Abstract
Emerging infectious diseases are of great concern for both wildlife and humans. Several highly virulent fungal pathogens have recently been discovered in natural populations, highlighting the need for a better understanding of fungal-vertebrate host-pathogen interactions. Because most fungal pathogens are not fatal in the absence of other predisposing conditions, host-pathogen dynamics for deadly fungal pathogens are of particular interest. The chytrid fungus Batrachochytrium dendrobatidis (hereafter Bd) infects hundreds of species of frogs in the wild. It is found worldwide and is a significant contributor to the current global amphibian decline. However, the mechanism by which Bd causes death in amphibians, and the response of the host to Bd infection, remain largely unknown. Here we use whole-genome microarrays to monitor the transcriptional responses to Bd infection in the model frog species, Silurana (Xenopus) tropicalis, which is susceptible to chytridiomycosis. To elucidate the immune response to Bd and evaluate the physiological effects of chytridiomycosis, we measured gene expression changes in several tissues (liver, skin, spleen) following exposure to Bd. We detected a strong transcriptional response for genes involved in physiological processes that can help explain some clinical symptoms of chytridiomycosis at the organismal level. However, we detected surprisingly little evidence of an immune response to Bd exposure, suggesting that this susceptible species may not be mounting efficient innate and adaptive immune responses against Bd. The weak immune response may be partially explained by the thermal conditions of the experiment, which were optimal for Bd growth. However, many immune genes exhibited decreased expression in Bd-exposed frogs compared to control frogs, suggesting a more complex effect of Bd on the immune system than simple temperature-mediated immune suppression. This study generates important baseline data for ongoing efforts to understand differences in response to Bd between susceptible and resistant frog species and the effects of chytridiomycosis in natural populations.
Introduction
Emerging infectious diseases are receiving increasing attention for their role in precipitous population declines and extinctions in natural populations (e.g., [1]). Historically, fungal pathogens of wildlife have been understudied because they are rarely lethal for vertebrates without other immuno-compromising conditions [2]. However the recent discovery of deadly fungal skin infections in natural vertebrate populations (e.g., [3]) raises a number of questions about how a pathogen localized to the skin can lead so quickly to death and threaten entire populations.
One of the most dramatic examples of the impact of pathogenic fungi on vertebrate populations is the chytrid Batrachochytrium dendrobatidis (hereafter Bd), which has been implicated in global amphibian declines. This pathogen, discovered and described only 10 years ago [4], infects hundreds of amphibian species worldwide [5]. Batrachochytrium dendrobatidis is an aquatic fungus and attacks the keratinized epithelial cells of adult amphibian skin [6]. Chytridiomycosis, the disease caused by Bd is often fatal, and is the clear cause of catastrophic reductions in many frog populations around the world (e.g., [7], [8]).
Although some clinical, physiological, and immunological responses of frogs suffering from chytridiomycosis have been described [6], [9], [10], remarkably little is known about the mechanisms involved in amphibian response to Bd. At the clinical level, frogs suffering from chytridiomycosis exhibit hyperkeratosis with epidermal hyperplasia and excessive skin shedding [6], [11]. Recent work has demonstrated that frogs with severe Bd infections exhibit decreases in electrolyte concentrations, plasma osmolality and blood pH [9]. Osmotic and electrolyte imbalances help explain the proximate cause of frog mortality due to chytridiomycosis, but much remains to be discovered about the physiological effects of Bd infection. Similarly, the immune response to Bd is not well understood. A number of studies have shown that the anti-microbial peptides (AMPs) secreted by amphibians can inhibit Bd growth in vitro and may play a role in resistance to infection (e.g. [10]). Although AMPs, as an important component of the innate immune system, represent a potential first line of defense against Bd, their direct role in combating Bd infection in nature remains to be demonstrated. More broadly, there is no published evidence for an adaptive immune response in frogs exposed to Bd.
Many other fungal pathogens elicit a robust adaptive immune response in their vertebrate hosts (see review in [12], [13]), and there is no reason to suspect a priori a diminished role for adaptive immunity to Bd. For example, the cell types involved in mammalian response to fungal skin pathogens (dermatophytes) are similarly found in the skin of Xenopus laevis [14]–[16] and would be expected to mount a similar localized inflammatory response. Several recent reviews have called for further investigation of the role of adaptive immunity in frog response to Bd [17], [18]. Moreover, given the considerable cross-talk between elements of innate and acquired immunity, it is productive to take a comprehensive approach to studying frog immune response to Bd. Such a comprehensive characterization of host response to pathogen exposure can be achieved with genome-wide expression studies [19], [20].
Here we present a global approach to understanding frog responses to chytrid exposure by using whole genome transcription assays in Silurana tropicalis (formerly Xenopus tropicalis, [21]). Xenopus and Silurana frogs have been used for many decades in immunological investigation (e.g., reviewed by [14], [22]). Additionally, the complete genome of S. tropicalis has now been sequenced, enabling genome-scale approaches to studying the response to pathogen exposure. We document gene expression changes in multiple tissues at multiple time-points for frogs experimentally exposed to Bd under controlled laboratory conditions. We present data on the genetic pathways perturbed in response to Bd and discuss the implications for understanding disease-related amphibian declines.
Materials and Methods
Animal husbandry
Adult Silurana tropicalis were used for infection experiments, and frogs were housed individually for the duration of the experiment. Twice per week, frogs were fed ad libitum, and tank water was changed. Stock colonies of S. tropicalis were maintained at 25°C. However, experimental inoculations and observations were executed at 18°C. It is important to note that this temperature regime may be suboptimal for amphibian immune response; however, there are several reasons that this first study was conducted at this temperature. First, we were interested in the physiological changes associated with fatal Bd infections, and we found a stronger reaction to Bd exposure at cooler temperatures during preliminary trials. Second, many catastrophic Bd-related declines in natural populations occur during cooler months and in cooler montane habitats [23], [24], therefore understanding the response to Bd exposure at non-preferred temperatures is critical. Third, we prioritized conducting experiments within the optimal growth temperature for Bd [25] to support ongoing work connecting gene expression patterns of both host and pathogen.
Experimental frogs were acclimated to progressively cooler temperatures over the course of one week and then allowed to acclimatize for an additional week at 18°C. A swabbing method was used for RT-PCR tests to ensure that all experimental animals were chytrid-free prior to inoculations [26]. Inoculum was prepared by growing Bd on 1% agar, 1% tryptone plates for 2 weeks. Chytrid plates were then flooded with water, and frogs in the exposed group were held overnight in a 100 mL water-bath inoculation. Exposures were repeated with each tank water change. Frogs in the control group were treated identically but received only sham inoculations (no Bd exposure). Each control frog was sacrificed at the same time as an infected frog from the same clutch, of the same sex, and of similar size and weight. Resulting paired control-infected samples were handled simultaneously throughout the experiment to ensure comparability. Frogs were swabbed prior to euthanasia to quantify Bd load.
Frogs were euthanized by decapitation at two time-points after initial Bd exposure. The “early” group was euthanized 72 hours after first exposure to Bd. The “late” group was euthanized once clinical symptoms of chytridiomycosis were observed between 10 and 21 days following initial exposure. We chose to sample the “late” group based on near-death symptoms rather than at a fixed time-point because we were interested in the genetic perturbations associated with the final stages of disease progression. Many of these late group frogs exhibited excessive skin shedding and extreme lethargy. The late group frogs exhibited higher infection loads than early group frogs (see Results), so the different time points correspond with infection intensity. Dorsal skin, liver, and spleen tissues were flash frozen and stored at −80°C.
Microarray Design
We used a custom NimbleGen microarray for S. tropicalis based on full length coding sequence (CDS) and expressed sequence tags (ESTs) from the Joint Genome Institute (JGI; available at http://genome.jgi-psf.org/Xentr4) and Gurdon Institute (available at http://informatics.gurdon.cam.ac.uk/online/xt-fl-db.html). The microarray contained 380,856 60-mer probes representing a possible 113,986 expressed transcripts (probe sets). Fifty-six percent of probes were designed from CDS and 44% were designed from ESTs. Probe sets were defined as all probes that mapped to a single transcript, which in turn mapped to a single gene. Each probe set consisted of 1 (44% of probe sets), 3 (31% of probe sets), or 8 (25% of probe sets) probes. Although probe sets consisting of only one probe were included on the array, none met our statistical criteria for differential expression (see Results). Both JGI and Ensembl (http://www.ensembl.org/Xenopus_tropicalis) annotation of the S. tropicalis genome were used to assign functional categories to genes.
Experimental design
For each microarray, a competitive hybridization was performed, in which two samples were labeled with different Cy dyes and hybridized to the same chip to provide a direct comparison between control and infected frogs. A total of 12 chips were analyzed from 24 samples comprising 2 conditions (control and infected), 2 tissues (dorsal skin and liver) and 2 time-points (early and late). Three biological replicates (i.e., individual frogs) were used for each condition and tissue at each time-point. Cy3 and Cy5 dyes were alternately used for labeling each tissue at each time-point (i.e., dye balanced) in order to account for potential dye bias.
In addition to the 12 liver and skin chips, a single chip was run to gain a preliminary understanding of gene expression patterns in the spleen. Because S. tropicalis spleens are small and a large amount of RNA is needed for whole genome assays, our ability to assay whole genome patterns of expression in the spleen was limited. We were able to compare expression patterns for control versus infected frogs early in infection by pooling spleens from frogs sampled at the early time-point. However frogs late in infection were often so dehydrated that we could not collect their spleens, and thus we did not analyze gene expression in the spleen at the late time-point.
Molecular Methods
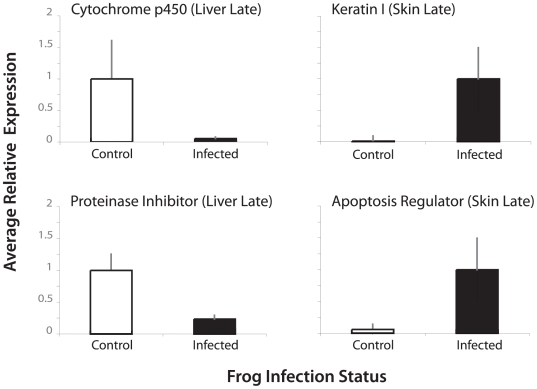
RNA was extracted with a Qiagen RNeasy Mini Kit (Qiagen,Valencia, CA) with the standard protocol and a DNase digestion. Double-stranded cDNAs were synthesized using Invitrogen's SuperScript cDNA Synthesis Kit (Invitrogen, Carlsbad, CA). cDNA samples were fluorescently labeled with custom Cy3 and Cy5 labeled oligonucleotides from TRILink BioTechnologies (TRILink BioTechnologies, San Diego, CA). A 16-hour hybridization was conducted at 42 degrees in a Hybex hybridization chamber (SciGene, Sunnyvale CA). Chips were washed and then scanned on a GenePix Professional 4200 Scanner (MDS Analytical Technologies, Sunnyvale, CA). We corroborated chip-based gene expression patterns for a subset of genes with RT- PCR using SYBR Green (Qiagen, Valencia, CA) and an ABI 7300 (Applied Biosystems, Foster City, CA). We quantified gene expression of target genes of interest relative to reference genes that exhibited constitutively high expression. Reference genes used were beta actin (295226) and proteasome activator 28 (294819) (throughout parenthetical gene numbers correspond to JGI gene IDs). Results for four genes of interest [cytochrome P450 (448177), keratin (178963), proteinase inhibitor (345734), and apoptosis regulator (416168)] are shown here (see Results). Mean and standard deviation of at least three control and three exposed individuals with technical replicates are presented. Results were the same using both reference genes; in all cases fold changes between naïve and exposed frogs estimated using RT-PCR were in the same direction and at least the same magnitude (often greater) than the chip-based estimates.
Data Analysis
NimbleScan software (NimbleGen. Madison, WI) was used to align a chip-specific grid to control features and extract raw intensity data for each dye and probe. Raw intensity data was read into the R computing environment using the LIMMA package for two-color array analysis and checked for quality. Chip pseudo-images were created for each array and dye and visually verified not to contain any spatial artifacts. Further, chip intensity distributions and MA-plots were compared and checked for any unusual global patterns. Each array was then normalized using loess normalization and then across arrays using the A-quantile normalization procedure [27]. Each probe set was then summarized using the median polish procedure as described in Irizarry et al. [28]. The median polish procedure is a robust method for summarizing all probes contained within each probe set to a single expression value for each gene taking into account individual probe effects. Probe sets with low levels of expression variation across all samples (IQR<0.5) were removed from further analysis, leaving 19,719 probe sets remaining in the study. Differential expression was then assessed using a linear model with an empirical Bayesian adjustment to the variances [29], and comparisons of interest were extracted using contrasts. The Benjamini and Hochberg (BH) method was used to control for the expected false discovery rate given multiple tests [30]. Probe sets were considered differentially expressed with a BH adjusted p-value of <0.05 and a raw-fold change greater than 1.5. Because only one chip was run for the pooled spleen samples, only preliminary analyses were performed for the spleen. The single spleen array was preprocessed in the same manner as the dorsal skin and liver arrays. Relative gene expression differences were then calculated for each probe set as the log difference in signal between infected spleens and control spleens. Genes with at least 2-fold differences in expression were considered the most likely candidates for further study. All microarray data is publicly available in accordance with MIAME (GEO accession numbers GPL8767, GSE16852), and the complete list of differentially expressed probe sets is provided in Table S1.
Results
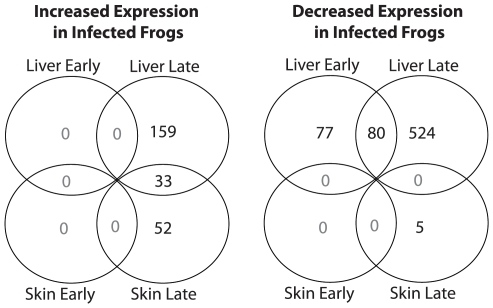
There were 923 probe sets corresponding to unique S. tropicalis transcripts with statistically significant patterns of differential expression in at least one tissue for at least one time-point (Fig. 1). Of these probe sets 42% contained 3 probes and 58% contained 8 probes. RT-PCR confirmations of gene expression patterns for a subset of genes of interest are shown in Figure 2. At the early time-point (72 hours after the first Bd exposure) the transcriptional response to Bd exposure was surprisingly weak. At this early time-point a per-swab average of 3,500 Bd zoospore equivalents was estimated by RT-PCR. There were 157 genes with decreased expression in exposed frogs (relative to naïve frogs) in the liver 3 days after initial exposure, but no genes showed significant increases in expression in the liver or skin early in infection. In the spleen early in infection, 120 genes showed significant differences in expression, with 99 genes showing increased expression and 21 genes showing decreased expression. At the late sampling point (when clinical symptoms of chytridiomycosis were manifest) there was a stronger global transcriptional response in exposed frogs. At the late sampling point a per-swab average of 51,500 Bd zoospore equivalents was estimated by RT-PCR. In the liver, 604 genes had significant decreases in expression, while only 192 showed increases. In the skin, 85 genes showed increases, while only 5 genes showed decreased expression.
Figure 1. Summary of significant changes in gene expression between Bd-exposed and naïve control frogs.
Numbers of genes with increased and decreased expression in infected frogs are shown for the skin (the site of infection) and the liver (an immunologically and physiologically important organ). Data are summarized for two time-points: early (3 days after initial exposure to Bd) and late (when clinical symptoms of chytridiomycosis are evident).
Figure 2. Validation of chip-based gene expression patterns with RT-PCR.
Average expression is shown for several genes of interest relative to an endogenous control (beta actin). Mean and standard deviation are shown for biological replicates. In all cases RT-PCR results were in the same direction as array-based results and of comparable (or greater) magnitude.
Immune response
To gain an overall picture of the expression changes observed in immune-related genes, the probe set annotations were queried with the keyword “immune”. Significant changes in gene expression were observed in only 3 out of 62 probe sets containing immune function Gene Ontology (GO) annotations. To identify additional genes related to immune function that did not contain GO immune annotation, the dataset of differentially expressed genes was manually interrogated to categorize genes related to immunological responses.
The expression changes in immune related genes mirrored the overall patterns of expression differences across the genome, with decreased expression of immune-related genes in both the skin and liver of Bd-exposed frogs (Table 1). Notably, a number of genes in the complement pathway showed decreased expression in exposed frogs. At the early time-point, 10 complement pathway genes (of 38 total) showed decreased expression in exposed frogs including C1r, C4, C9, factor H, factor I, and alternative pathway C3/C5 convertase (Table 1). At the late time-point, 5 complement pathway genes showed decreased expression including C1r, C9, and classical pathway C3/C5 convertase (Table 1). No complement-related genes showed increased expression in exposed frogs.
Table 1. Summary of immunological and physiological genes of interest with significant differences in expression between control and infected frogs.
| Function | Tissue | Time-Point | Average Fold Change | JGI Gene ID | Ensembl Gene ID |
| Coagulation factor | liver | early | −4 | 279909, 293803, 330213, 341521, 341530, 341561 | ENSXETG00000002964, ENSXETG00000002769 |
| Coagulation factor | liver | late | −5 | 147254, 279909, 292258, 292259, 293803, 330213, 341521, 341530, 341561 | ENSXETG00000002964, ENSXETG00000002769, ENSXETG00000003472, ENSXETG00000025492, ENSXETG00000025493 |
| Complement component (alternative pathway) | liver | early | −6 | 308546, 327877, 420558 | ENSXETG00000005261, ENSXETG00000010935, ENSXETG00000024110 |
| Complement component (membrane attack complex) | liver | early | −6 | 282192, 295244 | ENSXETG00000003532, ENSXETG00000006766 |
| Complement component (classical pathway) | liver | early | −3 | 399987, 471107 | ENSXETG00000005203, ENSXETG00000027636 |
| Complement component (classical pathway) | liver | late | −2 | 399976, 399987,400010, 420551 | ENSXETG00000010930, ENSXETG00000023914, ENSXETG00000027636 |
| Cytochrome Cyp450 | liver | early | −3 | 113474, 297805, 325065, 448177, 464259 | ENSXETG00000005868, ENSXETG00000006924, ENSXETG00000013846, ENSXETG00000016349, ENSXETG00000024804, ENSXETG00000027605 |
| Cytochrome Cyp450 | liver | late | −4 | >50 genes, see Table S1 | >50 genes, see Table S1 |
| Epoxide hydrolase | liver | late | −4 | 126998, 152658, 398826 | ENSXETG00000017581, ENSXETG00000022654, ENSXETG00000028064 |
| Heat shock protein 70 | liver | late | 4 | 268191, 287936, 361977, 456096 | ENSXETG00000006714, ENSXETG00000008828, ENSXETG00000010075, ENSXETG00000016838 |
| Heat shock protein 70 | skin | late | 6 | 361977, 456096 | ENSXETG00000006714, ENSXETG00000008828 |
| Keratin, type I | skin | late | 10 | 178963 | ENSXETG00000012482 |
| NF-kappa inhibitor | liver | late | 3 | 309572, 396637, 309732 | ENSXETG00000006998, ENSXETG00000012114 |
| Ornithine decarboxylase | skin | late | 18 | 196068, 284956, 295202 | ENSXETG00000003519, ENSXETG00000007373 |
| Small chemokine, interleukin 8 | liver | late | −3 | 295905 | NA |
Because multiple genes with similar function were often perturbed, fold change is given as an average for all differentially expressed genes in each category. Genes with increased and decreased expression in Bd-exposed frogs show positive and negative fold changes, respectively. See text for additional genes of interest and Table S1 for a complete gene list of differentially expressed genes.
A small number of immune-related genes were up-regulated in Bd-exposed frogs, but the up-regulation of most of these genes would have a net-inhibitory effect on immune response. Specifically, the function of immune genes with increased expression were generally modulatory (or regulatory) in signal transduction pathways including suppressor of cytokine signaling 3, NF-kappa-B inhibitor zeta, and A20-binding inhibitor of NF-kappa-B activation 2 (Table 1). Inhibition of the transcription factor NF-kappa-B is noteworthy given its important role in regulating many genes involved in the production of an immune response.
Several genes with increased expression in Bd-exposed frogs provide evidence for some activation of innate immune responses against Bd (Table 1). We observed increased expression in phagocyte-associated genes including neutrophil cytosolic factor 4, neutrophil collagenase precursor, cytochrome b-245 light chain, arginase I, and lysozyme (Table 1). However, nearly all of these increased expressions occurred at the late time-point rather than early in infection when activation of innate defenses would be expected. In addition, some evidence for innate and adaptive immune response at the early time-point was obtained from our preliminary spleen data. We observed increased expression in the spleens of Bd-exposed frogs for a small number of chemokine (295161, 457013) and neutrophil (449373) genes. Some additional immune function genes had weak evidence of activation in the spleen early in infection. Although fold-changes were lower than our cutoff, all probes in the following probe sets showed increased expression in the spleen: interleukin-17A/F-like (146326) and its precursors (e.g., 144803), MHC complex proteins (e.g., 443195), myd88 (a key molecule in relaying the signal from toll-like receptors, 294414), and complement (e.g., 296194, 194000). It is important to note that the signature of increased expression in sick frogs was not consistent across all genes in these categories. Furthermore, a large number of immune function genes that we expected to be differentially expressed during an immune response to Bd, did not show any significant changes in expression in Bd-exposed frogs. Table 2 illustrates examples of genes and gene classes that we expected to be differentially expressed but were not.
Table 2. Examples of genes expected to be involved in a robust immune response that showed unchanged expression between infected and control frogs.
| Description | JGI Gene_ID | Ensembl.Gene.ID |
| Anti-microbial peptide (AMP) | 287907 | NA |
| Anti-microbial peptide (AMP2) | 109728 | ENSXETG00000022894 |
| Cathelicidin (AMP) | 193370 | NA |
| Heat shock protein 90 family (HSP90) | e.g., 124362, 201402, 293016, 295840, 300776, 350432, 378380, 468257 | ENSXETG00000005359, ENSXETG00000010251, ENSXETG00000010896, ENSXETG00000019484, ENSXETG00000001738 |
| Indoleamine 2,3-dioxygenase | 312615 | ENSXETG00000025783 |
| Interferon gamma (IFNgamma) | 177216 | NA |
| Interleukin 1 (IL-1) | 119533 | NA |
| Interleukin 17 (IL-17) | 300117 | ENSXETG00000020860 |
| Interleukin 8 (IL-8) | 295161, 295905, 460999 | NA |
| Major histocompatibility complex (MHC) class II alpha and beta | 443195, 151191, 371515, 446178, 152977 | ENSXETG00000025586, ENSXETG00000025590, ENSXETG00000017066 |
| Major histocompatibility complex (MHC) class I | 386203, 295817, 386304, 386189, 386261 | ENSXETG00000014445, ENSXETG00000017065, ENSXETG00000009077 |
| T cell and B cell marker (cd45) | 308517 | ENSXETG00000000316 |
| T cell marker (cd28) | 149891 | NA |
| T cell marker [Thy1 (CD90)] | 452827 | NA |
| Toll-like receptor (TLR5) | 459490 | ENSXETG00000015027 |
| Tumor necrosis factor alpha (TNFalpha) | 379518 | NA |
Physiological Response
One of the most dramatic and consistent signatures observed in our experiment was the decreased expression of a large number of cytochrome p450 (P450) superfamily genes in exposed frogs (Table 1). Fifty-one unique P450 genes (out of 70) showed decreased expression in the liver of infected frogs late in infection. Six of these genes already showed decreased expression in the liver early in infection, although the average fold-change between sick and healthy frogs was more dramatic later in infection. Aside from changes in P450 genes, we observed decreased expression of several epoxide hydrolase genes (126998, 152658, 398826) late in infection in the liver, which are similarly involved in cellular detoxification. One of these genes (398826) also showed decreased expression in the liver at the early time-point. Another class of genes with a net decrease in expression is related to blood coagulation. We observed decreased expression at both the early and late time-point in the liver of coagulation factors, thrombin, and plasmin genes (Table 1). Other than fibrinogen-related genes (which had a mixed pattern), coagulation-related genes with higher expression in infected frogs were generally inhibitory (e.g. 287154) so the net effect of deactivation of coagulation genes appears consistent. Additionally, one important marker of hepatic anoxia; HIF-1 (hypoxia induced factor, 151770) showed increased expression in the late liver samples of infected frogs, suggesting a reduction in oxygen delivery and permeation.
We were also interested in determining whether Bd-exposed frogs exhibited signs of acute and/or chronic stress. All of the heat shock proteins we observed with significant changes in expression (9 out of 69 annotated genes) showed higher levels of expression in infected frogs. Increased expression of HSP70, HSP20 and HSP DnaJ were evident at the late time-point in both the liver and skin (Table 1). In the spleen early in infection we also observed increased expression of genes with HSP70 (174350, 287936, 445165) and HSP90 (468257) domains. Other than HSP72 and HSP90, we did not observe the up-regulation of many other HSPs typically associated with acute stress response (i.e., HSP9, HSP23, HSP26, HSP27) in any tissue at any time-point.
We also observed perturbations in genes involved in cellular integrity in the skin. There was more than a 10-fold increase in expression in the keratin I gene (178963) in frog skin late in infection. Additionally, late in infection we observed higher expression in Bd-exposed frogs of genes coding for matrix proteins (165447) growth factors (163909), connexin (175537), collegen and fibrinogen (335826, 317405, 336019, 166058), and ornithine decarboxylase genes (284956, 295202, 196068). There were no genes in these categories with decreased expression in the skin, nor were there any changes in these genes early in infection. Somewhat surprising was the fact that no members of the caspase family were significantly altered in the infected frog skin; this would seem to indicate that neither the intrinsic nor the extrinsic apoptotic pathways were activated in response to cutaneous colonization by Bd.
Discussion
We have evaluated whole genome expression profiles of S. tropicalis exposed to the pathogenic chytrid fungus, Bd. We characterized changes in gene expression globally for predicted transcripts throughout the genome at two time-points after Bd exposure (early and late) at the site of infection (skin) and important immune organs (liver and spleen). We observed a very weak transcriptional response 3 days after exposure (Fig. 1). Notably, there was not a single gene in the liver or skin with a significant increase in expression at the 3-day time-point suggesting that frogs were not recognizing and mounting an appropriate early response to infection in our experiment.. The transcriptional response late in infection was more dramatic (Fig. 1). But again the vast majority of perturbed genes showed decreases in expression in sick frogs compared to uninfected control frogs kept at the same temperature. This extensive negative effect on gene expression only in infected animals is likely linked to infection rather than just an effect of suboptimal temperature on host immune defenses. RT-PCR validations confirm the array results, and the consistent expression of transcriptional regulators indicates that we are not merely observing global transcriptional suppression. Below we discuss the implication of our findings with particular emphasis on the a) strength and nature of the immune response and b) physiological changes during disease progression.
Immune Response
A typical robust immune response to a skin infection would include both innate and adaptive components of the immune system at the site of infection as well as in main immune organs such as the liver and the spleen. The initial innate response would be expected to include activation of macrophages, neutrophils, and Langerhans cells as well as pro-inflammatory signaling to recruit more effector cells (leukocytes). The local immunological signals in the skin would be expected to involve activation of cell surface receptors (e.g., toll-like receptors and MHC molecules), inflammatory cytokines, chemokines, cytokine-induced proteins, and anti-microbial peptides. During the early phase of immune activation we would expect to observe increased hepatic gene expression of acute phase gene-encoded proteins including C-reactive protein, complement pathway genes, and coagulation genes. As the infection progresses, the adaptive component of the immune response would be expected to be activated. The signature of an adaptive response would entail cellular markers of lymphocyte maturation/activation (e.g., B and T cell receptors and co-receptors, activation-induced cytidine deaminase, antibodies), antigen presentation (e.g., LMP7, TAP1, cathepsines, MHC molecules) as well as cytokine production (e.g., IL-2, IL-4).
In mammals, the immune response to dermatophytes (pathogenic fungi that cause ailments such as ringworm) may present an analogous scenario from which to derive expectations for amphibian response to Bd. The response to dermatophytes is characterized by a delayed-type hypersensitivity response (reviewed in [13]). To initiate the process, Langerhans cells gather antigens and move to peripheral lymphoid tissue (e.g. spleen) and present the antigens to T lymphocytes. At the site of infection, T lymphocytes mediate the recruitment of phagocytes via cytokine-driven induction of a localized inflammatory response. Studies on Xenopus laevis have shown the presence of these cell types in the skin, so it is reasonable to expect a comparable response [14], [15].
Our results show that very few components of a typical innate or adaptive immune response occurred in S. tropicalis exposed to Bd, in fact most immune related genes showed decreased expression in infected frogs. Nearly all of the genes that we hypothesized to play a role in controlling Bd infection showed no expression change between exposed and naïve frogs (Table 2). For example in the liver and skin, there was no activation of toll-like receptors (which could play a role in pathogen recognition), no change in known antimicrobial peptide genes (although not all AMPs were represented on our chip because some sequences were shorter than our 60-mer probes), and no change in MHC class I or II genes, or genes involved in antigen presentation (e.g., LMP7, TAP1 and 2, cathepsines). Only phagocyte-associated genes exhibited increased expression in infected frogs, and only at the late time-point (Table 1). It is dramatic that of dozens of classes of immune function genes that are usually activated in the liver, phagocyte-associated genes are the only class showing a positive response. The activation of phagocytosis itself is not evidence of a robust immune response given that these genes could be activated in response to other cellular changes in the liver, especially at late stages of infection. There were a handful of immune-relevant genes with increased expression in the spleen. For example, several MHC and interleukin-related genes showed higher expression in Bd-exposed frogs. Although the trends in the spleen data may suggest some activated immune responses, there were only a handful of immune-function genes with consistent signature of increased expression, and robust statistical statements are precluded by the lack of replication for the spleen chip. It is therefore critical for future studies to further dissect patterns of immunogenetic response in replicated spleen samples.
In contrast to the few immune genes showing increased expression, a large number of immune genes showed decreased expression in exposed frogs. The strongest negative signal for immune-related genes was observed in the complement pathway. At both early and late sampling time-points we observed reduced expression of complement pathway genes in the liver. The consistency of the observation may suggest that complement suppression is a characteristic feature of chytridiomycosis in S. tropicalis. The cascade of events leading to complement suppression remains to be determined but may result from several causes. First, if one complement pathway is up-regulated, a compensatory down-regulation may be expected in an alternate pathway. However, we see decreased expression in complement genes in both the classical and alternate pathways suggesting this explanation is unlikely. Second, it is possible that the down-regulation of complement represents exhaustion from an earlier up-regulation, but again this seems unlikely given that we observed decreased expression of complement-related genes as early as 72 hours after the initial exposure. Finally, decreased expression of complement could be a direct result of an inhibitory signal from Bd. As of yet we have no direct evidence for active immune suppression by Bd, but other fungal pathogens have been associated with complement evasion. For example, human pathogenic fungi Aspergillus fumigatus and Candida albicans evade immune attack by binding complement regulators in order to suppress complement effector molecules [31]. Regardless of the mechanism by which complement and other elements of the immune system are suppressed, it is clear that the net effect will be detrimental for amphibian immunity given the importance of complement in B cell and antibody responses. The decreased expression of complement could also affect complement-enhanced phagocytosis, particularly given the expression changes observed in phagocyte-associated genes. Overall, we expect the observed reduction in complement activity to be problematic, not only for mounting an appropriate response to an existing assault from Bd, but also for responding to future pathogenic insults.
There are three non-mutually exclusive hypotheses for the lack of robust immune response observed in our experiments: 1) immune evasion/suppression by Bd, 2) reduced immune response in highly susceptible frog species, and 3) reduced immune response under certain environmental conditions. First, the lack of a robust immune response may indicate immune evasion/suppression strategies by Bd. For example, Bd may be able to sequester itself from immune receptors during infection in order to avoid immune attack. During intracellular stages of infection, Bd may be able to avoid immune recognition in the protected environment of the skin layer (keratinocytes). For example, several other intracellular pathogens are known to employ this strategy including Histoplasma capsulatum, which is a dimorphic fungal pathogen that proliferates inside mammalian macrophages [32]. In another example, fungal pathogen Coccidioides posadasii secretes a metalloproteinase that mediates immune evasion by digesting the pathogen's own surface antigens in order to avoid recognition [33]. Another possibility is that Bd suppresses the immune response by interfering with immune signaling. Other fungal pathogens are known to display or secrete molecules that disable host defense pathways. For example, dimorphic fungal pathogen Blastomyces dermatitidis produces an adhesion molecule (BAD1) that binds to a host complement receptor in order to suppress production of pro-inflammatory cytokine TNF-α [34]. Prior studies of Bd gene expression throughout development identified the differential expression of several genes with sequence similarity to genes thought to be involved in pathogen evasion of vertebrate immune systems [35], but further research is necessary to evaluate which Bd secreted proteins may interact with the host immune system.
Second, the reduced immune response may be characteristic of highly susceptible frog species. There is a wide variation of susceptibility of frogs to chytridiomycosis and different outcomes to Bd exposure have been observed in both laboratory and natural settings. In lab infection experiments mortality can range from zero to 100% depending on the species [36], [37]. Similarly, in nature, some species experience dramatic Bd related declines while others persist with moderate Bd loads while apparently suffering no ill effect. Silurana tropicalis is a susceptible species, so our observations are likely not to be generalized across amphibian species. Differences in species susceptibility to Bd are hypothesized to be related to differences among species in the production of antimicrobial peptides [10], [38], the occurrence of bacterial commensals with anti-fungal properties [39], and behavioral characteristics [40]. However, there is scant data exploring differences in innate or adaptive immune response among species with different susceptibilities, and it is possible that some susceptible species do not mount effective innate and/or adaptive immune responses, while other more resistant species do. Immunological studies on closely related resistant/susceptible species pairs (e.g., X. laevis and S. tropicalis) are a priority for continued research.
Third, the damped immunogenetic response observed for S. tropicalis in this study is unlikely to be generalized across all environmental conditions. Importantly amphibian immune responses can be strongly affected by temperature (e.g., [41]). Further, laboratory infection experiments (not in Silurana) suggest that differences in temperature on the order of 5–10 degrees C can affect mortality rate [24]. Although our study was not conducted at the low temperatures known to have dramatic impacts on frog immune function (e.g., 5 degree C [41]), the temperature regime we used was outside the preferred temperature profile of S. tropicalis. In X. laevis 18 degree C is cold enough to have inhibitory effects on T cell proliferation [42] and IgM-IgY switching [43]. Although it may be argued that the temperature at which the frogs were infected here was not optimal for a potent immune response, the fact we observed many immune genes down-regulated and not just unchanged compared to control animals (at the same temperature) is indicative of a more complex phenomenon linked to the infection. Additionally, because Bd related die-offs often occur in colder environments and colder seasons [23], [24], it is biologically relevant to understand frog response to Bd when not operating under optimal immunity. Follow-up studies should consider immunogenetic response of S. tropicalis under different temperature regimes and endeavor to further understanding the interaction between chytridiomycosis and environmental conditions that may decrease immunocompetence.
Physiological Response
While no specific physiological effect, outside of immunological response, was predicted from Bd exposure, we expected a pronounced transcriptional response in the hepatic and cutaneous tissues considering the lethality of Bd colonization. The overall patterns observed for physiologically relevant genes were complex but point to several important physiological changes in Bd infected frogs. First, we observed a strong signature of chronic stress in Bd-exposed frogs. Genes coding for heat shock proteins were up-regulated early in infection in the spleen (HSP70, HSP90) and late in infection in both the liver and skin (HSP70, HSP20, HSP90 and HSP DnaJ). The chronic stress response observed late in infection could be due to any number of physiological changes in frogs close to death, and does not necessarily reflect a signature particular to chytridiomycosis. However, the consistent increase in expression for HSPs just three days after Bd exposure does suggest a direct stress response to Bd. It is interesting to note that we observed the perturbation of only some of the stress-related HSP genes and that some of the classical markers of acute stress response were unaffected. However some inducible markers like HSP72, which is often increased during microbial infection, and HSP90 were perturbed. We are confident that the increase in stress-related genes was above and beyond any stress due to experimental conditions (e.g., captivity, handling, temperature) as the stress related genes showed on average four-fold greater expression in Bd-exposed versus control frogs.
We also observed genetic changes that can help explain some of the clinical symptoms of chytridiomycosis in the skin. The large increase in expression in the type I keratin gene in skin late in infection is particularly interesting. This gene is an ortholog of human keratin 1, which is known to be perturbed in humans with the ailment epidermolytic hyperkeratosis [44]. Although this disease and chytridiomycosis have different etiologies, it is intriguing that similar phenotypes result (e.g., expansion and irregularity of the stratum corneum). There is additional molecular evidence that the skin is damaged and trying to repair with increased expression of genes involved in cellular remodeling of the skin and tissue integrity (e.g., matrix proteins, connexins, collagens, fibrinogens). The up-regulation of genes involved in cell growth and proliferation (e.g., growth factors and ornithine decarboxylase), may contribute to the observed expansion of the stratum corneum and the sloughing response of frogs with chytridiomycosis. Whether this skin shedding is a physiological strategy of frogs to shed Bd infection or a response to Bd toxins requires further study. We did not observe perturbations of genes that could be involved in cytoxic or antigenic response in the skin, and thus our skin data do not directly indicate the production of toxic compounds by Bd. Further, the absence of significant changes in the skin in the expression of caspase genes, typically altered in response to cell damage and important mediators of the apoptotic cascade, was particularly surprising given the colonization by Bd.
However, changes in another class of genes important in cellular detoxification may point to the possibility of Bd toxicity. We observed the down-regulation of a large number (N>50) of cytochrome P450 genes (and relatedly epoxide hydrolase genes) in the liver. P450 genes are known to be important in detoxification [45], and their down-regulation may be indicative of exhaustion (although we did not observe an early up-regulation of these genes). Alternatively, their down-regulation may be indicative of Bd-derived toxins, as perturbation of liver P450 function can be endotoxin-mediated in other systems [46]. Perhaps more importantly than the cause of decreased expression is the effect of decreased expression of this gene family, which is expected to have physiological consequences in pathways related to cellular metabolism and detoxification. Systemic mycoses have been shown to alter the expression of P450 genes in mammalian livers, in some instances facilitating host defense while in others associated with the etiology of the disease [47]. Given the diversity of metabolic pathways that P450 gene products are involved with, we can suggest a broad-spectrum alteration in the hepatic capacity for oxidative metabolism associated with the down-regulation of these genes. The consequences of changes in Cyp450 function, particularly for amphibians, are likely to be severe because they are in close contact with environmental contaminants, and perturbations in this gene family may affect the ability of frogs to fight off future insults.
Finally, we observed a number of changes in blood coagulation factors in the hepatic transcriptome. It is possible that these are related to the subjective assessment that the Bd infected frogs were dehydrated, but we did not have any direct measures of hydration such as packed cell volume, osmolality or hematocrit to directly confirm this observation. Although the cause of alteration in expression of these hemostatic markers is uncertain, it is interesting to note that similar changes are observed in scenarios of known diminished liver function in mammals, such as cirrhosis [48] and post-transplantation [49]. Thus there is some indication that the livers of infected frogs were not fully functioning, however we did not observe perturbation of any classical markers of liver failure. Another important change in the livers of infected frogs was the up-regulation of hypoxia-induced factor-1. This gene is a potent and acute marker of low oxygen delivery and may indicate alteration in the efficacy of cutaneous blood gas exchange in the Bd infected late stage frogs. Prior studies have not reported any significant changes in measures of pCO2 in Bd infected frogs [9], but induction of hypoxia-induced factor-1 may indicate changes in skin gas exchange associated with Bd colonization. Of course transcriptome data should not be used as the sole index of response to infection, and additional work is necessary to link gene expression data to organismal level observations. But hypoxia-induced factor-1 is a sensitive marker for alterations in oxygen utilization or delivery and has broad-spectrum downstream targets [i.e., implicated in activating genes involved in angiogenesis, apoptosis, mitochondrial biogenesis, anaerobic metabolism (e.g. [50])]. Therefore patterns observed at this gene are worth further study.
Although our data do not determine definitively the proximate cause of Bd morbidity, we provide insight into which genetic systems are perturbed during infection. First, we show that the activation of the immune system is weaker than expected. Second, we provide information about the molecular mechanisms underlying stress response and skin sloughing. Finally we provide some insight into Bd pathogenesis with data suggesting that Bd may compromise immune response/detection. These baseline observations make possible future studies to understand response of frogs to Bd in natural settings and to compare populations and species with different levels of resistance to Bd.
Supporting Information
Complete list of genes with significant differences in expression between naïve and Bd-exposed frogs. Table columns correspond to gene identifiers (NimbleGen, Joint Genome Institute, and Ensembl), number of probes in probe set, summary of tissues and time points for differential expression, log2 fold-changes for tissues and time points with significant differences in expression, and functional annotation (e.g., InterPro, Gene Ontology, euKaryotic Orthologous Groups).
(0.40 MB XLS)
Acknowledgments
We thank Maria Lin for assistance with live animals experiments, Stewart Macarthur and Karen Vrazian for discussion about data analysis, Heidi Morales for discussion of results, and Jamie Voyles for comments on the manuscript. All live animal work was conducted in accordance with approved Animal Care and Use Protocols.
Funding Statement
The work was supported by NSF funding to EBR (Postdoctoral Fellowship in Bioinformatics and Ecology of Infectious Disease Grant EF-0723871), funds from the Hellman Fund to EBR and MBE, and NIH funding to JR (R24-AI-059830-01). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Smith KF, Acevedo-Whitehouse K, Pedersen AB. The role of infectious diseases in biological conservation. Anim Conserv. 2009;12:1–12. [Google Scholar]
- 2.Schaechter M, Medoff G, Schlessinger D, Popoff CM. Baltimore, MD: Williams & Wilkins; 1993. Mechanisms of Microbial Disease. [Google Scholar]
- 3.Blehert DS, Hicks AC, Behr M, Meteyer CU, Berlowski-Zier BM, et al. Bat White-Nose Syndrome: An Emerging Fungal Pathogen? Science. 2009;323:227–227. doi: 10.1126/science.1163874. [DOI] [PubMed] [Google Scholar]
- 4.Longcore JE, Pessier AP, Nichols DK. Batrachochytrium dendrobatidis gen et sp nov, a chytrid pathogenic to amphibians. Mycologia. 1999;91:219–227. [Google Scholar]
- 5.Skerratt LF, Berger L, Speare R, Cashins S, McDonald KR, et al. Spread of chytridiomycosis has caused the rapid global decline and extinction of frogs. EcoHealth. 2007;4:125–134. [Google Scholar]
- 6.Berger L, Speare R, Daszak P, Green DE, Cunningham AA, et al. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc Natl Acad Sci U S A. 1998;95:9031–9036. doi: 10.1073/pnas.95.15.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lips KR, Brem F, Brenes R, Reeve JD, Alford RA, et al. Emerging infectious disease and the loss of biodiversity in a Neotropical amphibian community. Proc Natl Acad Sci U S A. 2006;103:3165–3170. doi: 10.1073/pnas.0506889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rachowicz LJ, Knapp RA, Morgan JAT, Stice MJ, Vredenburg VT, et al. Emerging infectious disease as a proximate cause of amphibian mass mortality. Ecology. 2006;87:1671–1683. doi: 10.1890/0012-9658(2006)87[1671:eidaap]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 9.Voyles J, Berger L, Young S, Speare R, Webb R, et al. Electrolyte depletion and osmotic imbalance in amphibians with chytridiomycosis. Dis Aquat Org. 2007;77:113–118. doi: 10.3354/dao01838. [DOI] [PubMed] [Google Scholar]
- 10.Woodhams DC, Ardipradja K, Alford RA, Marantelli G, Reinert LK, et al. Resistance to chytridiomycosis varies among amphibian species and is correlated with skin peptide defenses. Anim Conserv. 2007;10:409–417. [Google Scholar]
- 11.Parker JM, Mikaelian I, Hahn N, Diggs HE. Clinical diagnosis and treatment of epidermal chytridiomycosis in African clawed frogs (Xenopus tropicalis). Comparative Med. 2002;52:265–268. [PubMed] [Google Scholar]
- 12.Romani L. Immunity to fungal infections. Nat Rev Immunol. 2004;4:11–23. doi: 10.1038/nri1255. [DOI] [PubMed] [Google Scholar]
- 13.Blanco JL, Garcia ME. Immune response to fungal infections. Vet Immunol Immunopathol. 2008;125:47–70. doi: 10.1016/j.vetimm.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 14.Du Pasquier L, Schwager J, Flajnik MF. The Immune System of Xenopus. Ann Rev Immunol. 1989;7:251–275. doi: 10.1146/annurev.iy.07.040189.001343. [DOI] [PubMed] [Google Scholar]
- 15.Mescher AL, Wolf WL, Ashley Moseman E, Hartman B, Harrison C, et al. Cells of cutaneous immunity in Xenopus: Studies during larval development and limb regeneration. Dev Comp Immunol. 2007;31:383–393. doi: 10.1016/j.dci.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Ramanayake T, Simon DAL, Frelinger JG, Lord EM, Robert J. In vivo study of T-cell responses to skin alloantigens in Xenopus using a novel whole-mount immunohistology method. Transplantation. 2007;83:159–166. doi: 10.1097/01.tp.0000250562.35175.06. [DOI] [PubMed] [Google Scholar]
- 17.Richmond JQ, Savage AE, Zamudio KR, Rosenblum EB. Toward Immunogenetic Studies of Amphibian Chytridiomycosis: Linking Innate and Acquired Immunity. Bioscience. 2009;59:311–320. [Google Scholar]
- 18.Kurtz J, Scharsack JP. Resistance is skin-deep: innate immunity may help amphibians to survive a deadly fungus. Anim Conserv. 2007;10:422–424. [Google Scholar]
- 19.Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Micro. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- 20.van der Sar AM, Spaink HP, Zakrzewska A, Bitter W, Meijer AH. Specificity of the zebrafish host transcriptome response to acute and chronic mycobacterial infection and the role of innate and adaptive immune components. Mol Immunol: In Press, Corrected Proof. 2009 doi: 10.1016/j.molimm.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 21.Evans BJ. Ancestry Influences the Fate of Duplicated Genes Millions of Years After Polyploidization of Clawed Frogs (Xenopus). Genetics. 2007;176:1119–1130. doi: 10.1534/genetics.106.069690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robert J, Ohta Y. Comparative and developmental study of the immune system in Xenopus. Dev Dyn. 2009;238:1249–1270. doi: 10.1002/dvdy.21891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kriger KM, Hero JM. Large-scale seasonal variation in the prevalence and severity of chytridiomycosis. J Zool. 2007;271:352–359. [Google Scholar]
- 24.Berger L, Speare R, Hines HB, Marantelli G, Hyatt AD, et al. Effect of season and temperature on mortality in amphibians due to chytridiomycosis. Aust Vet J. 2004;82:434–439. doi: 10.1111/j.1751-0813.2004.tb11137.x. [DOI] [PubMed] [Google Scholar]
- 25.Piotrowski JS, Annis SL, Longcore JE. Physiology of Batrachochytrium dendrobatidis, a chytrid pathogen of amphibians. Mycologia. 2004;96:9–15. [PubMed] [Google Scholar]
- 26.Boyle DG, Boyle DB, Olsen V, Morgan JAT, Hyatt AD. Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis Aquat Org. 2004;60:141–148. doi: 10.3354/dao060141. [DOI] [PubMed] [Google Scholar]
- 27.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 28.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 29.Smyth GK. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer; 2005. limma: Linear Models for Microarray Data. pp. 397–420. [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B-Methodol. 1995;57:289–300. [Google Scholar]
- 31.Vogl G, Lesiak I, Jensen DB, Perkhofer S, Eck R, et al. Immune evasion by acquisition of complement inhibitors: The mould Aspergillus binds both factor H and C4b binding protein. Mol Immunol. 2008;45:1485–1493. doi: 10.1016/j.molimm.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woods JP. Knocking on the right door and making a comfortable home: Histoplasma capsulatum intracellular pathogenesis. Curr Opin Microbiol. 2003;6:327–331. doi: 10.1016/s1369-5274(03)00080-8. [DOI] [PubMed] [Google Scholar]
- 33.Hung C-Y, Seshan KR, Yu J-J, Schaller R, Xue J, et al. A Metalloproteinase of Coccidioides posadasii Contributes to Evasion of Host Detection. Infect Immun. 2005;73:6689–6703. doi: 10.1128/IAI.73.10.6689-6703.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmid-Hempel P. Immune defence, parasite evasion strategies and their relevance for a macroscopic phenomena such as virulence. Philos Trans R Soc London [Biol] 2009;364:85–98. doi: 10.1098/rstb.2008.0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenblum EB, Stajich JE, Maddox N, Eisen MB. Global gene expression profiles for life stages of the deadly amphibian pathogen Batrachochytrium dendrobatidis. Proc Natl Acad Sci U S A. 2008;105:17034–17039. doi: 10.1073/pnas.0804173105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daszak P, Strieby A, Cunningham AA, Longcore JE, Brown CC, et al. Experimental evidence that the bullfrog (Rana catesbeiana) is a potential carrier of chytridiomycosis, an emerging fungal disease of amphibians. Herpetolog J. 2004;14:201–207. [Google Scholar]
- 37.Berger L, Marantelli G, Skerratt LL, Speare R. Virulence of the amphibian chytrid fungus Batrachochytrium dendrobatidis varies with the strain. Dis Aquat Org. 2005;68:47–50. doi: 10.3354/dao068047. [DOI] [PubMed] [Google Scholar]
- 38.Woodhams DC, Rollins-Smith LA, Carey C, Reinert L, Tyler MJ, et al. Population trends associated with skin peptide defenses against chytridiomycosis in Australian frogs. Oecologia. 2006;146:531–540. doi: 10.1007/s00442-005-0228-8. [DOI] [PubMed] [Google Scholar]
- 39.Harris RN, James TY, Lauer A, Simon MA, Patel A. Amphibian pathogen Batrachochytrium dendrobatidis is inhibited by the cutaneous bacteria of Amphibian species. EcoHealth. 2006;3:53–56. [Google Scholar]
- 40.Rowley JJL, Alford RA. Behaviour of Australian rainforest stream frogs may affect the transmission of chytridiomycosis. Dis Aquat Org. 2007;77:1–9. doi: 10.3354/dao01830. [DOI] [PubMed] [Google Scholar]
- 41.Maniero GD, Carey C. Changes in selected aspects of immune function in the leopard frog, Rana pipiens, associated with exposure to cold. J Comp Physiol [B] 1997;167:256–263. doi: 10.1007/s003600050072. [DOI] [PubMed] [Google Scholar]
- 42.Meier E. CA: University of Alberta; 2003. T cell development and the diversification and selection of the Xenopus T cell recepetor beta chain repertoire. p. 336. [Google Scholar]
- 43.Wabl MR, Du Pasquier L. Antibody patterns in genetically identical frogs. Nature. 1976;264:642–644. doi: 10.1038/264642a0. [DOI] [PubMed] [Google Scholar]
- 44.Smith FF. The molecular genetics of keratin disorders. Am J Clin Dermatol. 2003;4:347–364. doi: 10.2165/00128071-200304050-00005. [DOI] [PubMed] [Google Scholar]
- 45.Guengerich FP. Cytochrome Cyp450 and Chemical Toxicology. Chem Res Tox. 2008;21:70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- 46.Masubuchi Y, Horie T. Endotoxin-mediated disturbance of hepatic cytochrome Cyp450 function and development of endotoxin tolerance in the rat model of dextran sulfate sodium-induced experimental colitis. Drug Metab Dispos. 2004;32:437–441. doi: 10.1124/dmd.32.4.437. [DOI] [PubMed] [Google Scholar]
- 47.Letterio JJ, Lehrnbecher T, Pollack G, Walsh TJ, Chanock SJ. Invasive candidiasis stimulates hepatocyte and monocyte production of active transforming growth factor beta. Infect Immun. 2001;69:5115–5120. doi: 10.1128/IAI.69.8.5115-5120.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dymock IW, Tucker JS, Woolf IL, Poller L, Thomson JM. Coagulation Studies as a Prognostic Index in Acute Liver Failure. Br J Haematol. 1975;29:385–395. doi: 10.1111/j.1365-2141.1975.tb01836.x. [DOI] [PubMed] [Google Scholar]
- 49.Lewis JH, Bontempo FA, Awad SA, Kang YG, Kiss JE, et al. Liver-Transplantation - Intraoperative Changes in Coagulation-Factors in 100 1st Transplants. Hepatology. 1989;9:710–714. doi: 10.1002/hep.1840090509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dery MAC, Michaud MD, Richard DE. Hypoxia-inducible factor 1: regulation by hypoxic and non-hypoxic activators. Int J Biochem Cell Biol. 2005;37:535–540. doi: 10.1016/j.biocel.2004.08.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete list of genes with significant differences in expression between naïve and Bd-exposed frogs. Table columns correspond to gene identifiers (NimbleGen, Joint Genome Institute, and Ensembl), number of probes in probe set, summary of tissues and time points for differential expression, log2 fold-changes for tissues and time points with significant differences in expression, and functional annotation (e.g., InterPro, Gene Ontology, euKaryotic Orthologous Groups).
(0.40 MB XLS)