Abstract
For the cell-division cycle to progress, hundreds of genes and proteins must be coordinately regulated. Systems-level studies of this cycle show that positive-feedback loops help to keep events in sync.
The cell cycle is a complex but orderly sequence of events that culminates in the production of two cells from one. In eukaryotes, the cycle is divided into four phases: cell growth in G1 phase, DNA replication in S phase, more growth in G2 phase, and cell division in mitosis or M phase. The system of regulators that drives transitions between phases is centred on the cyclin-dependent kinases (CDKs), enzymes that are activated when regulatory proteins called cyclins bind to them. The CDK network directly or indirectly orchestrates coordinated regulation of proteins and genes involved in essentially every aspect of cell function. The complexity of these regulatory events raises the question of what systems-level strategies keep the process temporally coherent — how does the maestro of the cell cycle generate a definitive downbeat? Writing in this issue, Skotheim et al.1 and Holt et al.2 examine different phases of the cell cycle in the budding yeast Saccharomyces cerevisiae, and their findings converge on the same answer: positive feedback.
In budding yeast, the cell cycle begins with the synthesis of the initiator cyclin Cln3, which binds to and activates Cdk1. Substrates of the Cln3-Cdk1 complex include the SBF and MBF gene transcription factors (activated through phosphorylation), and the transcriptional inhibitor Whi5, which is translocated out of the nucleus (and so inactivated) after phosphorylation. The reciprocal regulation of SBF/MBF and Whi5 brings about the transcription of hundreds of genes that collectively constitute the G1/S regulon.
Among the targets of SBF/MBF are the genes that encode the G1 cyclins Cln1 and Cln2. Like Cln3, these two cyclins can activate Cdk1. And like Cln3-Cdk1, Cln1/2-Cdk1 complexes can activate SBF/MBF and inhibit Whi5. Thus, the Cdk1 system has a pair of interlinked positive-feedback loops that could, in principle, function as an irreversible, bistable trigger, with Cln1 and Cln2 promoting their own accumulation in an ever-accelerating cycle (Fig. 1a). The ‘explosive’ kinetics of the positive-feedback system could provide the definitive downbeat that keeps the G1/S regulon coherent.
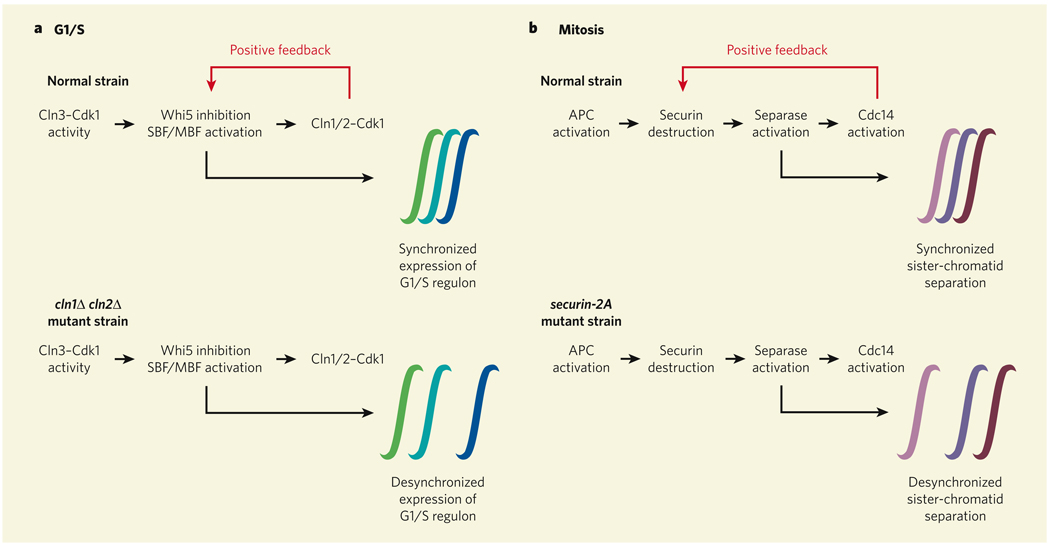
Figure 1. Cell-cycle synchronizers.
a, Skotheim et al.1 show that positive feedback between Cln1/2-Cdk1 and the transcriptional regulators Whi5 and SBF/MBF is essential for synchronous expression of the G1/S regulon, which is disrupted when positive feedback is brought to a halt in mutant yeast strains that lack the genes CLN1 and CLN2 (cln1Δ cln2Δ). b, Holt and colleagues2 find that positive feedback between Cdc14 and securin helps to make another step in the cell cycle — separation of sister chromatids at the end of mitosis — happen in unison. In the securin-2A mutant, where this feedback mechanism is lost, sister-chromatid separation does not occur as synchronously.
This attractive idea was tested more than a decade ago in cell populations3,4 and found to be (apparently) incorrect: cells lacking the CLN1 and CLN2 genes (cln1Δ cln2Δ cells) seemed to activate the promoter sequence for CLN2 just as quickly as normal cells. But sometimes studying individual cells can reveal things that are masked by averaging over a population, and Skotheim et al.1 (page 291) show that this is the case here.
Examining normal cells individually by live-cell fluorescence microscopy, these authors find that Whi5 can abruptly translocate out of the nucleus some 40–50 minutes after the start of the G1 phase, and that the CLN2 promoter is turned on at about the same time. By contrast, in cln1Δ cln2Δ cells, the redistribution of Whi5 to the cytoplasm occurs slowly and gradually, and activation of the CLN2 promoter is delayed by around 40 minutes. These observations indicate that Cln1/2-mediated positive feedback is required for Whi5 to be rapidly switched off and for timely induction of CLN2 expression.
Skotheim and colleagues also find1 that the expression of the G1/S regulon is desynchronized and incoherent in cln1Δ cln2Δ cells. Normally, the CLN2, RFA1 and RAD27 genes are turned on within a few minutes of each other. In cln1Δ cln2Δ cells, however, they splutter on one by one. Moreover, the most strongly in coherent yeast cells almost invariably arrest as unbudded G1-phase cells, never progressing into the S and M phases. Together, these results suggest that positive feedback makes the redistribution of Whi5 abrupt, which allows the G1/S regulon to be expressed synchronously and the cell cycle to proceed.
A similar story emerges from studies of another irreversible transition step in the cell cycle: the separation of sister chromatids at the onset of anaphase during mitosis. This event is mediated by the enzyme separase, which is normally inhibited by another protein called securin. At the onset of anaphase, the anaphase-promoting complex (APC) brings about securin’s destruction, allowing separase to be activated. Sister-chromatid separation generally seems to occur with near-perfect synchrony, and Holt et al.2 (page 353) provide an explanation for why this is so: securin is part of a previously unrecognized positive-feedback loop.
The starting point for these authors’ work was the identification of a new Cdk1-mediated phosphorylation site in securin. Phosphorylation of this evolutionarily conserved site makes securin a poor destruction substrate for the APC. But once some securin has been destroyed (owing to a graded increase in APC activity), separase is activated and, in turn, activates Cdc14 — an enzyme that can dephosphorylate many CDK substrates, including securin. Securin dephosphorylation further increases the rates of its own breakdown, and of separase and Cdc14 activation. This ever-accelerating positive-feedback system could, in principle, translate a gradual increase in APC activity into an abrupt ‘switch off’ of securin activity (Fig. 1b).
To test this idea, Holt et al. tagged two chromosomal loci on chromosomes IV and V with green fluorescent protein, and followed the timing of sister-chromatid separation by fast live-cell imaging. They find that, under normal conditions, chromosome-IV sisters separate from each other an average of 90 seconds before chromosome-V sisters do. But when the feedback was compromised, either through expression of securin-2A — a phosphorylation-site securin mutant that is resistant to positive feedback — or by manipulation of the activities of Cdc14 or Cdk1, this lag period doubled. Moreover, the rate of chromosome mis-segregation rose dramatically in the securin-2A strain. This is consistent with the idea that positive feedback in securin destruction is crucial for the synchronicity of sister-chromatid separation, and that perhaps this synchronicity is essential for the fidelity of genomic segregation.
These studies1,2 underscore the importance of single-cell, real-time approaches for understanding the dynamics of molecular signalling networks. But insight can also be obtained from population-based approaches. Recently, Haase and co-workers5,6 looked at what links G1-phase events to those later in the cell cycle. One plausible idea is that control is handed from one cyclin-Cdk1 complex to the next in temporal succession (Cln3-Cdk1 to Cln1/2- Cdk1 to the six B-type cyclin (Clb1-6)-Cdk1 complexes), ultimately resulting in APC activation, degradation of mitotic cyclins, and the resetting of the cell cycle to G1 phase. But the authors show6 that, in a yeast strain lacking all six CLB genes, most of the cell-cycle-regulated genes are still periodically expressed. For many of the genes, the amplitudes of the oscillations are compromised, but, astonishingly, most of them still come and go with near-normal timing. The authors propose that yeast cells have an autonomous transcriptional oscillator as well as the more familiar CDK oscillator.
So how is this transcriptional oscillator wired? Haase and colleagues6 hypothesize that the circuit is a long, slow, positive-feedback loop, with CLN1 and CLN2 promoting their own expression through the intermediacy of a cascade of transcriptional regulators with shorter, quicker, negative-feedback loops that keep each wave of transcriptional activation pulsatile. In a sense, this is the opposite of the usual CDK-centred cell-cycle models7, which assume that a slow, negative-feedback loop (Cdk1 activation leading to APC activation leading to Cdk1 inactivation) is coupled to several short, positive-feedback loops (such as the Whi5 and securin subcircuits1,2). In support of their idea, Haase and colleagues show6 that a Boolean model (a model that assumes all genes and proteins flip-flop between digital ‘on’ and ‘off’ states) of this network exhibits robust oscillations.
The positive-feedback loops examined in these studies1,2,6 are not the only ones involved in cell-cycle regulation. Experiments in frog egg extracts and human cell lines have established8–10 the importance of a positive-feedback loop that regulates the mitotic activation of Cdk1 in establishing stable, robust oscillations. And in yeast, the securin-mediated positive-feedback loop sits within another positive-feedback loop in which Cdk1 inactivation promotes activation of a form of the APC that feeds back to inactivate more Cdk1 molecules11. Positive feedback can help to make transitions between states decisive and irreversible12–14, suppress chatter during transitions15, and endow oscillator circuits with robustness and tunability16. The work of Skotheim et al.1 and Holt and colleagues2 adds another performance characteristic of positive feedback to this list: it can help to keep things in sync. This powerful organizational strategy may be essential for a process as beautiful and as complicated as the cell cycle.
References
- 1.Skotheim JM, Di Talia S, Siggia ED, Cross FR. Nature. 2008;454:291–296. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holt LJ, Krutchinsky AN, Morgan DO. Nature. 2008;454:353–357. doi: 10.1038/nature07050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dirick L, Böhm T, Nasmyth K. EMBO J. 1995;14:4803–4813. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stuart D, Wittenberg C. Genes Dev. 1995;9:2780–2794. doi: 10.1101/gad.9.22.2780. [DOI] [PubMed] [Google Scholar]
- 5.Haase SB, Reed SI. Nature. 1999;401:394–397. doi: 10.1038/43927. [DOI] [PubMed] [Google Scholar]
- 6.Orlando DA, et al. Nature. 2008;453:944–947. doi: 10.1038/nature06955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tyson JJ, Csikasz-Nagy A, Novak B. BioEssays. 2002;24:1095–1109. doi: 10.1002/bies.10191. [DOI] [PubMed] [Google Scholar]
- 8.Cross FR, Archambault V, Miller M, Klovstad M. Mol. Biol. Cell. 2002;13:52–70. doi: 10.1091/mbc.01-05-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pomerening JR, Kim SY, Ferrell JE., Jr Cell. 2005;122:565–578. doi: 10.1016/j.cell.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 10.Pomerening JR, Ubersax JA, Ferrell JE., Jr Mol. Biol. Cell. 2008 doi: 10.1091/mbc.E08-02-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cross FR. Dev. Cell. 2003;4:741–752. doi: 10.1016/s1534-5807(03)00119-9. [DOI] [PubMed] [Google Scholar]
- 12.Ferrell JE, Jr, Machleder EM. Science. 1998;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- 13.Gardner TS, Cantor CR, Collins JJ. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 14.Xiong W, Ferrell JE., Jr Nature. 2003;426:460–465. doi: 10.1038/nature02089. [DOI] [PubMed] [Google Scholar]
- 15.Thron CD. Biophys. Chem. 1996;57:239–251. doi: 10.1016/0301-4622(95)00075-5. [DOI] [PubMed] [Google Scholar]
- 16.Tsai TY-C, et al. Science. 2008;321:126–129. doi: 10.1126/science.1156951. [DOI] [PMC free article] [PubMed] [Google Scholar]