Abstract
Previous studies have indicated that the Ca2+-dependent protease, calpain, is activated in platelets within 30–60 s of thrombin stimulation, but specific roles of calpain in platelets remain to be identified. To directly test the functions of calpain during platelet activation, a novel strategy was developed for introducing calpain’s specific biological inhibitor, calpastatin, into platelets prior to activation. This method involves treatment of platelets with a fusion peptide, calpastat, consisting of the cell-penetrating signal sequence from Kaposi’s fibroblast growth factor connected to a calpain-inhibiting consensus sequence derived from calpastatin. Calpastat specifically inhibits thrombin peptide (SFLLR)-induced α-granule secretion (IC50 = 20 µm) during the first 30 s of activation, thrombin-induced platelet aggregation (IC50 = 50 µm), and platelet spreading on glass surfaces (IC50 = 34 µm). Calpastat-Ala, a mutant peptide in which alanine is substituted at conserved calpastatin residues, lacks calpain inhibitory activity and fails to inhibit secretion, aggregation, or spreading. The peptidyl calpain inhibitors calpeptin, MDL 28,170 (MDL) and E64d also inhibit secretion, aggregation and spreading, but require 3–10-fold higher concentrations than calpastat for biological activity. Together, these findings demonstrate that calpain regulates platelet secretion, aggregation, and spreading and indicate that calpain plays an earlier role in platelet activation following thrombin receptor stimulation than had been previously detected.
Reorganization of the actin cytoskeleton is required for a number of platelet activation processes, including shape change, aggregation and spreading. In rapid response to platelet agonists, intracellular Ca2+ transients regulate the early platelet activation events of shape change (1, 2) and α-granule secretion (3–5), occurring within seconds of platelet stimulation, as well as aggregation (3, 6) and spreading, occurring within minutes. Since the discovery of the Ca2+ -activated protease, calpain, in platelets (7), there has been considerable interest in determining whether the Ca2+ transients occurring early in platelet activation function, in part, through calpain-mediated cytoskeletal remodeling. Calpain has been found to modulate the late event of platelet-mediated fibrin clot retraction (8–10). We have recently found that calpain regulates actin remodeling events of endothelial wound healing and fibroblast spreading, including lamellipodial and filopodial protrusion formation (11, 12). Calpain has also been implicated in cell motility through its effects on release of rear integrin contacts (13). Calpain translocates to the plasma membrane within seconds of platelet activation, where it is activated and may cleave cytoskeletal proteins (14–16). These findings together suggest a regulatory role for calpain in early cytoskeletal remodeling, including secretion, aggregation, and spreading.
There has been growing interest in the role of calpain in the site-specific cleavage of actin-associated proteins such as talin, actin-binding protein 280 (ABP 2801 or filamin) (7, 17), and the cytoplasmic domain of β3 integrin (18). Regulatory kinases and phosphatases, including protein kinase C (PKC) (19), pp60c-src (20), inositol polyphosphate 4-phosphatase (4-phosphatase) (21), focal adhesion kinase (22), and protein-tyrosine phosphatase 1-B (23) are also cleaved by calpain during platelet activation. Nonetheless, despite the identification of cytoskeletal and kinase/phosphatase targets for calpain in platelets, a functional role for calpain in aggregation and spreading has not been demonstrated.
A major obstacle to determining the physiological functions of calpain in platelets is lack of specific cell-permeant inhibitors. Peptidyl calpain inhibitors inhibit other cysteine proteases, including cathepsins (24). In addition, peptidyl calpain inhibitors require high micromolar concentrations to inhibit calpain in platelets (25), in contrast to fibroblasts (12). The lack of efficacy of peptidyl calpain inhibitors in platelets may be due to the high calpain content of platelets, where calpain represents 2% of platelet protein (26). While calpain’s reversible biological inhibitor, calpastatin, is the only known specific inhibitor of calpain (24, 27–32) simple strategies for increasing calpastatin in platelets have not been available. Calpastatin is cell-impermeant, and thus unsuitable for conventional studies of platelet activation (24). To solve this problem, we developed a cell-permeant specific inhibitor of calpain by joining the minimal inhibitory sequence of calpastatin to the cell-penetrating signal sequence of k-FGF, which confers cellular peptide importation (33–35). Using this specific cell-penetrating inhibitor and a set of peptidyl calpain inhibitors, with appropriate specificity controls, we demonstrate that calpain regulates platelet degranulation, aggregation, and spreading.
EXPERIMENTAL PROCEDURES
Materials
Sepharose 2B was from Amersham Pharmacia Biotech (Upsala, Sweden); pure μ-calpain (porcine erythrocyte) was from Chemicon (Temecula, CA); succinyl-LLVY-AMC was from Bachem (King of Prussia, PA); ZLLYCHN2 was a gift of Herbert Angliker (Friedrich Miescher Institut, Basel, Switzerland); A23187 was from LC Labs (Woburn, MA); human α-thrombin was from Hematologic Technologies (Essex Junction, VT); anti-P-selectin antibodies conjugated to phycoerythrin were from Becton Dickinson (San Jose, CA); Oregon Green-phalloidin was from Molecular Probes (Eugene, OR). All other chemicals were ACS grade or better and were obtained from Sigma.
Preparation of Gel-filtered Platelets
Blood from healthy aspirin-free volunteers was collected by venipuncture into 4% sodium citrate (9:1 v/v, final concentration 0.4%). Blood was centrifuged at 200 × g for 20 min to prepare platelet-rich plasma. Following incubation with apyrase (4 units/ml, 5 min, room temperature), platelets were purified from platelet-rich plasma by gel filtration (room temperature) using a Sepharose 2B column equilibrated in PIPES buffer (PIPES, pH 6.8, 25 mm/NaCl, 140 mm/KCl, 4 mm 0.1% glucose) (36). Final gel-filtered platelet concentrations were about 1.5 × 108 cells/ml.
Peptide Synthesis and Purification
Synthetic peptides, including calpastat, calpastat-Ala and SFLLR, were synthesized using solid phase Fmoc (N-(9-fluorenyl) methoxycarbonyl/N-methylpyrrolidone) chemistry (37). Calpastat, calpastat-Ala, and the 24-amino acid calpastatin peptide were greater than 95% pure by high performance liquid chromatography and had the correct molecular mass and protein sequences when assayed by mass spectrometry and Edman degradation. Concentrations of the pure solutions were determined by quantitative amino acid analysis.
Calpain Assays
Calpain inhibition assays were performed in assay buffer (50 mm KCl, 10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 1 mm DTT) with CaCl2 added to a final concentration of 3 mm. Purified μ-calpain (50 nm) was used in the assays. The substrate, suc-LLVY-AMC, was used at a concentration of 200 µm. The concentrations of calpastat tested ranged from 25 nm to 10 µm. Calpastat-Ala mutant peptide was tested at 10 µm. To assay calpain activity, cleavage of a fluorogenic substrate, suc-LLVY-AMC, was measured in an SLM Aminco 8000 fluorescence spectrometer using a temperature-controlled cuvette (37 °C). The buffer was prewarmed in the cuvette, and CaCl2, substrate, and inhibitor were added following temperature equilibration. At t = 0 min, μ-calpain was added and the cuvette was mixed by inversion. Excitation and detection wavelengths were 360 and 460 nm, respectively. The initial rate of substrate cleavage, which was linear, was measured at t = 15 to t = 30 s. AMC standard solutions were used to determine picomoles of AMC generated, from emission data.
Assay of Actin-binding Protein and Talin Proteolysis by Calpain
Gel-filtered platelets (1.0 ml at 2 × 108 platelets/ml) were treated with 50 µm ZLLYCHN2 dissolved in Me2SO or 100 µm calpastat dissolved in HEPES-buffered saline (pH 7.4) for 2 h at 37 °C. Following incubation with drug or vehicle control, the platelets were recalcified (2 mm CaCl2, final concentration) and stimulated with calcium ionophore A23187 (1 mm). At the indicated time points, 250-µl aliquots of platelets were removed and immediately lysed with 50 µl of 6× SDS-PAGE loading buffer containing EDTA and EGTA (10 mm each). Lysates were immediately incubated at 95 °C for 3 min. For each time point, a 25-µl sample was removed, heated, and electrophoresed by SDS-PAGE (6% gel). Platelet proteins were visualized by staining the gel with Coomassie Blue.
Measurement of Peptide Uptake by Platelets
Calpastat and the calpastatin peptide 24-mer lacking the signal sequence (Fig. 1A) were separately labeled by reductive tritiation, using NaB(3H)4. Each peptide has the same number of lysine residues and was labeled to a similar specific activity (calpastat 2.64 × 106 cpm/mg and the calpastatin 24-mer 2.06 × 106 cpm/mg). Gel-filtered platelets (approximately 6 × 107) were incubated with tritiated peptides for 60 min at 50 µm concentration, under similar incubation conditions used in secretion, aggregation and spreading experiments. Following the incubation period, the platelets were treated with trypsin/EDTA to remove any surface adherent peptide. There are five potential trypsin cleavage sites in the calpastatin consensus sequence.
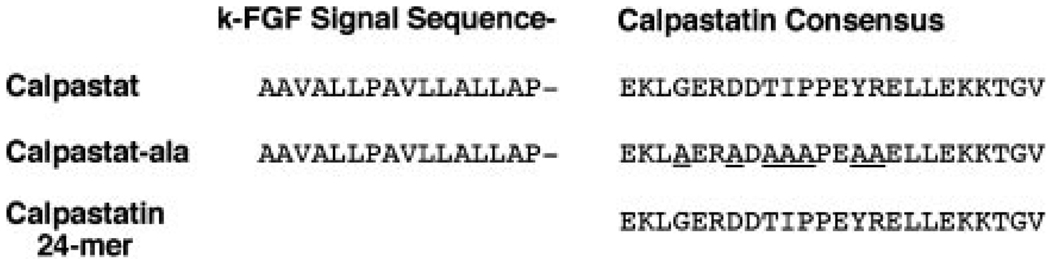
FIG. 1. Amino acid sequence of calpastat and related peptides.
Calpastat, 40 amino acids in length, consists of amino acids 7–22 of human k-FGF at the amino terminus, followed by the 24-amino acid calpastatin consensus sequence of Croall and McGrody (42). Calpastat-Ala is a control peptide, in which the most highly conserved amino acids of the calpastatin consensus sequence are replaced with alanine. The calpastatin 24-mer is the calpastatin consensus sequence (42).
P-selectin Surface Expression Assay
Gel-filtered platelets (1.8 × 108 platelets/ml) were aliquoted (20 µl) into 1.5-ml microcentrifuge tubes. Vehicle (100 mm HEPES, pH 7.4) or inhibitory agent (calpastat, calpastat-Ala, or calpastatin 24-mer) was delivered in 1.0-µl volumes to the indicated final concentration (Fig. 3, A and B). The platelets were then incubated for 2 h at 37 °C. Experiments utilizing the rapid acting peptidyl inhibitors were performed with 10-min pre-incubations with drug. Following treatment with the inhibitors, platelets were recalcified, to 2 mm CaCl2, and then exposed to either SFLLR (50 µm) or phosphate-buffered saline, delivered in a 1-µl volume. The concentration of SFLLR peptide used in these studies (50 µm) resulted in maximal P-selectin surface expression in control studies. After a 30-s exposure, an aliquot of platelets (7.5 µl) was transferred to a microfuge tube containing monoclonal anti-P-selectin antibody (AC1.2) conjugated to phycoerythrin (20-µl volume). To monitor nonspecific binding, a second aliquot of platelets (7.5 µl) was transferred to an Eppendorf tube containing an IgG1 non-immune antibody conjugated to phycoerythrin (20-µl volume). Platelets were incubated with the antibodies for 20 min at room temperature. The mixture was subsequently diluted with 1 ml of paraformaldehyde (2%) and incubated at 4 °C for 2 h. Antibody binding to platelet surface P-selectin was quantified using a FACScan flow cytometer (Becton Dickinson, San Jose, CA). The geometric mean of platelet fluorescence intensity was used to determine surface P-selectin expression.
FIG. 3. P-selectin surface expression is calpain-dependent.
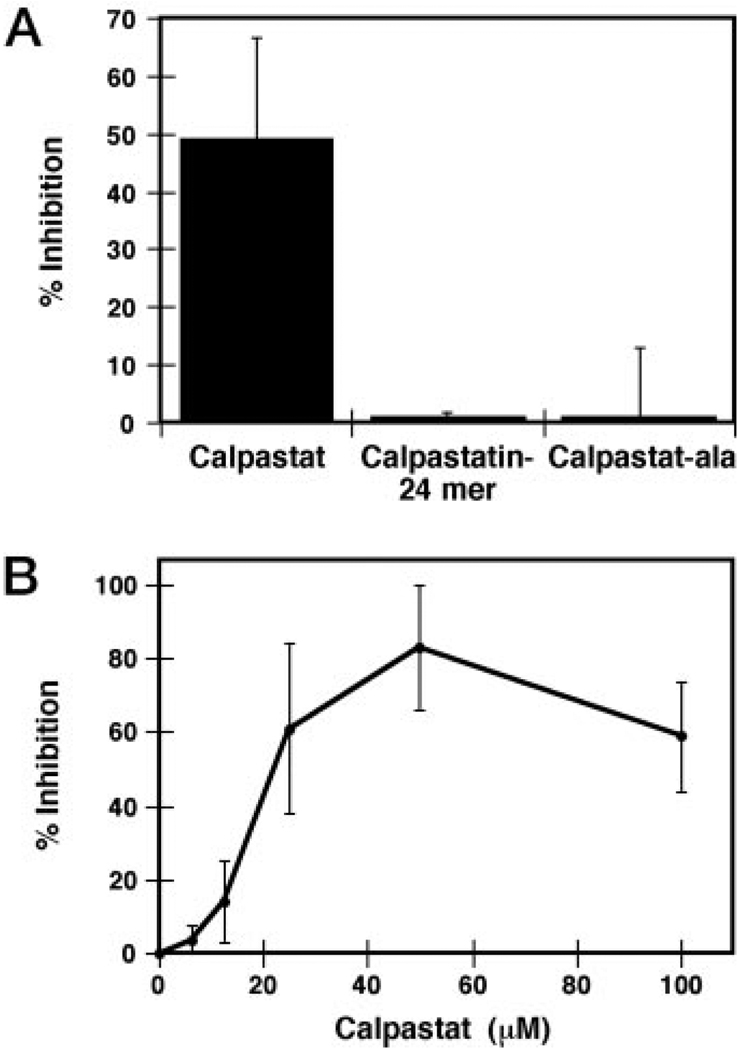
A, calpastat, but not calpastat-Ala nor the calpastatin 24-mer, inhibits P-selectin surface expression on gel-filtered platelets. The extent of P-selectin surface expression from SFLLR-treated platelets was determined from the geometric mean of P-selectin fluorescence, measured by flow cytometry (see “Experimental Procedures”). The SFLLR concentration utilized, 50 µm, results in maximal P-selectin expression. B, the dose response curve for inhibition of platelet P-selectin surface expression by calpastat. The IC50 for calpastat inhibition of SFLLR-induced P-selectin presentation on gel-filtered platelets is 20 µm. Measurement of P-selectin surface expression was performed as in A.
Quantitation of Platelet Aggregation
Gel-filtered platelets (250-µl aliquots at 1.6 × 108/ml) were incubated with the indicated concentration of calpastat or mutant calpastat-Ala for either 20 or 30 min at 37 °C in the absence of calcium and stirring. To test peptidyl calpain inhibitors, platelets were preincubated with the indicated concentration of calpeptin, MDL, E64d, Me2SO (vehicle control), or NH4Cl (cathepsin control) for 10 min at 37 °C. Following incubation, platelets were recalcified (CaCl2, 2 mm) and stimulated with 0.05–1.0 unit/ml thrombin, while being stirred (1400 rpm) at 37 °C. The sensitivity of platelet preparations to thrombin decreased over the course of a given experiment. The thrombin concentration required to initiate maximal aggregation varied between 0.05 and 1.0 unit/ml, but in most cases was 0.05 unit/ml. Aggregation was measured using a Bio-Data lumi-aggregometer. Percentage of inhibition of aggregation was measured at 4 min following the onset of aggregation. The initial rate of aggregation during the first 15 s was compared between treated and untreated samples to calculate the IC50 for each inhibitor.
Assay of Platelet Spreading
Gel-filtered platelets were preincubated with either calpastat or calpastat-Ala mutant (100 µm concentration for 30, 60, and 90 min at 37 °C) or with the low molecular weight inhibitors calpeptin, MDL, or E64d (50–500 µm, for 10 min at 37 °C). An aliquot of the platelet suspension (80 µl) was plated on a glass coverslip, and the platelets were allowed to settle and spread for 20 min at 37 °C in a tissue culture incubator (Revco Ultima Incubator; Revco Scientific, Inc. Asheville, NC). The platelets were then fixed in formaldehyde (3.7%), permeabilized with Triton X-100 (0.1% for 4 min), and blocked in PIPES-buffered saline with 3% bovine serum albumin, as described (12). The coverslips were stained with Oregon Green-phalloidin and photographed on Kodak Tri-X film by photomicrography using a Nikon Optiphot 2 fluorescence microscope (100× objective). Cell area of spread platelets was determined by computer-assisted image analysis using NIH Image 1.61.
Treatment of platelets with 3% Me2SO led to a complete inhibition of spreading and provided base-line cell area for unspread platelets. Percentage of inhibition of spreading was calculated from the increase in mean cell area for untreated and treated samples compared with unspread Me2SO controls.
RESULTS
Design and Characterization of Calpastat and Control Peptides
To investigate the role of calpain in platelet activation, a novel calpain inhibitor was derived from the specific calpain inhibitor, calpastatin. Calpastatin is a modular protein, containing 4 conserved calpain-inhibiting motifs. Short peptide sequences derived from calpastatin effectively inhibit calpain at submicromolar concentrations (38–42). A 24-amino acid calpastatin consensus peptide has been shown to specifically inhibit calpain (42) and not other proteases. Transfection studies have recently demonstrated that this consensus sequence inhibits calpain function in vivo in myocytes (43). To directly import the calpastatin consensus peptide into cells, a hybrid peptide denoted calpastat was designed, which combines the 24-mer consensus sequence of calpastatin (42) with a fragment of the cell-penetrating signal sequence of k-FGF (44) (Fig. 1). This k-FGF sequence facilitates passive entry of peptide sequences into intact cells. Similar hybrid peptide approaches have been used to inhibit NF-κB and FGF-1 nuclear localization and the function of the cytoplasmic tail of glycoprotein IIb IIIa (33–35).
Two control peptides, calpastat-Ala and the calpastatin 24-mer (Fig. 1), were designed to determine the functional importance of the calpastatin consensus sequence and the signal sequence, respectively. Because the calpastatin residues involved in its interaction with calpain are incompletely defined, it was not possible to design an inhibitor peptide containing a single point mutation that would render calpastat inactive. The control peptide calpastat-Ala was thus generated by substituting seven of the conserved amino acids within the calpastatin sequence with alanine residues (Fig. 1). It was predicted that the calpastat-Ala peptide would penetrate cell membranes, but not inhibit calpain. Calpastat-Ala was used to control for any unexpected effects of the k-FGF peptide on platelet function. It was predicted that the calpastatin 24-mer lacking the k-FGF sequence would not penetrate the cell but could inhibit calpain in in vitro assays (Fig. 1). The 24-mer was used to assess the ability of the k-FGF signal sequence to direct cellular uptake of the calpastatin peptide (see below).
Calpastat Inhibits Calpain in Vitro
A fluorometric calpain assay of suc-LLVY-AMC cleavage was used to quantitate the effectiveness of calpastat as a calpain inhibitor (12, 45). Purified μ-calpain was incubated with increasing concentrations of calpastat, and the initial rate of fluorescence increase was measured as an index of calpain activity. The IC50 of calpastat for inhibition of calpain in this assay is 50 nm (Fig. 2A). Calpastat is also a potent inhibitor of μ-calpain-mediated in vitro caseinolysis; an IC50 of 500 nm was measured (data not shown). Calpastat-Ala has no inhibitory activity in either assay. These results demonstrate that calpastat is a potent in vitro inhibitor of calpain activity.
FIG. 2. Calpastat inhibits calpain in vitro.
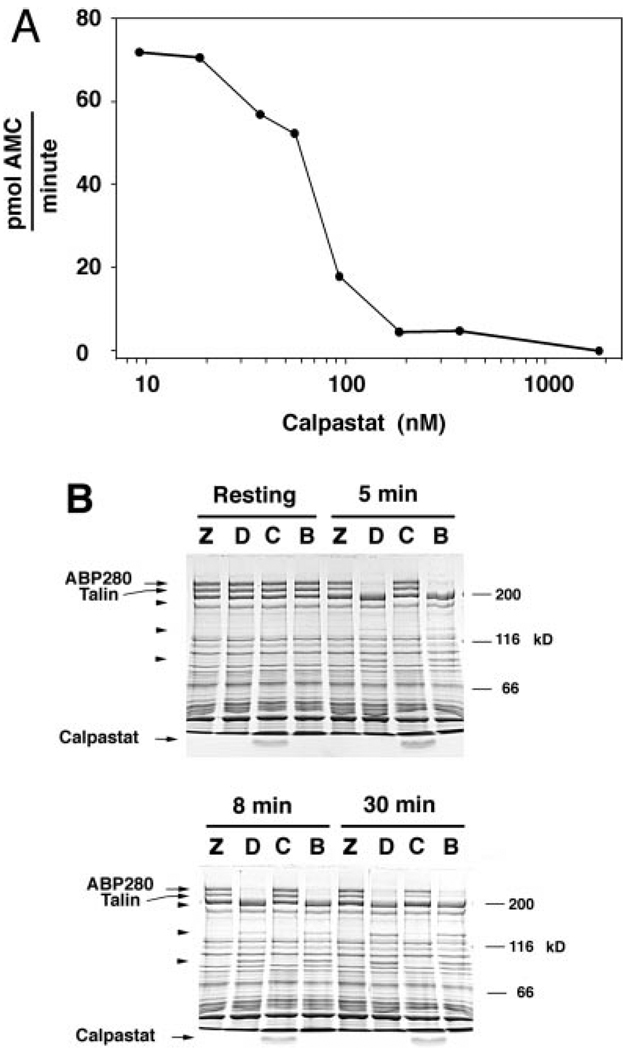
A, the dose-response curve of calpastat for inhibition of μ-calpain cleavage of the substrate suc-LLVY-AMC, assayed as described (see “Experimental Procedures”). The IC50 of calpastat for inhibition of μ-calpain is 70 nm. The calpastat-Ala mutant peptide has no inhibitory activity in this assay. B, the cell-penetrating calpastatin peptide inhibits calpain in vivo. Platelets were preincubated with ZLLYCHN2 (Z), Me2SO vehicle (D), calpastat (C), or HEPES vehicle for calpastat (B), and treated with the ionophore A23187 for 0, 5, 8, or 30 min. Platelet proteins were analyzed by SDS-PAGE, as described (see “Experimental Procedures”). Arrowheads designate the 135- and 93-kDa calpain breakdown products of ABP280 and the 190-kDa calpain breakdown product of talin. The 49-kDa calpain breakdown product of talin is not visualized in this gel system. Free calpastat is visualized at the bottom of lanes corresponding to conditions where the peptide was added.
Platelet Uptake of Calpastat
To assess the ability of the k-FGF-derived signal sequence to direct cellular entry of calpastat, we examined the platelet uptake of tritiated forms of calpastat and the calpastatin 24-mer. Both peptides were labeled by reductive tritiation and incubated with gel-filtered platelets under conditions identical to those used in all other platelet assays. In these experiments, the k-FGF sequence leads to an increase in the uptake of calpastat over the 24-mer by more than 14-fold, from 2.8 fmol/plateletzh to 41 fmol/plateletzh, respectively.
Calpastat Inhibits Calpain in Platelets
To establish the efficacy of calpastat as an inhibitor of intracellular calpain in platelets, we measured the ability of calpastat to inhibit ionophore-induced calpain proteolysis of the cytoskeletal proteins ABP 280 and talin. ABP 280 and talin are calpain substrates and have been used to measure the effectiveness of peptidyl calpain inhibitors such as the suicide substrate ZLLYCHN2 (46, 47). Detectable calpain proteolysis of ABP 280 and talin is stimulated by the calcium ionophore A23187 (Fig. 2B), but not by thrombin or the thrombin receptor agonist SFLLR (data not shown). The ionophore-induced degradation of ABP 280 and talin is seen in control lanes at 5, 8, and 30 min (Fig. 2B, lanes B and D). ABP 280 breakdown products are seen at 135 and 93 kDa, while the predominant talin breakdown product is seen at 190 kDa. Pre-incubation with either calpastat (100 µm) or ZLLYCHN2 (50 µm) effectively inhibits the calpain proteolysis of ABP 280 and talin for up to 30 min following activation with A23187 (Fig. 2B). The inhibition of ionophore-induced ABP 280 and talin cleavage by calpastat establishes its efficacy as an inhibitor of calpain in intact platelets.
Calpain Inhibition Blocks α-Granule Secretion
To determine whether calpain plays a role in α-granule secretion, calpastat was tested for inhibition of thrombin peptide (SFLLR)-induced P-selectin expression. P-selectin surface expression is a result of α-granule fusion with the plasma membrane and exposure of the P-selectin extracellular domain within 30 s of platelet activation by thrombin or SFLLR. A concentration of SFLLR that leads to a maximal increase in P-selectin surface expression (50 µm) was used in these experiments. Pre-incubation of platelets with calpastat (100 µm, 2 h) prior to stimulation results in greater than 50% inhibition of maximal P-selectin surface expression (Fig. 3A). In contrast, pre-incubation with the control peptides, calpastat-Ala (100 µm) or the calpastatin 24-mer (100 µm) has no effect on P-selectin expression (Fig. 3A). Control experiments demonstrated that calpastat did not inhibit P-selectin antibody binding to surface P-selectin, since incubation of platelets with calpastat following exposure to SFLLR has no effect on the binding of antibody to surface expressed P-selectin (data not shown).
Calpastat inhibits the surface expression of P-selectin in a dose-dependent manner, with half-maximal inhibition occurring at a concentration of 20 µm (Fig. 3B). Maximal inhibition of P-selectin surface expression occurs at 50 µm calpastat (80%). Calpastat also inhibits ionophore A23187-induced P-selectin expression in a similar fashion (data not shown).
To further confirm that calpain regulates the surface expression of P-selectin, the inhibitory effects of the rapidly acting, but less specific, peptidyl calpain inhibitors calpeptin, MDL, and E64d were tested. Platelets were pre-incubated with increasing concentrations of calpeptin, MDL, E64d, or Me2SO vehicle, for 10 min prior to stimulation with SFLLR. Calpeptin, MDL, and E64d each inhibit P-selectin surface expression with IC50 values between 200 and 300 µm. These results, together with those derived using the specific inhibitor calpastat, demonstrate a novel role for calpain in the regulation of α-granule exocytosis.
Calpain Inhibition Blocks Platelet Aggregation
The role of calpain in platelet aggregation is controversial because prior studies using the highly specific peptidyl calpain inhibitor, ZLLYCHN2, have suggested that calpain is not required for aggregation, secretion, or spreading (46). We have established in prior work that ZLLYCHN2 entry into cells is delayed, requiring incubation times of 2–16 h to observe the effects of this calpain inhibitor on the actin cytoskeleton (12). We therefore used calpastat to reevaluate the role of calpain in platelet aggregation.
Gel-filtered platelets were preincubated with calpastat or control peptides and recalcified (2 mm Ca2+) just prior to treatment with thrombin. In initial experiments, the thrombin concentration was titrated to the lowest dose capable of inducing aggregation within 15 s (range: 0.05–1.0 unit/ml). In four experiments, calpastat effectively inhibited the extent of platelet aggregation by 60–95% at 4 min following the onset of aggregation (Fig. 4). In contrast, calpastat-Ala (Fig. 4) or the calpastatin 24-mer has no measurable effect on platelet aggregation (Table I). In subsequent experiments, the thrombin concentration used was 0.5 unit/ml and the rate of aggregation was determined in the presence and absence of inhibitor (Table I). Comparing the initial rate of aggregation in calpastat-treated platelets versus controls, the IC50 of calpastat for inhibition of platelet aggregation was determined to be 50 µm (Table I).
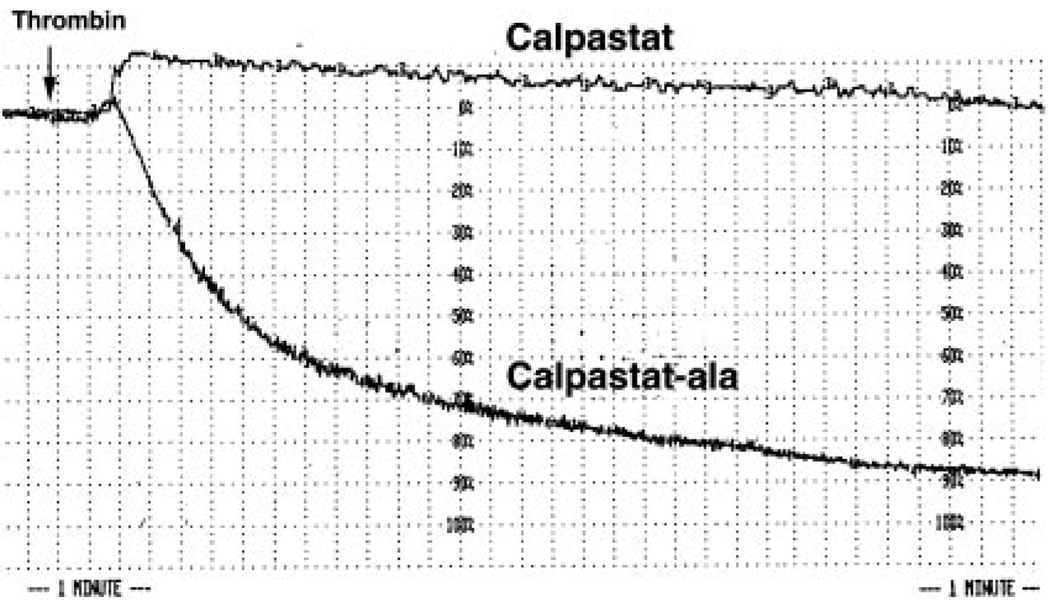
FIG. 4. Aggregation of thrombin-stimulated platelets is calpaindependent.
Calpastat (50 µm) but not calpastat-Ala (50 µm) inhibits thrombin-induced (1.0 unit/ml) platelet aggregation. Aggregation of stirred platelets was initiated by thrombin treatment, as described (see “Experimental Procedures”). The abscissa, indicating time, is marked by broken lines in intervals of 15 s. The ordinate, indicating percentage of light transmission, is marked by broken lines in intervals of 10%. 100% and 0% light transmission were defined by buffer and unaggregated platelets in buffer, respectively. The time of thrombin addition is indicated with an arrow. Gel-filtered platelets were preincubated with calpastat or calpastat-Ala for 20 min before recalcification and subsequent addition of thrombin.
TABLE I. Calpain inhibition blocks platelet aggregation.
The initial slope of thrombin-induced platelet aggregation was compared at varying concentrations of inhibitor. The uninhibited rate of aggregation was determined from peptide vehicle (150 mm HEPES-KOH, pH 7.4), in the case of calpastat and calpastat-Ala, or Me2SO vehicle, in the case of calpeptin, MDL, and E64d. NI indicates that no inhibition of aggregation occurred with calpastat-Ala, at the concentrations used for calpastat, and that no inhibition of aggregation occurred with NH4Cl lysosomal inhibition. Rates of thrombin-induced (0.05 units/ml) aggregation of gel-filtered platelets were measured during the first 15 s of aggregation, immediately following shape change. Thrombin-induced shape change was not affected by any of the inhibitors.
| Inhibitor | IC50 |
|---|---|
| µm | |
| Calpastat | 50 |
| Calpastatin 24-mer | NI |
| Calpastat-Ala | NI |
| Calpeptin | 150 |
| MDL | 240 |
| E64d | 340 |
| NH4Cl (10 mm) | NI |
The immediately acting peptidyl calpain inhibitors calpeptin, MDL, and E64d also inhibit thrombin-induced platelet aggregation, following a 10-min pre-incubation period. Calpeptin, MDL, and E64d inhibit the initial rate of platelet aggregation with IC50 values of 150, 240, and 340 µm, respectively (Table I). At high micromolar concentrations, these peptidyl compounds also inhibit members of the cathepsin family of lysosomal proteases. To test whether the peptidyl calpain inhibitors prevent aggregation by inhibiting cathepsins as well as calpain, platelets were pre-incubated with 10 mm NH4Cl, which maximally inhibits cathepsin function (48). Pre-treatment of platelets with 10 mm NH4Cl does not affect platelet aggregation (Table I), demonstrating that inhibition of aggregation by peptidyl calpain does not occur through inhibition of cathepsins. Inhibition of aggregation by calpastat and by peptidyl calpain inhibitors together demonstrate that calpain plays a regulatory role in the intracellular events leading to platelet aggregation.
Calpain Inhibition Blocks Platelet Spreading, and Filopodial and Lamellipodial Protrusion Formation
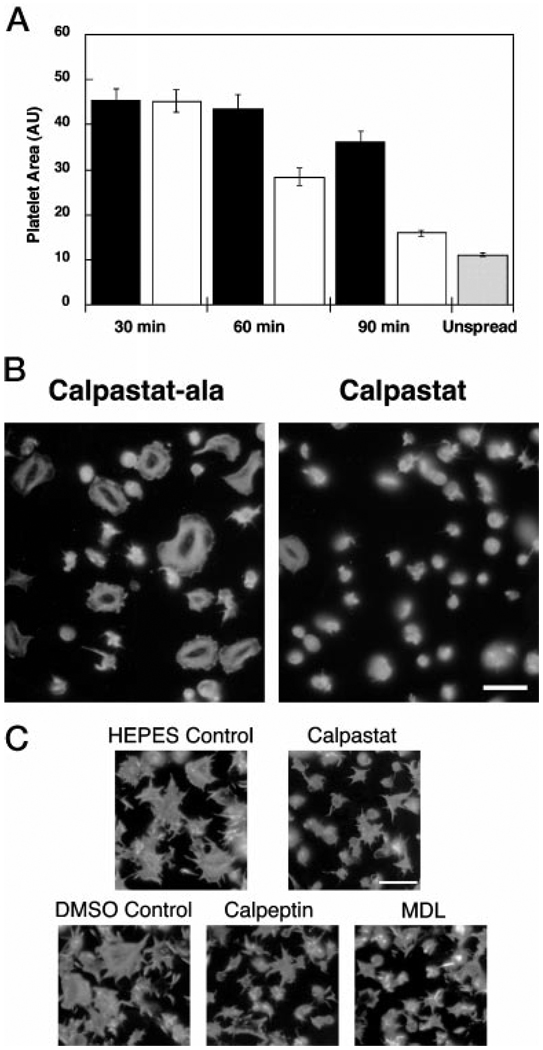
In platelets and fibroblasts, spreading progresses through formation of filopodia and extension of intervening lamellae. Because calpain regulates filopodial and lamellipodial protrusion formation in fibroblasts (12), we tested whether platelet spreading is also calpain-regulated. Spreading, measured by the increase in mean platelet area, was compared between calpastat and calpastat-Ala-treated platelets spread on glass. Pre-incubation of platelets with calpastat (100 µm) for 30, 60, or 90 min results in 0, 46, or 80% inhibition of platelet spreading, or increase in mean platelet area, respectively, relative to the calpastat-Ala control (Fig. 5A). Calpastat-Ala results in minimal inhibition of platelet actin remodeling, including filopodial and lamellipodial protrusion formation, relative to calpastat (Fig. 5B), indicating that the calpastatin motif is responsible for inhibition of spreading. These results indicate that the extent of inhibition of spreading by calpastat is dependent upon the pre-incubation time, and that 30–60 min of pre-incubation is required for activity in this assay. Calpastat thus does not act as rapidly as the peptidyl calpain inhibitors calpeptin, MDL, and E64d (see below). In contrast, ZLLYCHN2, which takes up to 14 h to inhibit spreading in fibroblasts (12), has no inhibitory activity in this assay (data not shown), consistent with prior results (46). The IC50 of calpastat for inhibition of platelet spreading is 35 µm (Table II).
FIG. 5. Spreading of platelets on glass is calpain-dependent.
A, calpastat inhibits platelet spreading, dependent upon the calpastatin consensus sequence, and requires a pre-incubation period of between 30 and 60 min. Platelets were preincubated with calpastat-Ala (black bar) or calpastat (white bar) peptide (100 µm) for 30, 60, or 90 min and then spread for 20 min on glass coverslips. The coverslips were fixed, stained with Oregon Green-phalloidin, and photographed by fluorescence microscopy. The cell areas of the spread platelets were measured by computerized image analysis (NIH Image 1.61). One area unit (AU) is 0.625 µm2. Although calpastat-Ala demonstrated a 20% nonspecific inhibition of spreading relative to a vehicle control at 90 min of preincubation (data not shown), this inhibition was 4-fold less than the 85% inhibition observed for calpastat, compared with the vehicle control. Unspread platelets (gray bar) were obtained by treating platelets with 3% Me2SO, which completely blocks spreading, and allowing them to adhere to glass coverslips for 20 min as above. B, calpastat inhibits platelet actin remodeling during spreading, dependent upon the calpastatin consensus sequence. Platelets were incubated with peptide for 90 min and then spread on glass for 20 min. The coverslips were processed, stained with Oregon Green-phalloidin, and photographed by fluorescence microscopy. The size bar is 10 µm. C, similar to calpastat, MDL and calpeptin inhibit platelet actin remodeling during spreading. Calpastat (100 µm), MDL (400 µm), and calpeptin (300 µm) were used at their respective IC60 concentrations for spreading inhibition. Preincubation times were 90 min for calpastat and HEPES vehicle control and 10 min for MDL, calpeptin, and the Me2SO vehicle control. NH4Cl (10 µm) treatment for 10 min had no detectable effect on spreading (data not shown). The coverslips were processed, stained with Oregon Green-phalloidin, and photographed by fluorescence microscopy. The size bar is 10 µm. The IC60 concentrations of inhibitors were chosen to demonstrate the morphologies of the actin cytoskeleton for calpastat and peptidyl calpain inhibitor-treated platelets, under conditions of partial and equal spreading.
TABLE II. Calpain inhibition blocks platelet spreading.
The quantitation of platelet spreading on glass coverslips was compared at varying concentrations of inhibitor. The uninhibited extent of spreading was determined from peptide vehicle (150 mm HEPES-KOH, pH 7.4), in the case of calpastat and calpastat-Ala, or Me2SO vehicle, in the case of calpeptin, MDL, and E64d. NI indicates that no inhibition of spreading occurred following treatment of platelets with calpastat-Ala, or following lysosomal inhibition with NH4Cl. The distribution of spread and unspread platelets is bimodal under these conditions.
| Inhibitor | IC50 |
|---|---|
| µm | |
| Calpastat | 35 |
| Calpastat-Ala | NI |
| Calpeptin | 200 |
| MDL | 200 |
| E64d | >250 |
| NH4Cl (10 mm) | NI |
Notably absent in the calpastat-treated cells is the peripheral rim of F-actin, at the leading edge of each lamellae (Fig. 5B). Half of the platelets pre-treated with calpastat (100 µm) for 1.5 h are round and unspread on glass, having a smooth cortical layer of F-actin (Fig. 5B). The disclike unspread platelets demonstrate few or no filopodia and lack lamellipodial protrusions. This finding strongly suggests that calpain is involved in an early process in spreading, rather than retraction following spreading, because the unspread platelets lack the pattern of retraction fibers together with extended filopodia, seen in platelets undergoing cytoskeletal retraction.
To confirm the role of calpain in platelet spreading, the peptidyl calpain inhibitors calpeptin, MDL, or E64d were tested. Platelets were pre-incubated for 10 min with varying concentrations of calpeptin, MDL, and E64d (50–500 µm) and then spread on glass as above. All three compounds effectively inhibit platelet spreading (Table II). The IC50 values for inhibition of spreading by calpeptin, MDL, and E64d are 200, 200, and >250 µm, respectively (Table II), concentrations at least 6-fold higher than the IC50 of calpastat. Calpeptin and MDL demonstrate minimal inhibitory concentrations (MICs) similar to those previously observed for inhibition of fibroblast spreading, in the 50–80 mm range (12).
The spreading morphology of peptidyl calpain inhibitor treated platelets is similar to that of calpastat-treated platelets, when compared at concentrations resulting in equivalent inhibition of spreading. There is a notable absence of the peripheral rim of F-actin at the leading edges of the lamellae, when platelets are spread at the IC60 concentrations of calpastat, calpeptin, or MDL (Fig. 5C). When platelets are spread at the IC50 concentrations for calpeptin or MDL (200 µm), 30% are round and unspread, with no filopodia or lamellipodia, indicating complete inhibition of actin protrusion formation. In contrast, none of the control platelets lack filopodia or lamellipodia under identical spreading conditions. About 20% of platelets pretreated at the IC20 of E64d (250 µm) are round and unspread (data not shown). As in assays of fibroblast spreading (12), 10 mm NH4Cl has no effect on platelet spreading (Table II), indicating that inhibition of platelet spreading by peptidyl calpain inhibitors does not occur through inhibition of lysosomal cathepsins. Blocking of platelet spreading by calpastat and by the peptidyl calpain inhibitors, with similar inhibitor effects on spreading-associated actin remodeling, together demonstrate that calpain regulates platelet spreading. The morphology of the unspread cells suggests that calpain plays an important role in the actin remodeling events of lamellipodial and filopodial protrusion formation in platelets.
DISCUSSION
Although calpain is activated in agonist-stimulated platelets, the precise roles of calpain in platelet activation are unknown. The present study demonstrates the Ca2+ and calpain dependence of platelet secretion, aggregation, and spreading. A cell-permeant calpastatin motif, calpastat, and three distinct peptidyl calpain inhibitors, calpeptin, MDL, and E64d, block platelet secretion, aggregation, and spreading. These inhibitors block the expression of P-selectin on the platelet surface, remodeling of the actin cytoskeleton during filopodial and lamellipodial protrusion formation, and cleavage of the membrane-associated proteins ABP 280 and talin during platelet activation events. These results implicate calpain in Ca2+-associated secretion events occurring during the first 30 s of platelet activation by the thrombin receptor agonist peptide, SFLLR. This suggests a novel and direct connection between thrombin receptor signaling and early activation of calpain. Blocking of platelet aggregation and spreading by calpain inhibition also indicates that calpain regulates later platelet activation events involving integrin function, as well as actin remodeling. These results suggest that calpain plays a critical, rate-limiting role in the activation of platelets by thrombin, promoting remodeling of actin networks and the platelet membrane cytoskeleton.
Prior studies of thrombin-induced platelet aggregation have demonstrated that an early sign of calpain activation, autolysis, occurs within 30–60 s of agonist stimulation, but not before (15). Calpain also rapidly translocates to the plasma membrane within 10 s of thrombin stimulation, but it has been proposed that this event alone is not sufficient for calpain activation (15). Activation of calpain was reported to be dependent upon both engagement of integrin IIb/IIIa (15) and stirring (16, 17, 49). Nonetheless, α-granule secretion due to thrombin receptor stimulation is independent of integrin IIb/IIIa function (50, 51). The results presented here indicate that calpain is functionally active in thrombin peptide-stimulated α-granule secretion during the first 30 s of activation, suggesting that rapid translocation of calpain to the platelet membrane may be of functional significance earlier in platelet activation than had been previously anticipated. Of interest, activation of calpain, measured by autolysis, is greatly reduced in Glanzmann’s thrombasthenia platelets (15), despite the fact that α-granule secretion is not defective (50, 51). It is possible that secretion may require activation of only a small fraction of the calpain pool, not detectable by Western blotting. We hypothesize that there may be a direct connection between thrombin receptor engagement and initial calpain activation that may be sufficient to activate secretion and precede integrin IIb/IIIa mechanisms. Further studies will address this possibility.
While Ca2+ transients regulate secretory processes in platelets and other cells, its precise role in platelet degranulation has yet to be elucidated. Our investigations have demonstrated that inhibition of calpain by calpastat or by peptidyl inhibitors resulted in 50–80% inhibition of P-selectin surface expression and α-granule secretion. Consistent with these findings are reports that calpain regulates secretion in type II alveolar cells (52–54). At present it is unknown whether resting platelet α-granules exist both pre-docked at the plasma membrane and free in the cytoplasm. The ability of the calpain inhibitors to block 50–80%, but not all α-granule secretion may result from a specific role for calpain in initial events enabling the priming of undocked rather than pre-docked granule stores.
Recently, it was demonstrated that calpain cleaves spectrin, α-SNAP, syntaxin I, and synaptobrevin II during activationdirected degranulation of type II alveolar cells (52, 55). Our recent finding that SNARE family members are involved in α-granule secretion raises the possibility that calpain proteolysis facilitates granule fusion via cleavage of α-granule associated SNARE family members (5). The role of proteolysis in vesicle fusion is nonetheless unknown, and future investigations will examine the cleavage of these proteins during platelet activation. At present, our experiments also have not eliminated the possibility that calpain is involved in upstream signaling events required for α-granule secretion.
Platelet aggregation draws on coordinated regulation of numerous signaling, cytoskeletal and adhesive processes. Calpain may be involved in the proteolytic cleavage of signaling kinases and phosphatases to active or inactive forms. Calpain cleavage of protein kinase C generates a constitutively active isoform, protein kinase M (PKM), which no longer requires diacyl glycerol for activity (24). Calpain cleavage of phosphatidylinositol 4-phosphatase results in inactivation of this enzyme, which may facilitate platelet activation by elevating levels of phosphatidylinositol 3,4-bisphosphate (21, 56). Alternatively, calpain may facilitate the activation of phosphatidylinositol 3-phosphate 4-kinase, through mechanisms that remain to be fully elucidated (57). It is also possible that calpain regulates the clustering and ligand engagement of platelet integrins, which are involved both in signaling process and in adhesive events. Thrombin-induced cleavage of the cytoplasmic domain of β3 integrin is dependent on calpain (18), and calpain is known to regulate integrin clustering in other cell types (8).
In the present study, inhibition of calpain with calpastat or the peptidyl calpain inhibitors blocks cell spreading. This finding is consistent with our previous report that calpain regulates spreading in fibroblasts (12). Notably, the MICs of calpeptin, MDL, and E64d for spreading are comparable to the MICs of these inhibitors for spreading of fibroblasts (12). When calpain is inhibited in platelets, filopodial and lamellipodial protrusion formation is significantly attenuated, demonstrating a role for calpain in remodeling of the cortical actin cytoskeleton in platelets. Together, the platelet and fibroblast results suggest that modulation of cell spreading by calpain may be a general mechanism used by a variety of different cell types.
There are at least three mechanisms by which calpain could modulate platelet spreading. First, calpain cleavage of actin filament capping complexes may expose barbed ends for polymerization and/or untethering the filamentous actin cytoskeleton from the plasma membrane. Although there is evidence to support a role for calpain in the uncapping of β-actin filaments through cleavage of the cytoskeleton-membrane linking protein, ezrin (11, 58), ezrin is not abundant in platelets (59). The dominant member of the ezrin/radixin/moesin family found in platelets is moesin, which is not a calpain substrate (60). Recently, adducin, a calpain substrate, has been identified as an actin capping protein in a variety of cell types (61–65). It is possible that adducin may be a calpain target during actin remodeling events in platelets. Calpain might also facilitate platelet spreading by raising intracellular levels of the 4-phosphate phosphatidylinositols (PIs) PI (4,5)P and PI (3,4)P, which can directly uncap β-actin filaments (56, 66). Finally, calpain could facilitate platelet spreading by activating PKC (19), which is functionally active during platelet activation and is known to facilitate cell spreading by an undetermined mechanism (67).
Regulated proteolysis plays an important role in the regulation of intracellular signal transduction and cytoskeletal remodeling events. Our data provide strong evidence that calpain is a critical Ca2+ triggered molecular switch that regulates a series of complex processes during platelet activation. Identification of the critical physiologic substrates of calpain during platelet activation will add considerably to our understanding of calpain’s specific role in cell activation.
Acknowledgments
We are grateful to Drs. Dorothy E. Croall, John S. Elce, Kevin K. W. Wang, Ronald L. Mellgren, Yao-Zhong Lin, J. Fred Dice, Jr., Anna Maria Cuervo, John H. Hartwig, Gail W. V. Johnson, Herbert Angliker, Jun-ichi Kambayashi, Shujaath Mehdi, Un-Jin P. Zimmerman, Athan Kuliopulos, and Michael E. Mendelsohn, for reagents and/or helpful discussions. A. M. Cuervo and J. F. Dice, Jr. performed reductive tritiation of peptides. We thank Margaret Jacobs and Michael Berne for expert technical assistance in peptide synthesis.
Footnotes
This work was supported by National Institutes of Health Grants K08-CA 1562 and P30 DK34928 (GRASP Digestive Disease Research Center grant to D. A. P. and I. M. H.), GM 55110, EY09033 and P30 DK34928 (GRASP Imaging Core grant) (to I. M. H.), and HL42443 and HL51926 (to B. F. and B. C. F.), and by a New England Medical Center grant (to D. A. P.).
The abbreviations used are: ABP 280, actin-binding protein 280; AMC, aminomethylcoumarin; k-FGF, Kaposi’s fibroblast growth factor; MDL, MDL28,170; MIC, minimal inhibitory concentration; SFLLR, thrombin agonist peptide; suc-LLVY-AMC, succinyl-leucyl-leucyl-valyltyrosyl-7-amino-4-methylcoumarin; ZLLYCHN2, benzyloxycarbonylleucyl-leucyl-tyrosyl-diazomethane; PI, phosphatidylinositol; PKC, protein kinase C; PAGE, polyacrylamide gel electrophoresis; PIPES, 1,4-piperazinediethanesulfonic acid.
REFERENCES
- 1.Yoshida K, Dubyak G, Nachmias VT. FEBS Lett. 1986;206:273–278. doi: 10.1016/0014-5793(86)80995-4. [DOI] [PubMed] [Google Scholar]
- 2.Winokur R, Hartwig JH. Blood. 1995;85:1796–1804. [PubMed] [Google Scholar]
- 3.White JG, Rao GH, Gerrard JM. Am. J. Pathol. 1974;77:135–149. [PMC free article] [PubMed] [Google Scholar]
- 4.Gartner TK, Gerrard JM, White JG, Williams DC. Blood. 1981;58:153–157. [PubMed] [Google Scholar]
- 5.Flaumenhaft R, Croce K, Chen E, Furie B, Furie BC. J. Biol. Chem. 1999;274:2492–2501. doi: 10.1074/jbc.274.4.2492. [DOI] [PubMed] [Google Scholar]
- 6.Bottecchia D, Fantin G, Gruppo F, Nassuato G. Haemostasis. 1976;5:176–188. doi: 10.1159/000214133. [DOI] [PubMed] [Google Scholar]
- 7.Phillips DR, Jakabova M. J. Biol. Chem. 1977;252:5602–5605. [PubMed] [Google Scholar]
- 8.Stewart MP, McDowall A, Hogg N. J. Cell Biol. 1998;140:699–707. doi: 10.1083/jcb.140.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hato T, Pampori N, Shattil SJ. J. Cell Biol. 1998;141:1685–1695. doi: 10.1083/jcb.141.7.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoenwaelder SM, Yuan Y, Cooray P, Salem HH, Jackson SP. J. Biol. Chem. 1997;272:1694–1702. doi: 10.1074/jbc.272.3.1694. [DOI] [PubMed] [Google Scholar]
- 11.Shuster CB, Herman IM. J. Cell Biol. 1995;128:837–848. doi: 10.1083/jcb.128.5.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potter DA, Tirnauer JS, Janssen R, Croall DE, Hughes CN, Fiacco KA, Mier JW, Maki M, Herman IM. J. Cell Biol. 1998;141:647–662. doi: 10.1083/jcb.141.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huttenlocher A, Palecek SP, Lu Q, Zhang W, Mellgren RL, Lauffenburger DA, Ginsberg MH, Horwitz AF. J. Biol. Chem. 1997;272:32719–32722. doi: 10.1074/jbc.272.52.32719. [DOI] [PubMed] [Google Scholar]
- 14.Fox JE, Reynolds CC, Morrow JS, Phillips DR. Blood. 1987;69:537–545. [PubMed] [Google Scholar]
- 15.Fox JE, Taylor RG, Taffarel M, Boyles JK, Goll DE. J. Cell Biol. 1993;120:1501–1507. doi: 10.1083/jcb.120.6.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saido TC, Suzuki H, Yamazaki H, Tanoue K, Suzuki K. J. Biol. Chem. 1993;268:7422–7426. [PubMed] [Google Scholar]
- 17.Fox JE, Reynolds CC, Phillips DR. J. Biol. Chem. 1983;258:9973–9981. [PubMed] [Google Scholar]
- 18.Du X, Saido TC, Tsubuki S, Indig FE, Williams MJ, Ginsberg MH. J. Biol. Chem. 1995;270:26146–26151. doi: 10.1074/jbc.270.44.26146. [DOI] [PubMed] [Google Scholar]
- 19.Pontremoli S, Melloni E, Sparatore B, Michetti M, Salamino F, Horecker BL. J. Biol. Chem. 1990;265:706–712. [PubMed] [Google Scholar]
- 20.Oda A, Druker BJ, Ariyoshi H, Smith M, Salzman EW. J. Biol. Chem. 1993;268:12603–12608. [PubMed] [Google Scholar]
- 21.Norris FA, Atkins RC, Majerus PW. J. Biol. Chem. 1997;272:10987–10989. doi: 10.1074/jbc.272.17.10987. [DOI] [PubMed] [Google Scholar]
- 22.Cooray P, Yuan Y, Schoenwaelder SM, Mitchell CA, Salem HH, Jackson SP. Biochem. J. 1996;318:41–47. doi: 10.1042/bj3180041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frangioni JV, Oda A, Smith M, Salzman EW, Neel BG. EMBO J. 1993;12:4843–4856. doi: 10.1002/j.1460-2075.1993.tb06174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carafoli E, Molinari M. Biochem. Biophys. Res. Commun. 1998;247:193–203. doi: 10.1006/bbrc.1998.8378. [DOI] [PubMed] [Google Scholar]
- 25.Inomata M, Hayashi M, Ohno-Iwashita Y, Tsubuki S, Saido TC, Kawashima S. Arch. Biochem. Biophys. 1996;328:129–134. doi: 10.1006/abbi.1996.0152. [DOI] [PubMed] [Google Scholar]
- 26.Schmaier AH, Bradford HN, Lundberg D, Farber A, Colman RW. Blood. 1990;75:1273–1281. [PubMed] [Google Scholar]
- 27.Crawford C. In: Intracellular Calcium Dependent Proteolysis. Mellgren R, Murachi T, editors. Boca Raton, FL: CRC Press; 1990. pp. 75–89. [Google Scholar]
- 28.Croall DE, DeMartino GN. Physiol. Rev. 1991;71:813–847. doi: 10.1152/physrev.1991.71.3.813. [DOI] [PubMed] [Google Scholar]
- 29.Nishiura I, Tanaka K, Yamato S, Murachi T. J. Biochem. (Tokyo) 1978;84:1657–1659. doi: 10.1093/oxfordjournals.jbchem.a132296. [DOI] [PubMed] [Google Scholar]
- 30.Nishiura I, Tanaka K, Murachi T. Experientia (Basel) 1979;35:1006–1007. doi: 10.1007/BF01949908. [DOI] [PubMed] [Google Scholar]
- 31.Waxman L, Krebs EG. J. Biol. Chem. 1978;253:5888–5891. [PubMed] [Google Scholar]
- 32.Murachi T, Tanaka K, Hatanaka M, Murakami T. Adv. Enzyme Regul. 1980;19:407–424. doi: 10.1016/0065-2571(81)90026-1. [DOI] [PubMed] [Google Scholar]
- 33.Lin YZ, Yao SY, Veach RA, Torgerson TR, Hawiger J. J. Biol. Chem. 1995;270:14255–14258. doi: 10.1074/jbc.270.24.14255. [DOI] [PubMed] [Google Scholar]
- 34.Liu KY, Timmons S, Lin YZ, Hawiger J. Proc. Natl. Acad. Sci. U. S. A. 1996;93:11819–11824. doi: 10.1073/pnas.93.21.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin YZ, Yao SY, Hawiger J. J. Biol. Chem. 1996;271:5305–5308. doi: 10.1074/jbc.271.10.5305. [DOI] [PubMed] [Google Scholar]
- 36.Hsu-Lin S, Berman CL, Furie BC, August D, Furie B. J. Biol. Chem. 1984;259:9121–9126. [PubMed] [Google Scholar]
- 37.Jacobs M, Freedman SJ, Furie BC, Furie B. J. Biol. Chem. 1994;269:25494–25501. [PubMed] [Google Scholar]
- 38.Maki M, Takano E, Osawa T, Ooi T, Murachi T, Hatanaka M. J. Biol. Chem. 1988;263:10254–10261. [PubMed] [Google Scholar]
- 39.Maki M, Bagci H, Hamaguchi K, Ueda M, Murachi T, Hatanaka M. J. Biol. Chem. 1989;264:18866–18869. [PubMed] [Google Scholar]
- 40.Kawasaki H, Emori Y, Imajoh-Ohmi S, Minami Y, Suzuki K. J. Biochem. (Tokyo) 1989;106:274–281. doi: 10.1093/oxfordjournals.jbchem.a122844. [DOI] [PubMed] [Google Scholar]
- 41.Maki M, Hatanaka M, Takano E, Murachi T. In: Intracellular Calcium-dependent Proteolysis. Mellgren R, Murachi T, editors. Boca Raton, FL: CRC Press; 1990. pp. 37–54. [Google Scholar]
- 42.Croall DE, McGrody KS. Biochemistry. 1994;33:13223–13230. doi: 10.1021/bi00249a008. [DOI] [PubMed] [Google Scholar]
- 43.Huang J, Forsberg NE. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12100–12105. doi: 10.1073/pnas.95.21.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delli-Bovi P, Curatola AM, Newman KM, Sato Y, Moscatelli D, Hewick RM, Rifkin DB, Basilico C. Mol. Cell. Biol. 1988;8:2933–2941. doi: 10.1128/mcb.8.7.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bronk SF, Gores GJ. Am. J. Physiol. 1993;264:G744–G751. doi: 10.1152/ajpgi.1993.264.4.G744. [DOI] [PubMed] [Google Scholar]
- 46.Anagli J, Hagmann J, Shaw E. Biochem. J. 1991;274:497–502. doi: 10.1042/bj2740497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anagli J, Hagmann J, Shaw E. Biochem. J. 1993;289:93–99. doi: 10.1042/bj2890093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hopgood MF, Clark MG, Ballard FJ. Biochem. J. 1977;164:399–407. doi: 10.1042/bj1640399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallace RW, Tallant EA, McManus MC. Biochemistry. 1987;26:2766–2773. doi: 10.1021/bi00384a017. [DOI] [PubMed] [Google Scholar]
- 50.Cramer EM, Berger G, Berndt MC. Blood. 1994;84:1722–1730. [PubMed] [Google Scholar]
- 51.Furman MI, Liu L, Benoit SE, Becker RC, Barnard MR, Michelson AD. Proc. Natl. Acad. Sci. U. S. A. 1998;95:3082–3087. doi: 10.1073/pnas.95.6.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zimmerman UJ, Speicher DW, Fisher AB. Biochim. Biophys. Acta. 1992;1137:127–134. doi: 10.1016/0167-4889(92)90193-f. [DOI] [PubMed] [Google Scholar]
- 53.Zimmerman UJ, Wang M, Liu L. Cell Calcium. 1995;18:1–8. doi: 10.1016/0143-4160(95)90040-3. [DOI] [PubMed] [Google Scholar]
- 54.Li HL, Feinstein SI, Liu L, Zimmerman UJ. Cell. Signal. 1998;10:137–142. doi: 10.1016/s0898-6568(97)00101-0. [DOI] [PubMed] [Google Scholar]
- 55.Zimmerman UJP, Malek SK, Liu L, Li HL. TUBMB Life. 1999;48:453–458. doi: 10.1080/713803537. [DOI] [PubMed] [Google Scholar]
- 56.Hartwig JH, Bokoch GM, Carpenter CL, Janmey PA, Taylor LA, Toker A, Stossel TP. Cell. 1995;82:643–653. doi: 10.1016/0092-8674(95)90036-5. [DOI] [PubMed] [Google Scholar]
- 57.Banfic H, Downes CP, Rittenhouse SE. J. Biol. Chem. 1998;273:11630–11637. doi: 10.1074/jbc.273.19.11630. [DOI] [PubMed] [Google Scholar]
- 58.Shuster CB, Lin AY, Nayak R, Herman IM. Cell Motil. Cytoskeleton. 1996;35:175–187. doi: 10.1002/(SICI)1097-0169(1996)35:3<175::AID-CM1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 59.Amieva MR, Furthmayr H. Exp. Cell Res. 1995;219:180–196. doi: 10.1006/excr.1995.1218. [DOI] [PubMed] [Google Scholar]
- 60.Shcherbina A, Bretscher A, Kenney DM, Remold-O’Donnell E. FEBS Lett. 1999;443:31–36. doi: 10.1016/s0014-5793(98)01674-3. [DOI] [PubMed] [Google Scholar]
- 61.Kuhlman PA, Hughes CA, Bennett V, Fowler VM. J. Biol. Chem. 1996;271:7986–7991. doi: 10.1074/jbc.271.14.7986. [DOI] [PubMed] [Google Scholar]
- 62.Kuhlman PA, Fowler VM. Biochemistry. 1997;36:13461–13472. doi: 10.1021/bi970601b. [DOI] [PubMed] [Google Scholar]
- 63.Li X, Matsuoka Y, Bennett V. J. Biol. Chem. 1998;273:19329–19338. doi: 10.1074/jbc.273.30.19329. [DOI] [PubMed] [Google Scholar]
- 64.Matsuoka Y, Li X, Bennett V. J. Cell Biol. 1998;142:485–497. doi: 10.1083/jcb.142.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scaramuzzino DA, Morrow JS. Proc. Natl. Acad. Sci. U. S. A. 1993;90:3398–3402. doi: 10.1073/pnas.90.8.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schafer DA, Jennings PB, Cooper JA. J. Cell Biol. 1996;135:169–179. doi: 10.1083/jcb.135.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vuori K, Ruoslahti E. J. Biol. Chem. 1993;268:21459–21462. [PubMed] [Google Scholar]