequent objective identification of subjects with connective tissue disorder from sixteen-phase 4D aortic MR images. Starting with a step of multi-view image registration, this automated CAD system allows simultaneous segmentation of a sequence of 3D luminal surfaces in a 4D aortic MR image, followed by calculation of quantitative indices of aortic shape and motion. The descriptive indices of shape and motion are capable of correctly distinguishing the normal and diseased subjects, which may help with early diagnosis of this congenital disorder.
2 Methods
The reported CAD method consists of two main stages – aortic segmentation and connective tissue disorder assessment. Surface segmentation of the aortic lumen is obtained with an automatic 4D segmentation method. Next, a quantitative method to detect the differences in the aortic 4D function between normal and connective tissue disorder patients is employed to provide quantitative descriptors used in a disease classification step.
2.1 Segmentation
An n-phase 4D (spatio-temporal) image I can be viewed as a discrete set of n volumetric images defined at n different time instants {It (x, y, z)}t∈[0,n−1]. The 4D aortic surface can be viewed as a sequence of surfaces {St}t∈[0,n−1]. During the segmentation stage, the 4D segmentation algorithm consists of the following steps:
Aortic surface presegmentation: A 4D fast marching level set method simultaneously yields approximate 4D aortic surfaces {appSt}t∈[0,n−1].
Centerline extraction: Aortic centerlines are determined from each approximate surface by skeletonization.
Accurate aortic surface segmentation: Accurate 4D aortic surface {St}t∈[0,n−1] is obtained simultaneously with the application of a novel 4D optimal surface detection algorithm.
2.1.1 Aortic Surface Presegmentation
In order to segment the 4D aortic image, 4D fast marching method was used [15]. Starting with an interactively identified seed point within the 4D aortic image, the initial surface propagates in an outward direction with speed F. Let Tq(i, j, k) be the arrival time at which the level set surface passes through point (i, j, k) in phase q of the 4D image. The gradient of this arrival time shall be inversely proportional to the speed function F [15]
| (1) |
The principal idea behind the fast marching methods is to trace the surface according to the solution function Tq(i, j, k) solved using Eq. (1). The upwind difference scheme [15] was used to facilitate numerical solution.
To minimize the tendency of the level set segmentation to leak into the extra-aortic space in the presence of imperfect border properties, a 3D tubular-structure enhancement filter is applied to the original image [16]. The filtering step enhances the appearance of tubular structures and suppresses the appearance of non-tubular structures. This filter is only used for the level set segmentation as follows. Let Gs be the Gaussian-smoothed image with standard deviation s. Let the eigenvalues of the Hessian matrix ∇2Gs be λ1, λ2 and λ3. The vesselness function is defined as:
| (2) |
where α, β and γ are parameters. The value of γ was experimentally determined as half of the value of the maximum Hessian norm; s denotes scale. The ratio RA helps distinguish between plate-like and line-like structures and the ratio RB helps distinguish between blob-like and line-like structures:
| (3) |
The second order index W represents object deviation from the background
| (4) |
For the multiscale vessel filter, the vesselness generates the maximum response at a scale which matches the diameter of the vessel to detect. The final vessel image { VIt (x, y, z)}t∈[0,n−1] can be formed as
| (5) |
Using the vesselness image VIt(x, y, z), the vessel enhanced image VEIt(x, y, z) can be defined by Eq. (6), where θ is a noise/background suppression threshold which suppresses the noise and background. The speed function F can be defined by Eq. (7), where Gσ * V EIt(x, y, z) represents the image smoothed by a Gaussian filter with a characteristic width σ. This definition guides the surface development to stop at a high gradient
| (6) |
| (7) |
Using a binary tree sorting technique, the fast marching method gets the solution with time complexity of O(N log N), where N is the number of visited image points [15]. The fast marching algorithm stops the surface in the vicinity of object boundaries, thus yielding the approximate object surface. The tubular structure enhancement filter limits the level set segmentation leaks to local areas and the resulting inaccuracy of the preliminary segmentation is resolved in the subsequent surface detection step.
In order to achieve an accurate segmentation, a 3D thinning algorithm [17] is applied to the result of presegmentation to extract an approximate aortic centerline for each phase. During each iteration, this method first marks all surface border voxels that are simple points and not line-end points as potential deletable points, then, rechecks those points and deletes the point which is still simple and not line-end point after the deletion of some previously visited marked points. This procedure is performed recursively until a one voxel-wide centerline is produced. A minimum length threshold is also set to prune the unwanted small branches.
2.1.2 Accurate Aortic Surface Segmentation
The 4D optimal surface detection algorithm uses a graph-based method consisting of a) coordinate transformation, b) multi-surface detection using 4D optimal surface detection algorithm, and c) mapping of the segmentation result back onto the original image.
2.1.2.1 Coordinate Transformation
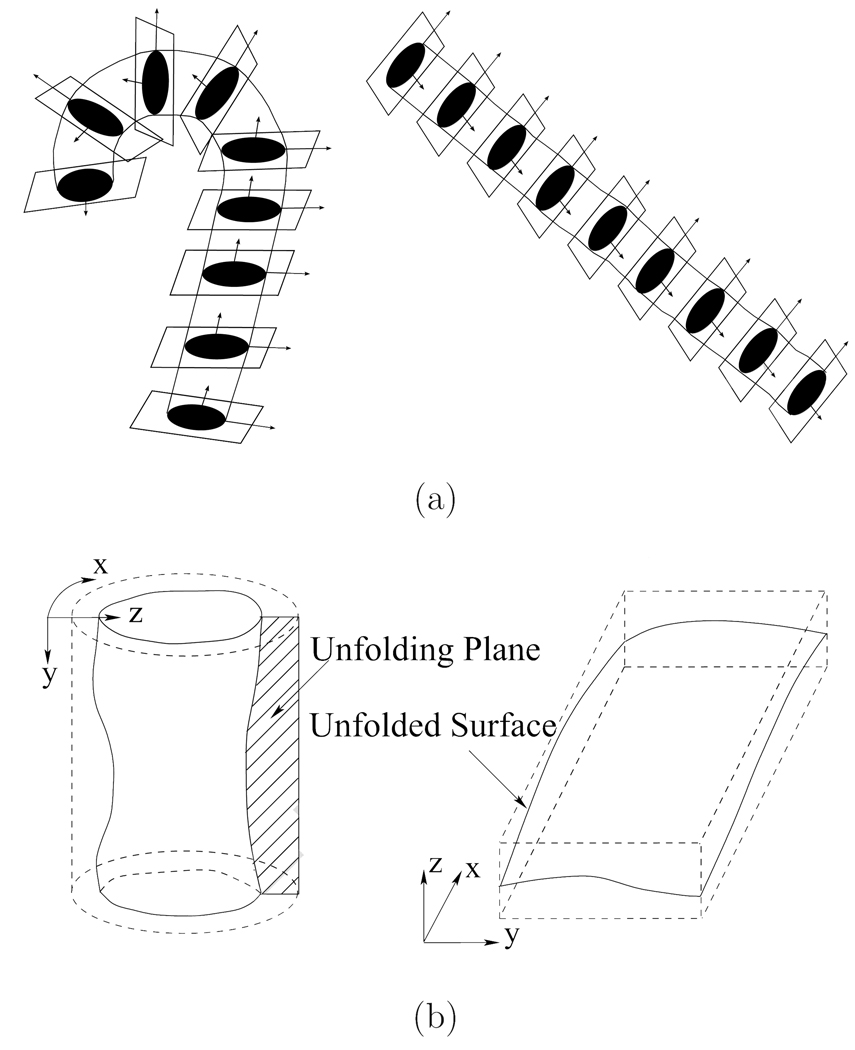
In order to construct the 4D aortic surface detection graph, a coordinate transformation is applied to each single phase image It. The cross sections are obtained by resampling the image using cubic B-spline interpolation in the plane perpendicular to the center-line [18,19]. The aorta is straightened by stacking the resampled cross sections to form a new volume. Each cross section in the resampled volume is unfolded into polar coordinates to transfer the cylindrical surface into a terrain-like surface [20]. This unfolded image is used for construction of the aortic surface detection graph. Fig. 2(a) shows the process of straightening the aorta into a cylindrical tube. Fig. 2(b) shows the unfolding process. Note that this coordinate transform is cylindrical-shape specific. For general shapes, a somewhat more involved coordinate transform has been developed and independently reported that starts with a pre-segmentation step yielding an approximate shape which is subsequently meshed [21]. Normal vectors, derived from the meshed surface, serve as columns of a graph that is used for optimal segmentation.
Fig. 2.
The unfolding process. (a) Transforming aorta into a straight cylinder. (b) Unfolding the cylindrical surface into a terrain-like surface.
2.1.2.2 Segmentation of 4D Aortic Surface
After coordinate transformation, the desired 3D surfaces in an n-phase 4D aortic image are determined using our multi-surface segmentation algorithm [22]. Instead of resorting to sequential segmentations of time series of 3D images, our segmentation method obtains a globally optimal 4D surface, where all 3D surfaces are segmented simultaneously and the temporal interrelationship of the 3D surfaces is considered in the global optimization. The two steps of this 4D algorithm, graph construction and optimization, are briefly described here.
Graph Construction
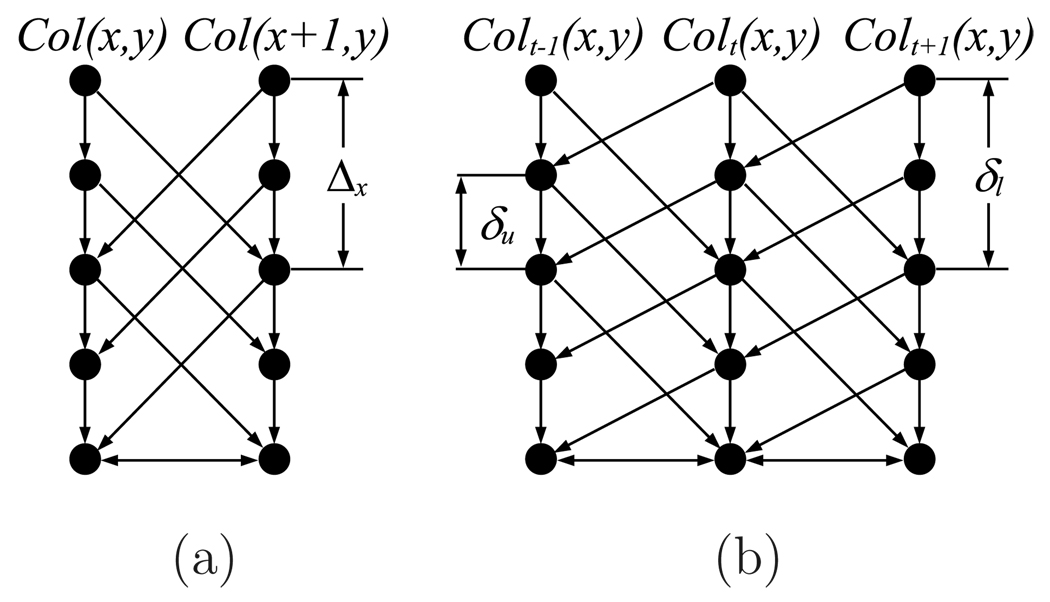
Let G = (V, E) be a connected, directed 4D graph constructed from an n-phase 4D image {It(x, y, z)}t∈[0,n−1]. The vertex set V is represented by {Vt(x, y, z)}t∈[0,n−1]. Considering each volumetric image as a 3D matrix with image sizes X, Y and Z in x, y and z direction, respectively, each vertex Vt(x, y, z) in the 4D graph G corresponds to one voxel It(x, y, z) of I. A cost, denoted by ct(x, y, z), is calculated for each vertex so that the lower the cost, the more likely it is that the vertex is on the sought surface. The arc set E consists of three arc types, intracolumn arcs Ea, intercolumn arcs Er and intersurface arcs Es.
Intracolumn arcs Ea
Let Colt(x, y) represent the vertex subset {Vt(x, y, z)|z ∈ [0, Z − 1]}. For each vertex in the column Colt(x, y), there is a directed arc connecting vertex Vt(x, y, z) to vertex Vt(x, y, z−1). Then, the set Ea is {〈Vt(x, y, z), Vt(x, y, z − 1)〉 |z > 0}.
Intercolumn arcs Er
The terrain-like surface intersects with one vertex of each column Colt(x, y). The surface continuity in 3D is constrained by two smoothness parameters Δx, Δy. More specifically, if vertices Vt(x, y, z) and Vt(x + 1, y, z′) belong to the surface, then |z − z′| ≤ Δx. Likewise, if vertices Vt(x, y, z) and Vt(x, y + 1, z′) belong to the surface, then |z − z′| ≤ Δy. Since the terrain-like surface is unfolded from a cylindrical surface along the x direction, the first and last rows along the surface shall also satisfy the smoothness constraints. These constraints are used to construct the complete set of intercolumn arcs Er.
Intersurface arcs Es
Considering aortic motion, the surfaces in consequent 3D images are interrelated. These temporal interrelationships can be introduced by adding intersurface arcs Es in the 4D graph. By doing so, a globally optimal 4D graph solution corresponds to an optimal 4D segmentation.
Intuitively, the terrain-like surfaces in two continuous phases can be viewed as a pair of interacting surfaces which cross each other within a specific distance range. Let St and St+1 be two surfaces in two continuous phases Gt and Gt+1 of the 4D graph G, and let Colt(x, y) and Colt+1(x, y) denote two corresponding columns in these two continuous 3D graphs. After assigning a pair of appropriate distance constraints δu and δl representing the maximum distance for the second surface with respect to the first surface, the intersurface arcs Es are constructed as
| (8) |
where t ∈ [0, n − 1]. The construction of Es guarantees that if the vertex Vt(x, y, z) is on St, then its corresponding vertex on St+1 must be no further than vertex Vt+1 (x, y, max(0, z − δl)). Similarly, if the vertex Vt+1(x, y, z) is on St+1, then its correspondence vertex on St must be no further than vertex Vt(x, y, max(0, z − δu)). The distance constraints δu and δl are determined by the aortic motion change in two continuous phases. Fig. 3(a) shows an example of intracolumn and intercolumn arcs, Fig. 3(b) shows an example of intersurface arcs.
Fig. 3.
Graph construction. (a) Intracolumn and intercolumn arcs, (b) Intersurface arcs.
Optimization
After constructing the 4D graph, the surface detection problem is transformed into a search for the minimum closed set [23] in a weighted graph by assigning the weight of each vertex as
| (9) |
The minimum closed set problem can be solved by using the minimum s–t cut algorithm as presented in [22].
2.1.2.3 Cost function design
The cost function designed for this application takes advantage of the edge information distinguishing the aortic surface from the aortic lumen and background [20]. Additionally, aortic wall is visible in portions of the aorta. To take advantage of this double-surface appearance, a combination of the edges of opposite orientations is used in parts of the aortic surface [24,25]. More specifically, the aortic wall appearance is different for the ascending and the descending aorta portions on MR images. Therefore, the aorta was divided in two parts – the first 20% of the length between the aortic valve and the diaphragm was labeled as the ascending aorta and the remaining part included the aortic arch and the descending aorta. To deal with the different image appearance of the aortic wall in these two aortic parts, two different cost functions were developed as outlined below. First, a 3D edge image of the aorta is computed from the straightened image data. To detect the accurate surface, a combination of first and second derivatives (3 × 3 Sobel edge detector and 5 × 5 Marr-Hildreth edge detector) was employed in the individual cross sections [26]. The edge image is therefore calculated as
| (10) |
where I2D is the 2D cross section of the original image, S is the Sobel operator, and M is the Marr-Hildreth operator. The parameter ω controls the relative weights of the first and second derivatives.
Descending Aorta and Aortic Arch
Let d(i, j) represent the edge direction of a pixel (i, j). The edge function for the descending aorta and the aortic arch is defined as
| (11) |
where ΔP is a constant penalty term.
Ascending Aorta
The ascending aorta surface segmentation requires a knowledge-based cost function [24]. In the ascending aorta, the aortic wall is typically visible in the MR data. The thickness of the wall ranges from 2 to 4 pixels in the analyzed data. Using this information, the edge function of the ascending aorta is calculated as a combination of two related edges:
| (12) |
The inner wall border edge function and outer wall border edge function are
| (13) |
| (14) |
The overall cost function is formed as:
| (15) |
where is the edge function value.
2.2 Disease Detection
The disease assessment method is directly based on the analysis of the 4D segmentation result. First, a point distribution model (PDM) is built representing the aortic shape and its motion during the cardiac cycle. Then, the modal indices of the PDM are used as input to a Support Vector Machine (SVM) classifier.
2.2.1 Point distribution models
Building the PDM consists of two stages: 1) Automatic generation of aortic landmarks representing the segmented aorta in 4D. 2) Capturing the shape variation by performing principal component analysis (PCA) on the 4D shape vectors of the aorta.
2.2.1.1 Landmark generation
To build the PDM, the aortic shape must be described by spatially corresponding landmarks. For a 4D segmentation result {BIt}t∈[0,n−1], the landmarks are automatically generated by applying a 3D landmark generation algorithm adapted from Frangi’s approach [27] in a phase by phase fashion. For each single phase segmentation result BIt, the automatic landmark generation algorithm consisted of the following 3 steps;
template shape generation,
template shape landmarks generation,
landmark mapping.
While these main steps were identical to those described in [27], the registration procedure and the landmark generation algorithm were tailored specifically for the aortic shape case by performing a surface-based registration [28] [29] instead of the multi-level B-spline transform used by Frangi et al. This resulted in a substantial speed-up of the landmarking process.
2.2.1.2 Shape variation
The shape vectors resulting from the previous step are first aligned into a common coordinate frame [30]. Isotropic scaling is used in the alignment since it preserves certain shape variations that are associated with scale of the objects and can be important for disease diagnosis. After the shape alignment, the PCA is applied to the aligned shape vectors. Let Y̅ be the mean shape and M represent the number of samples. The mean shape and covariance matrix C are calculated. With the resulting eigenvalues λi and the eigenvectors ϕi of C, a shape Y can be approximated as
| (16) |
where the matrix Φ = (ϕ1|ϕ2| … |ϕp) defines the first p principal components, whose eigenvalues are sorted such that λi > λi+1, and b is a weight coefficient vector or vector of modal indices
| (17) |
The employed number of variations p was determined by assigning a cutoff at some percentage of the total variance [31].
2.2.2 Discrimination model
With each 4D aortic instance represented by the modal indices b describing the observed shape and motion variations, an efficient classification algorithm was developed for pattern recognition using a support vector machine classifier (SVM) [32–35]. The classifier is used to classify 4D aortic instances into classes of normal and connective tissue disorder subjects. Given M input training samples d ∈ ℛn with class labels g ∈ {− 1,1}, the SVM maps a sample d into a high-dimensional space using a kernel function K(d, di) and constructs an optimal hyper-plane separating the 2 classes in this space. The optimal hyper-plane is identified so that it maximizes its distance from the training samples (maximal margin in the high dimensional space). The decision function given by the SVM is of the form
| (18) |
Here, η = (η1, η2, … ηn) is determined by optimizing the quadratic programming problem
| (19) |
where T is a predefined parameter controlling the number of admissible errors. In this study, radial basis functions K(d, di) = e−κ*|d−di|2 were used. The “grid-search” method, which is recommended in [35], was used to determine κ and T.
3 EXPERIMENTAL METHODS
3.1 Image Data Acquisition
The algorithm was developed and evaluated on a set of 104 MR image sequences acquired from 104 subjects (52 normal, 52 patients with genetically suspected or established connective tissue disorder aortic disease — the patient status was established based on familial history or 2D ultrasound-based clinical diagnosis, physical examination, and genetic testing when available). The MR imaging was performed either on a GE Signa or Siemens Avanto 1.5 T scanners, in both cases using a 2D cine true steady-state free precession imaging. Each image sequence was composed of 16–25 cardiac phases representing one cardiac cycle. For each subject, both the candy cane view and left ventricular outflow tract (LVOT) view were captured with voxel sizes ranging from 1.5 × 1.5 × 3.0 mm3 to 2.0 × 2.0 × 6.0 mm3. The two views are used since the LVOT best depicts the aortic root while the candy cane view is best for the rest of the aorta all the way to the diaphragm. The patients were selected due to their family history of connective tissue disorder. These patients had either normal aortic dimensions or clinically established connective tissue disease.
3.2 Image Data Preprocessing
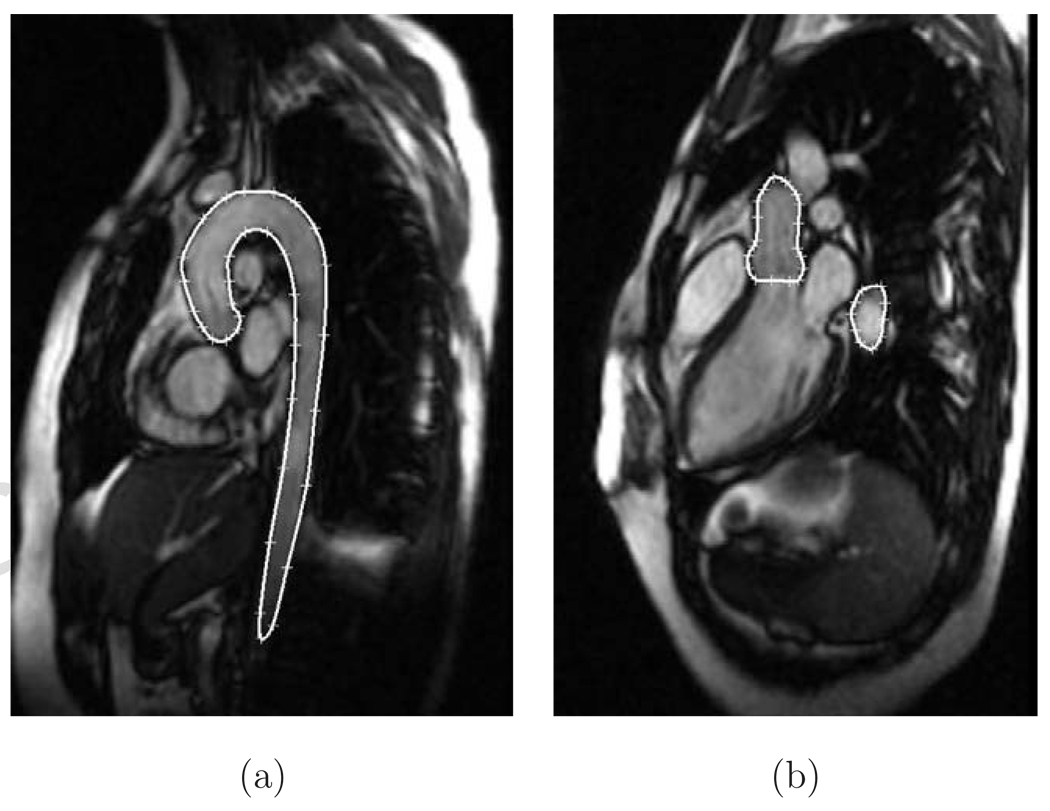
To obtain 4D image data that would have the same number of cardiac phases for all subjects and consisted of isometric voxels, the number of phases was normalized to 16. The LVOT and candy-cane view image data were registered using a mutual information registration algorithm and interpolated using B-splines. The registration/interpolation process achieves isotropic image voxel sizes from the combination of the two image views - LVOT and candy-cane. For some images, the aortic information that is not present in the candy-cane view is available in the LVOT view. Figs. 4(a, b) show sample candy cane and LVOT view slices.
Fig. 4.
MR image data with manual tracing of the aorta. (a) Candy cane view. (b) LVOT view.
3.3 Study Protocol and Data Analysis
From the available 104 image datasets, a subset of 21 datasets was randomly selected and used for validating the segmentation performance. Limiting the validation set to 21 datasets was motivated by the extraordinary labor intensity of manual tracing in 4D. Once validated, the aortic segmentation was performed on the entire set of 104 subjects for functional studies as described below. Therefore, 21 subjects (7 patients and 14 normal subjects) were used to assess the performance of the automated 4D segmentation. Aortic luminal surfaces were compared with the expert traced independent standard. The independent standard was defined by manual tracing of 4 randomly selected slices in each of 5 randomly selected phases in the 21 subjects (total of 420 manually traced slices). Surface positioning errors were defined as the shortest distances between the manually traced contours and computer-determined surfaces in the 4D aortic images. Signed and unsigned surface positioning errors are expressed as mean±standard deviation in voxels and millimeters.
To assess and compare the diagnostic performance of the modal indices of aortic shape and motion derived from (a) a single end-diastolic 3D segmentation, (b) 2-phase end-diastolic and end-systolic 3D segmentation, and (c) 16-phase 4D segmentation, the method parameter design and classification performance assessment must be separated from each other to maintain complete independence. Therefore, the available set of 104 MR images was divided in two randomly identified subsets, 52 data sets each, each consisting of 26 MR data sets from normal subjects and 26 data sets from patients. One of these subsets was used for identifying the optimal number of modal indices of aortic shape and motion with respect to the classification performance in each of the three classification tasks, respectively. For each of these three classification tasks, the optimal number of modal indices was determined as that maximizing the classification correctness. Expert-defined disease status derived from the clinical records formed the binary prediction output (normal/patient). The best normal/patient classification performance that was achieved in the training set called for the use of 8 modal indices for the single-phase case, 8 modal indices for the 2-phase case, and 10 modal indices for the 16-phase case. In all cases, the 8 to 10 most significant principal components of shape and motion corresponded to about 92% of the total variance detected in this design subset. The numbers of utilized principal components were determined using a leave-one-out train/test approach.
The second subset was never used for the method design and was solely used for performance assessment. This way, the determination of which and how many modal indices to use and the performance assessment were kept strictly independent of each other. The main goal of the performance assessment was to determine how well each of the three different classification tasks (using 1-phase, 2-phase, and 16-phase data) can distinguish between the patients (suffering from connective tissue disorder) and normal subjects. Similar to the design subset, leave-one-out performance assessment method was used to train and evaluate the predictive classifier performance in the second performance assessment subset. Performance was assessed in terms of the overall classification correctness and expressed as percentages.
4 RESULTS
In all results referenced below, the following method’s parameters were used [equations (2), (5), (7), (8), (10), (11)]: α = 0.5, β = 0.5, θ = 0.4, σ = 1.2, δu = 5, δl = 5, ω = 0.8, ΔP = 100. All these values were determined empirically.
4.1 Segmentation Results
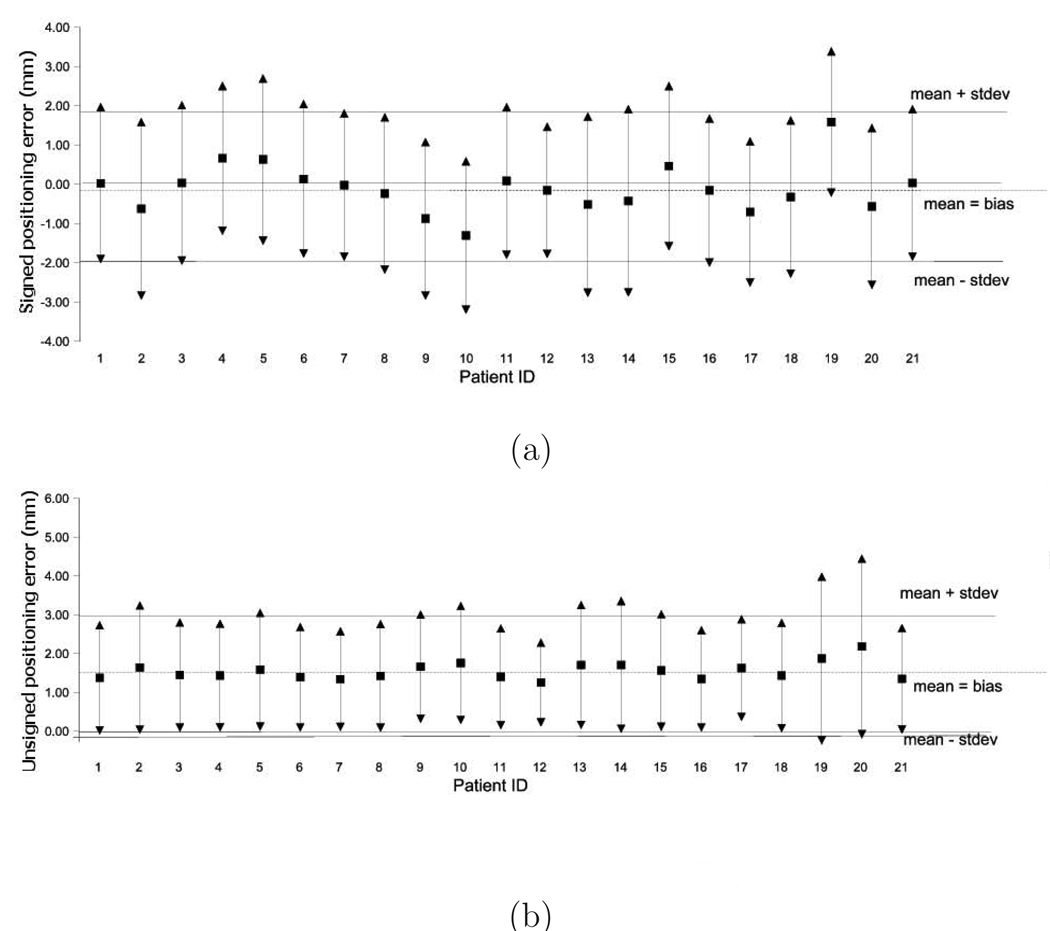
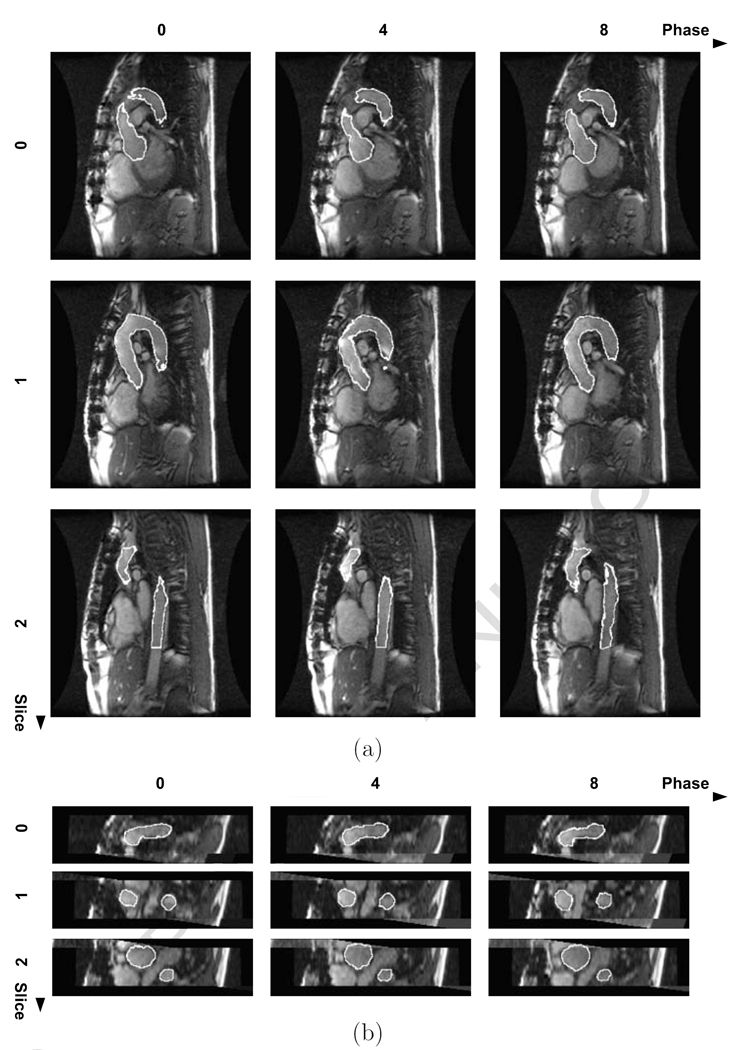
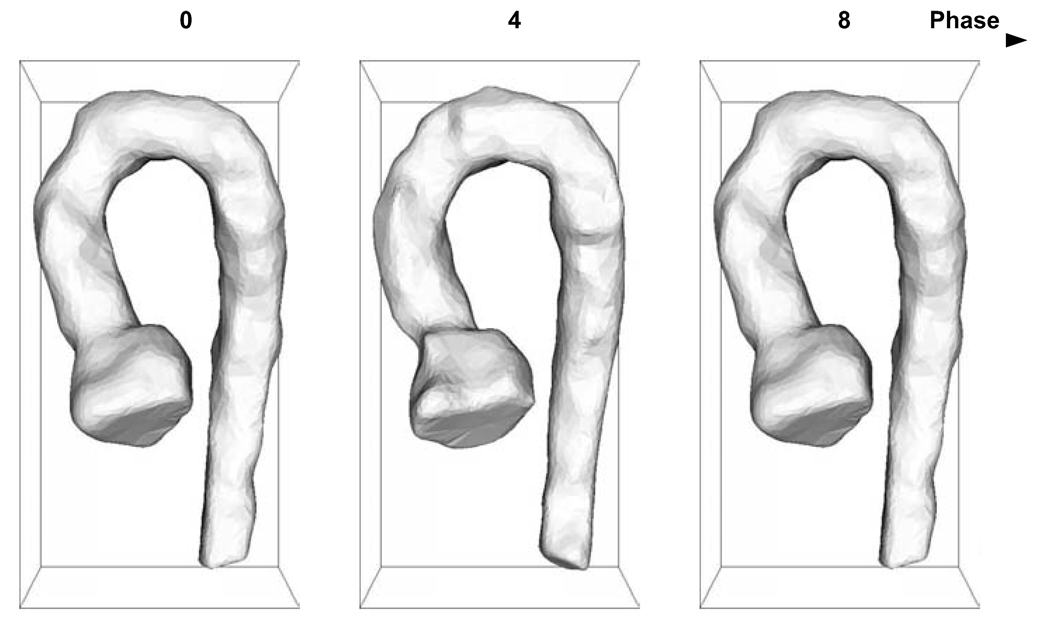
All 4D aortic MR images used for the segmentation performance assessment were successfully segmented by our 4D segmentation algorithm. Comparison of the computer-determined and expert-traced surfaces showed a good agreement. The segmentation produced aortic surfaces with subvoxel accuracy and a very minimal bias as judged by the signed surface positioning errors of −0.07 ± 1.16 voxel (−0.10 ± 2.05 mm) and unsigned positioning errors of 0.88 ± 0.81 voxel (1.55 ± 1.44 mm). Fig. 5 summarizes the segmentation performance in individual 4D datasets and shows that no major segmentation failures were experienced. An example of a typical 4D segmentation result is shown in Fig. 6 in transverse and coronal views. The volumetric representation of the achieved segmentation is shown in Fig. 7.
Fig. 5.
Surface positioning errors (in mm) for 21 4D datasets used for segmentation performance assessment. For each dataset, mean and 1 stdev error bars are shown. The overall mean (analysis bias) ± 1 standard deviation range is shown as dashed and solid lines, respectively. (a) Signed positioning errors. (b) Unsigned positioning errors.
Fig. 6.
Automated 4D segmentation result – diseased subject. Three slices and their three phases (0, 4, 8) were selected from the 4D image. The horizontal coordinates represent the phase number, the vertical coordinates represent the slice number. (a) Transverse view. (b) Coronal view.
Fig. 7.
Volumetric representation of computer-based segmentation of a diseased aorta.
4.2 Disease Detection Results
As described above, the classification performance was assessed in 52 data sets after the method’s parameters were independently optimized in a separate design data set. For the single-phase case, 300 landmarks were automatically generated on each aortic luminal surface. The classifier based on 8 most significant principal components (κ = 4, T = 4) exhibited an overall classification correctness of 82.7%. For the two-phase case (end-systole and end-diastole), the two cardiac phases were considered a single shape/motion instance and 600 landmarks were generated. The classifier based on 8 most significant principal components (κ = 1, T = 4) exhibited correctness of 88.5%. For the sixteen-phase 4D case, the sixteen cardiac phases were considered a single shape/motion instance and 4800 landmarks were generated. The first 10 principal components were selected as SVM input features. The classifier (κ = 0.25, T = 32) exhibited correctness of 90.4%.
The achieved results suggest that the functional (shape and motion) information is an important contributor to the ability to distinguish between the normal and connective tissue disorder subjects. Clearly, the functional information (shape changes, motion) missing in the single-phase 3D model is partly present in the two-phase model and more completely in the sixteen-phase model. The additional information is reflected in the corresponding improvement of classification performance. The overall results are summarized in Table 1 – Table 3.
Table 1.
Classification correctness of the one-phase model.
| Predicted | ||
|---|---|---|
| Disease Status | Diseased | Normal |
| Diseased | 25 | 1 |
| Normal | 7 | 19 |
Table 3.
Classification correctness of the sixteen-phase model.
| Predicted | ||
|---|---|---|
| Disease Status | Diseased | Normal |
| Diseased | 22 | 4 |
| Normal | 1 | 25 |
4.3 Time Requirements
Training of the method on 52 4D aortic MR datasets (landmarking, PDM construction, leave-one-out SVM classifier training) required approximately 90 minutes of computing time on a Pentium-4 2.4GHz machine. In the analysis mode, processing of a single 4D dataset required less than 5 minutes. The individual main steps contributed to the total run time as follows: level set presegmentation less than 2 minutes, skeletonization less than 10 seconds, accurate graph-based segmentation less than 1 minute, landmarking less than 1.5 minute, classification <1 s.
5 DISCUSSION AND CONCLUSION
This section will concentrate on four separate topics: a) utility of the developed methods for diagnosing patients at risk from connective tissue disorder, b) properties of the aortic segmentation method, c) limitations of this technique, and d) performance of the method to distinguish between normal and early-stage disease.
5.1 Connective Tissue Disorder Subjects
Patients with connective tissue disorders (CTD) are at high risk for developing aortic aneurysm and dissection, both of which are major causes of morbidity and mortality in this patient population. Patients with a family history of CTD may or may not have CTD, but they are certainly at risk for this disease. The usual clinical practice is to perform cine MRI of the aorta on patients who have an established diagnosis of CTD or who are at risk of having CTD (e.g., positive family history, echocardiography showing dilated aortic root, and certain findings on physical examination). MRI is currently not used to routinely establish a diagnosis of CTD. MRI is used to detect aortic dilatation. The presented work may allow the extraction of novel quantitative indices describing static and dynamic properties of the normal and diseased aorta. The clinical role of imaging in this population is to identify patients who need aortic surgery before they suffer a major complication, such as dissection, aortic rupture, or hemopericardium (tamponade). The current gold standard, static measurements of aortic diameter, may fail to identify patients who will suffer catastrophic complications.
5.2 Aortic Segmentation and Analysis Method
An automated 4D segmentation algorithm which combines a 4D fast marching level-set segmentation with an optimal graph-based multiple surface detection algorithm was presented and provided accurate and reliable aortic surfaces.
This method reports on a number of novel techniques, from the 4D aortic segmentation to the shape-based early diagnosis of the connective tissue disorder.
We consider the 4D aortic segmentation to be the most appropriate for complete segmentation over the entire cardiac cycle. Graph-based segmentation of the aortic lumen in 4D requires that the aorta is first approximately detected in 4D. The use of the 4D fast marching algorithm provides an automated presegmentation of the aorta allowing determination of the aortic centerline.
The general applicability of the proposed method to tubular object segmentation allows its use in a variety of vascular structures, intrathoracic airways, and other objects with cylindrical topology. Following an accepted division of image segmentation in two stages – detection and delineation, the graph-based segmentation method is very powerful for accurate delineation. Its overall performance, however, depends on the quality of the presegmentation step so that a proper approximate detection of the object is available prior to its delineation. In our case, approximate level set segmentation is used for aortic detection. Clearly, if the presegmentation step fails, so does the graph-search step. On the other hand, the presegmentation does not have to be perfect. What is required is that the centerline resulting from skeletonization of the presegmentation result is located fully inside the aorta and has no spurious branches. Therefore, even if the presegmentation is far from perfect, the graph-search based step yields highly accurate segmentation as long as the true surface location is included in the graph formed from the presegmentation.
The major accomplishment of the presented approach is a first successful optimal segmentation of a tubular surface in 4D – i.e., inherently considering object motion in a single optimization process. To further increase the method’s robustness and delineation accuracy, the 4D graph search utilizes a cost function containing surface information from both the inner and outer walls in the ascending aorta. This is another unique characteristic of the reported method. Note however, that our definition of the exact location where the arch and/or the descending aorta start is not very precise. The used value of 20% puts the dividing location to a roughly reproducible position. Most importantly, our segmentation is not sensitive to the exact location of this separating point. The important part is to deal with the difference between the aortic appearance in the proximity to the aortic valve (ascending aorta) and in the descending aorta. The way we are dealing with the appearance differences is that we utilize single luminal surface information to segment the surface of the arch and descending aorta and double surface information (inner/outer) to segment the ascending aorta. Since the cost function term of the arch/descending aorta is identical to the inner surface term for the ascending aorta (Eq. 13), the exact position of the “switch” point location is not critical. This common cost function property also guarantees that even if the outer surface information is missing in particular patient data or if this outer surface image information is not present in the ascending aorta, the cost function according to (Eq. 13) can be used to find the ascending aorta surface.
By comparing our previous single-phase slice-by-slice segmentation [36], we did not experience any worsening of the surface positioning errors in the end-diastolic phase. As such, the 4D technique adds the benefit of a temporal context without decreasing individual phase segmentation accuracy. In quantitative terms, method [36] achieved signed and unsigned surface positioning errors of −0.09±1.21 voxel and 0.93±0.76 voxel, respectively; compared with the signed and unsigned surface positioning errors of −0.07±1.16 voxel and 0.88±0.81 voxel reported here for the 4D method. As an additional benefit, the complete 4D segmentation is performed using a single optimization step and a single initialization.
The reported surface positioning errors combine several sources of error: 1) The accuracy of the independent standard is affected by partial volume effects in MR images and the lack of 3D/4D context during manual tracing (the absolute error values in millimeters may seem large but need to be assessed in context of the voxel sizes of the original image data, which were up to 2×2×6 mm3); 2) At some parts of the aorta, the aortic border is estimated by the human expert when there is no strong intensity evidence; this kind of human knowledge is not embedded in the segmentation method; 3) The inaccuracies of the computer segmentation. The main goal is therefore to minimize the segmentation bias (assessed by the average signed error), as well as minimize the unsigned mean errors and error standard deviations. Our method has succeeded in both of these metrics.
5.3 Limitations of the Presented Approach
An automatic algorithm was developed to generate the landmarks on the 4D surfaces. Principal component analysis applied to the landmarked 4D shapes resulting from the aortic segmentation formed a very efficient approach for capturing global information about the variation of the structure’s shape and motion. An SVM based classifier inducing nonlinear features through a nonlinear kernel function was appropriate for the complex high-dimensional classification task. The combination of the shape/motion model representation with the SVM classification is well suited for the disease diagnostic task at hand. As demonstrated, it yields very promising classification results, clearly showing the feasibility of quantitative assessment of aortic disease from cardiovascular MR images.
A question may however be raised whether the reported analysis truly uses motion information or rather utilizes shape differences across cardiac phases.
Our landmarking method attempts to obtain a good landmark registration across the 16 cardiac phases, and the landmarks seem to correspond very well over the entire cardiac cycle. Yet, we have no proof that this is indeed the case in all situations. For example, we cannot guarantee that no landmark sliding occurs along the aortic long axis, although we have attempted to minimize this behavior by the employed 3D template approach. Needless to say, we also lack an independent standard to validate this behavior. However, since we attempt to determine motion changes of locally registered landmarks, it seems appropriate to talk about motion analysis. Importantly, with 16-phase segmentation results available, dynamic behavior-describing indices can be calculated and utilized such as the rates of change of local aortic diameter, pulse wave propagation, etc., which are not available when using the 1- or 2-phase segmentations. In the future, we will furher concentrate on the improvements of 4D registration and analysis of 4D dynamic behavior.
Another limitation may arise from the global character of the shape and motion description. We have separately explored the use of Independent Component Analysis for identifying local changes of myocardial tissue properties over time and this work was reported in [37]. We are also developing what we call Active Index Model that will specifically focus on the local changes and local differences. Clearly, identifying local changes is an important issue and this topic continues to be a focus of the future research.
5.4 Role of Shape and Motion in CTD Assessment
The classification accuracy generated from the sixteen-phase 4D model is substantially better compared to that obtained by the single-phase 3D model (from 82.7% to 90.4%). This result demonstrates that the motion information which is captured in the 4D model contributes to the classification performance and is related to the connective tissue disorder disease. This motion information is difficult to observe by a human eye but it can apparently be detected by our 4D approach. The results demonstrate the importance of utilizing functional information about the aortic motion for the connective tissue disease diagnosis using shape modeling. It can also be noted that the performance difference between the 2-phase and the 16-phase approaches is not very large. Yet, the 16-phase approach exhibits a noticeable improvement.
After analyzing the data, it is clear that the 16-phase method is very specific with respect to correctly classifying normal controls. This brings a question why 3 of 26 suspected CTD subjects were not correctly classified by the 2-phase approach, and why 4 of 26 were not correctly identified using the 16-phase approach. The most likely explanation of this behavior comes from the subject selection. All subjects included in the patient group are established or suspected future CTD patients. Some of our study subjects were referred for an MRI because of a family history of aortic dissection. Due to the believed genetic basis of the connective tissue diseases that cause aortic dissection, first degree relatives of individuals who have had an aortic dissection undergo MRI screening of their aortic dimensions. The “patients” who were inaccurately classified fall into this category. They had family members who experienced dissection of the aortas (one had a daughter and one had three family members with dissection). However, both were found to have normal aortas and on follow-up clinical evaluations have (so far) not demonstrated any evidence of connective tissue disease. In the absence of a genetic test that would definitively classify subjects into normal or patient groups, we have elected to keep these subjects in the patient group. It is not known at this point whether or not they will indeed develop a connective tissue disease. While we have presented our results in terms of classification correctness, as with any other classification technique, the classifier can be further fine-tuned to increase either the sensitivity or the specificity of the disease status assessment.
The main promise of this approach is the expected ability to quantify the disease progression, identify and predict periods of rapid progression, and therefore predict the onset of aortic aneurysm and/or dissection prior to its development. Assessing the predictive ability of our method will be a topic of future work. If successful, such a method capable of predicting an aortic event for several months or years prior to its onset would have major implications on the management of patients with congenital connective tissue disease.
Table 2.
Classification correctness of the two-phase model.
| Predicted | ||
|---|---|---|
| Disease Status | Diseased | Normal |
| Diseased | 23 | 3 |
| Normal | 3 | 23 |
Acknowledgment
The authors would like to thank Drs. Nicholas E. Walker and Steven C. Mitchell, as well as Natalie R. Van Waning for their contribution to the project. This work was supported, in part, by the NIH grants R01HL071809 and R0lEB004640.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.The Centers for Disease Control and Prevention (CDC) http://www.cdc.gov/.
- 2.Atsuchi Y, Nagai Y, Komatsu Y, Nakamura K, Shibuya M, Hirosawa K. Echocardiographic manifestation of annuloaortic ectasia: its paradoxical motion of the aorta and premature systolic closure of the aortic valve. American Heart Journal. 1977;93:428–433. doi: 10.1016/s0002-8703(77)80404-3. [DOI] [PubMed] [Google Scholar]
- 3.Rueckert D, Burger P, Forbat S, Mohiaddin R, Yang G. Automatic tracking of the aorta in cardiovascular MR images using deformable models. IEEE Trans. Med. Imag. 1997;16(5):581–590. doi: 10.1109/42.640747. [DOI] [PubMed] [Google Scholar]
- 4.Behrens T, Rohr K, Stiehl H. Robust segmentation of tubular structures in 3D medical images by parametric object detection and tracking. IEEE Trans. Syst., Man, Cybern. 2003;33(4):554–561. doi: 10.1109/TSMCB.2003.814305. [DOI] [PubMed] [Google Scholar]
- 5.de Bruijne M, van Ginneken B, Viergever MA, Niessen WJ. Adapting active shape models for 3D segmentation of tubular structures in medical images; Proc. of IPMI, Vol. 2732 of LNCS; Springer-Verlag; 2003. pp. 136–147. [DOI] [PubMed] [Google Scholar]
- 6.Subasic M, Loncaric S, Sorantin E. 3D image analysis of abdominal aortic aneurysm; Proc. SPIE (Medical Imaging 2002: Image Processing); SPIE Press; 2002. pp. 1681–1689. [Google Scholar]
- 7.Cheng TO. Decreased aortic root distensibility rather than increased aortic root diameter as an important cardiovascular risk factor in the Marfan syndrome. The American Journal of Cardiology. 2006;97:1422. doi: 10.1016/j.amjcard.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Kardesoglu E, Ozmen N, Aparci M, Cebeei BS, Uz O, Dincturk M. Abnormal elastic properties of the aorta in the mitral valve prolapse syndrome. Acta Cardiologica. 2007;62:151–155. doi: 10.2143/AC.62.2.2020235. [DOI] [PubMed] [Google Scholar]
- 9.Vitarelli A, Conde Y, Cimino E, D’Angeli I, D’Orazio S, Stellato S, Padella V, Caranci F. Aortic wall mechanics in the Marfan syndrome assessed by transesophageal tissue Doppler echocardiography. The American Journal of Cardiology. 2006;97:571–577. doi: 10.1016/j.amjcard.2005.09.089. [DOI] [PubMed] [Google Scholar]
- 10.Sandor GG, Hishitani T, Petty RE, Potts MT, Desouza A, Desouza E, Potts JE. A novel Doppler echocardiographic method of measuring the biophysical properties of the aorta in pediatric patients. J Am Soc Echocardiogr. 2003;16:745–750. doi: 10.1016/S0894-7317(03)00407-3. [DOI] [PubMed] [Google Scholar]
- 11.Bardinet E, Cohen L, Ayache N. Tracking and motion analysis of the left ventricle with deformable superquadrics. Medical Image Analysis. 1996;1(2):129–149. doi: 10.1016/S1361-8415(96)80009-0. [DOI] [PubMed] [Google Scholar]
- 12.Chandrashekara R, Rao A, Sanchez-Ortiz GI, Mohiaddin RH, Rueckert D. Construction of a statistical model for cardiac motion analysis using nonrigid image registration; Proc. of IPMI, Vol. 2878 of LNCS; Springer-Verlag; 2003. pp. 599–610. [DOI] [PubMed] [Google Scholar]
- 13.McInerney T, Terzopoulos D. A dynamic finite element surface model for segmentation and tracking in multidimensional medical images with application to cardiac 4D image analysis. Computerized Medical Imaging and Graphics. 1995;19(1):69–83. doi: 10.1016/0895-6111(94)00040-9. [DOI] [PubMed] [Google Scholar]
- 14.Montagnat J, Delingette H. 4D deformable models with temporal constraints: application to 4D cardiac image segmentation. Medical Image Analysis. 2005;9(1):87–100. doi: 10.1016/j.media.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 15.Malladi R, Sethian JA. A real-time algorithm for medical shape recovery; Proc. of ICCV; Narosa Publishing House; 1998. pp. 304–310. [Google Scholar]
- 16.Frangi A, Niessen W, Vincken K, Viergever M. Multiscale vessel enhancement filtering; Proc. of MICCAI, Vol. 1496 of LNCS; Germany: Springer-Verlag; 1998. pp. 130–137. [Google Scholar]
- 17.Palagyi K, Tschirren J, Sonka M. Quantitative analysis of intrathoracic airway trees: Methods and validation; Proc. of IPMI, Vol. 2732 of LNCS; Springer-Verlag; 2003. pp. 222–233. [DOI] [PubMed] [Google Scholar]
- 18.Unser M, Aldroubi A, Eden M. B-spline signal processing: Part I - theory. IEEE Trans. Signal Processing. 1993;41(2):821–833. [Google Scholar]
- 19.Unser M, Aldroubi A, Eden M. B-spline signal processing: Part II - efficient design and applications. IEEE Trans. Signal Processing. 1993;41:834–848. [Google Scholar]
- 20.Sonka M, Hlavac V, Boyle R. Image Processing, Analysis, and Machine Vision. 3rd Edition. Thomson Engineering; 2007. [Google Scholar]
- 21.Li K, Millington S, Wu X, Chen DZ, Sonka M. Simultaneous segmentation of multiple closed surfaces using optimal graph searching. Proc. of the 18th Biennial International Conference on Information Processing in Medical Imaging (IPMI); 2005. pp. 406–417. [DOI] [PubMed] [Google Scholar]
- 22.Li K, Wu X, Chen DZ, Sonka M. Optimal surface segmentation in volumetric images - a graph-theoretic approach. IEEE Trans. Pattern Anal. Machine Intell. 2006;28(1):119–134. doi: 10.1109/TPAMI.2006.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X, Chen DZ. Optimal net surface problems with applications; Proc. of ICALP, Vol. 2380 of LNCS; Springer-Verlag; 2002. pp. 1029–1042. [Google Scholar]
- 24.Sonka M, Zhang X, Siebes M, Bissing MS, DeJong SC, Collins SM, McKay CR. Segmentation of intravascular ultrasound images: A knowledge-based approach. IEEE Trans. Med. Imag. 1995;14(4):719–732. doi: 10.1109/42.476113. [DOI] [PubMed] [Google Scholar]
- 25.Tschirren J, Hoffman EA, McLennan G, Sonka M. Segmentation and quantitative analysis of intrathoracic airway trees in low-dose CT scans. IEEE Transactions on Medical Imaging. 2005:1529–1539. doi: 10.1109/TMI.2005.857654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sonka M, Reddy G, Collins SM. Adaptive approach to accurate analysis of small diameter vessels in cineangiograms. IEEE Trans. Med. Imag. l997;16(1):87–95. doi: 10.1109/42.552058. [DOI] [PubMed] [Google Scholar]
- 27.Frangi A, Rueckert D, Schnabel J, Niessen W. Automatic construction of multiple-object three-dimensional statistical shape models: Application to cardiac modelling. IEEE Trans. Med. Imag. 2002;21:1151–1166. doi: 10.1109/TMI.2002.804426. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, Thomas MT, Walker NE, Stolpen AH, Wahle A, Scholz TD, Sonka M. Four-dimensional functional analysis of left and right ventricles using MR images and active appearance models; Proc. SPIE (Medical Imaging 2007: Physiology, Function, and Structure); SPIE Press; 2007. [Google Scholar]
- 29.Gu X, Wang Y, Chan TF, Thompson PM, Yau S-T. Genus zero surface conformal mapping and its application to brain surface mapping. IEEE Trans. Med. Imag. 2004;23(8):949–958. doi: 10.1109/TMI.2004.831226. [DOI] [PubMed] [Google Scholar]
- 30.Besl PJ, McKay ND. A method for registration of 3-D shapes. IEEE Trans. Pattern Anal. Machine Intell. 1992;14:239–256. [Google Scholar]
- 31.Jolliffe IT. Principal Component Analysis. Springer-Verlag; 1986. [Google Scholar]
- 32.Boser BE, Guyon I, Vapnik V. A training algorithm for optimal margin classifiers; Proc. of ACM Workshop on Computational Learning Theory; 1992. pp. 144–152. [Google Scholar]
- 33.Cortes C, Vapnik V. Support-vector networks. Machine Learning. 1995;20(3):273–297. [Google Scholar]
- 34.Cristianini N, Taylor JS. An Introduction to Support Vector Machines and Other Kernel-based Learning Methods. Cambridge University Press; 2000. [Google Scholar]
- 35.Chang C-C, Lin C-J. LIBSVM: A library for support vector machines. URL http://www.csie.ntu.edu.tw/~cjlin/libsvm.
- 36.Zhao F, Zhang H, Walker NE, Yang F, Olszewski ME, Wahle A, Scholz T, Sonka M. Quantitative analysis of two-phase 3D+time aortic MR images; Proc. SPIE (Medical Imaging 2006: Image Processing); SPIE Press; 2006. pp. 699–708. [Google Scholar]
- 37.Hansen MS, Zhao F, Zhang H, Ersboll BK, Wahle A, Scholz T, Sonka M. Diagnosis of connective tissue disorders based on independent component analysis of aortic shape and motion from 4D MR images; Proc. of Computer Vision for Intravascular and Intracardiac Imaging; 2006. pp. 154–161. [Google Scholar]