Abstract
Gain of chromosome 6p is a consistent feature of advanced melanomas. However, the identity of putative oncogene(s) associated with this amplification has remained elusive. The chromatin remodeling factor DEK is an attractive candidate as it maps to 6p (i.e. within common melanoma-amplified loci). Moreover, DEK expression is increased in metastatic melanomas, although the functional relevance of this induction remains unclear. Importantly, in other tumor types, DEK can display various tumorigenic effects, in part through its ability to promote proliferation and inhibit p53-dependent apoptosis. Here, we report a generalized upregulation of DEK protein in cells from aggressive melanomas. In addition, we provide genetic and mechanistic evidence to support a key role of DEK in the maintenance of malignant phenotypes of melanoma cells. Specifically, we show that long-term DEK downregulation by independent shRNAs resulted in premature senescence of a variety of melanoma cell lines. Short-term abrogation of DEK expression was also functionally relevant, as it attenuated the traditional resistance of melanomas to DNA damaging agents. Unexpectedly, DEK shRNA had no impact on p53 levels or p53-dependent apoptosis. Instead, we identified a new role for DEK in the transcriptional activation of the antiapoptotic MCL-1. Other MCL-1 related factors such as BCL-2 or BCL-xL were unaffected by changes in the endogenous levels of DEK, indicating a selective impact of this gene on the apoptotic machinery of melanoma cells. These results provide support for DEK as a long sought-after oncogene mapping at chromosome 6, with novel functions in melanoma proliferation and chemoresistance.
Keywords: chemoresistance, DEK, MCL-1, melanoma, senescence
Introduction
Metastatic melanomas are invariably chemoresistant and carry a grim prognosis (1). Complicating drug development, melanoma cells accumulate a plethora of changes in gene expression with the potential to unleash uncontrolled proliferation, evade senescence programs and inhibit death pathways at multiple levels (2). A hierarchical mapping of these alterations (to distinguish central nodes in tumor cell maintenance from inconsequential byproducts of tumor development) has remained elusive. Equally unclear are the mechanisms underlying common melanoma-associated events that have been ascribed neither to mutations nor to genetic or epigenetic alterations. This is particularly relevant for the antiapoptotic members of the BCL-2 family (i.e. BCL-2, BCL-xL and MCL-1). These proteins are invariably overexpressed in melanoma cells and play key roles in their chemoresistance (3). BCL-2 upregulation may be a consequence of MITF gene amplification (4). BCL-xL and MCL-1 can be under the control of the MAPK or NF-κB signaling cascades (5). However, neither MITF, MAPK nor NF-κB inhibition abrogates the expression of these BCL-2 family members (6-8), suggesting alternative, and yet unknown, mechanisms of regulation of the apoptotic machinery in melanoma cells.
Alteration of chromosome 6 is one of the most consistent cytogenetic changes in melanoma (9). In particular, gain of the 6p arm is prevalent in different manifestations of the disease, including cutaneous (both sun-exposed and non-sun-exposed), acral, mucosal, and uveal melanomas (10, 11). Furthermore, temporal clustering of karyotypic changes in melanoma indicates that 6p gain may represent an early event in the acquisition of chromosomal imbalances (12). Adding to the clinical relevance of chromosome 6, 6p copy gains have been associated with poor prognosis in ocular melanomas (11). Therefore, it has been long suggested that this genomic region could contain one or multiple pro-oncogenes (9, 13, 14). Although the minimal region of 6p gain has not been described in melanoma, several reports in retinoblastoma and bladder cancer have identified a narrow region of gain in 6p22-23 (15-18). This region notably contains the chromatin remodeling factor DEK.
DEK was originally discovered as the target of a chromosomal translocation event [t(6;9)(p23;q34)] in a subset of acute myeloid leukemias (19). Subsequent studies have repeatedly identified DEK as a frequently overexpressed gene in a number of neoplasms (16, 20-22). Furthermore, we and others have shown that DEK has effects on mRNA splicing, transcriptional control, DNA damage repair, differentiation, cell viability and cell to cell signaling (23-31). Pro-oncogenic roles of DEK are supported by its ability to inhibit p53-mediated apoptosis (32), cooperate with the viral oncogenes E6 and E7 to overcome senescence (29), and promote HRAS-driven keratinocyte transformation (28). The mechanism(s) through which DEK mediates its effects, particularly in the context of cellular oncogenes is not well understood. DEK is a structurally unique protein (33) that binds preferentially to euchromatin, although it can also be found within interchromatin granule clusters (34). Consequently, it is unclear which DEK targets have a causative role in tumor development and which are passengers of global changes of chromatin architecture.
The contribution of DEK to melanoma progression and chemoresistance has yet to be analyzed. Increased DEK mRNA and protein has been reported in a small number of melanoma cell lines or tissues (21, 35). However, DEK function is highly dependent on post-translational modifications (27, 34, 36). Therefore, it is unclear whether altered DEK expression is a generalized feature of advanced melanomas and whether this gene has a functional contribution to melanoma cell maintenance and drug response. To assess these questions we used tissue microarrays to compare the expression of DEK among benign nevi and malignant melanoma specimens. In addition, the role of DEK was defined in a panel of aggressive melanoma cell lines that recapitulate the most frequent melanoma-associated defects. Our studies establish DEK, a gene mapping to the genetically unstable 6p region, as the first chromatin remodeling factor with dual and selective roles in proliferation and apoptotic resistance in melanoma, acting independently of classical tumor suppressor pathways.
Materials and Methods
Cells and reagents
Human melanocytes and melanoma lines were isolated and cultured as previously described (6, 37). Doxorubicin hydrochloride, cycloheximide and 4′,6-diamidino-2-phenylindole (DAPI) were purchased from Sigma Chemical (St. Louis, MO). Bortezomib was obtained from Millennium Pharmaceuticals (Cambridge, MA). zVAD-fmk was purchased from Calbiochem (Cambridge, MA). TW-37 synthesis and validation has been described elsewhere (38).
Extract preparation and gene expression analyses
Total cell lysates were obtained by Laemmli extraction, separated by SDS-PAGE and transferred to Immobilon-P membranes (Millipore, Bedford, MA) for immunoblotting (antibodies used in this study are listed in the Supplementary Materials). Protein expression was quantified using Scion image densitometry software (Scion Corporation, Frederick, MD). Lentiviral-mediated modulation of DEK, MCL-1, Bcl-2 and BCL-xL, Tissue microarrays (TMAs), spectral karyotype (SKY) and comparative genomic hybridization (CGH) procedures are described in the Supplementary Information.
Cell proliferation and viability assays
Total cell numbers were estimated at the indicated times by manual counting, and cell death rates were determined by standard typan blue exclusion assays (38). For flow cytometry assays, cells were fixed in 70% ethanol, digested with 0.5 mg/mL RNaseA, and stained with 50 μg/mL propidium iodide prior to analysis in a FACSCalibur (Becton Dickinson Immunocytometry Systems, San Jose, CA, USA). Data were fitted using ModFit software (Verity Software, Topsham, ME, USA). Senescence-associated β-Galactosidase staining (SA-β-Gal) was performed as described previously (37). Nuclear condensation and fragmentation were assessed by fluorescence microscopy after DAPI staining, using an Olympus BX-51 upright microscope.
Quantitative real-time RT-PCR
RNA purification and real-time RT-PCR using a Bio-Rad iCycler (Bio-Rad, Hercules, CA) were performed essentially as previously described (37, 39) using the following primers: mcl1 forward - ATGCTTCGGAAACTGGACAT; mcl1 reverse - TCCTGATGCCACCTTCTAGG; β-actin forward - CGCCCAGCACGATGAAA; β-actin reverse - CCGCCGATCCACACAGA. The assay included a no-template control and a standard curve of six dilutions for both mcl-1 and β-actin. Results are given as the average of three experiments ± SE, and each sample was amplified and analyzed in triplicate.
Luciferase reporter gene assays
The human MCL-1 promoter reporter plasmid -203/+10 was a generous gift of Douglas Cress (40). Cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and the lysates were assayed for firefly and renilla luciferase activity using the Dual Luciferase Reporter Assay System (Promega Corp., Madison, WI). Luminescence was detected with a Tecan GENios plate reader (Phenix, Austria). Data are presented as the mean ± SE of three experiments.
Statistics
Statistical analyses of cell death, quantitative real-time RT-PCR, and luciferase reporter assays were done using a paired, two-tailed Student’s t-test.
RESULTS
DEK protein overexpression in metastatic melanoma
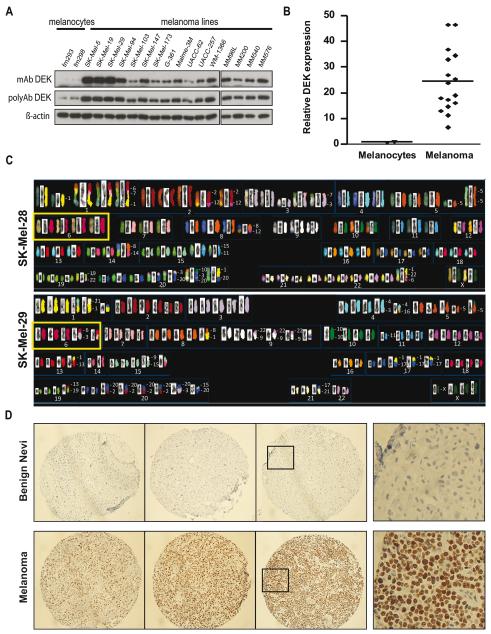
We hypothesized that gain of 6p in melanoma may function to promote DEK overexpression. To this end, a panel of sixteen metastatic melanoma lines was selected to reflect the most common melanoma-associated alterations in oncogenes or tumor suppressors (See Supplementary Table 1). In parallel, primary human foreskin-derived melanocytes were used as controls for normal cells. Immunoblotting revealed a generalized high expression of DEK, with an average 30-fold induction over the basal levels of normal melanocytes (Fig. 1A, B).
Figure 1. Overexpression of DEK in melanoma cell lines and tissue specimens.
A, immunoblotting of a panel of metastatic melanoma lines and primary foreskin-derived melanocytes (FM293 and FM298), with monoclonal (mAb) and polyclonal (polyAb) anti-DEK antibodies. B, Monoclonal anti-DEK band densities were quantified by densitometry, and relative DEK protein levels were normalized to FM293 melanocytes. The horizontal line indicates the mean for each group. C, SKY analysis showing increased copy number at chromosome 6 (yellow boxes) in SK-Mel-28, and SK-Mel-29, both expressing high levels of DEK (Fig. 1A and Fig. 2C). D, Representative examples of DEK staining in benign nevi (upper panels) and melanoma lesions (bottom panels) included in TMAs. The figures in the right column show higher magnification views of the indicated areas.
Interestingly, spectral karyotyping (SKY) confirmed that melanoma cell lines with high DEK expression (i.e. SK-Mel-28 and -29) contained, among other genetic abnormalities, an increased copy number of chromosome 6 (Fig. 1C). However, large-scale chromosome 6p amplifications are unlikely to be the sole mechanism underlying DEK upregulation. Thus, lines such as SK-Mel-103 or SK-Mel-147, with moderate levels of DEK protein (Fig. 1A), had a normal copy number of chromosome 6 defined by SKY (Supplementary Fig. S1A). Comparative genomic hybridization (CGH) showed focal gains at 6p22.3, but centromeric to DEK (Fig. S1B).
To validate the increased DEK protein expression found in cell lines, an extensive immunohistochemical analysis was performed on two tissue microarrays (TMAs), containing surgically removed melanocytic lesions, each spotted in triplicate. Specifically, these TMAs included 87 primary melanomas and 115 metastatic specimens. In addition, 3 dysplastic nevi, 7 benign nevi and 5 biopsies of normal skin were included as reference. Notably, 90% of the tumors analyzed showed clear positive DEK expression (10% being very marked and 80% with moderate DEK levels; see representative examples in Fig. 1D, bottom panels). Normal skin melanocytes and benign nevi showed no detectable DEK expression (Fig. 1D, upper panels, and results not shown). Therefore, these results support a broad spectrum upregulation of DEK during melanoma progression.
Long-term downregulation of the endogenous DEK expression in melanoma induces premature senescence
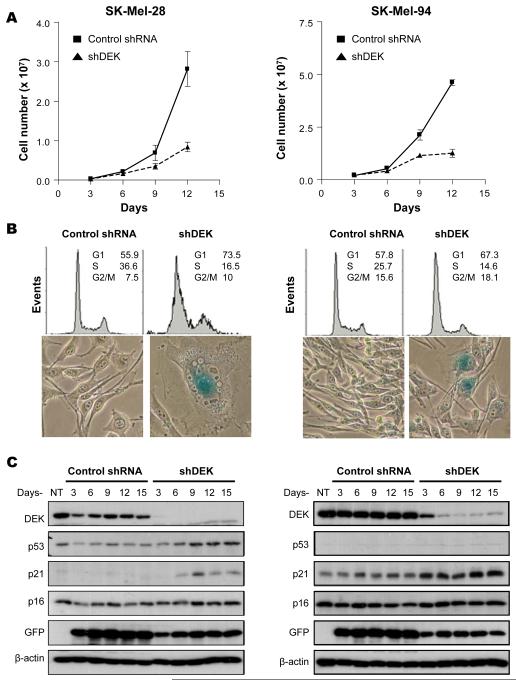
Lentiviral vectors were used to transduce short hairpin interfering RNAs (shRNAs) and downregulate DEK in a stable manner. To avoid complications of off-target effects, three independent shRNAs were tested (See supplementary Fig. S2). In 6 of 12 lines analyzed (SK-MEL-28, SK-MEL-94, G-361, UACC-62, UACC-257, and MM576), DEK depletion led to a progressive cell cycle arrest, eventually resulting in a near complete inhibition of cell proliferation as determined by total cell counting (Fig. 2A) and flow cytometry (Fig. 2B, upper panels).
Figure 2. Induction of senescence-like cell cycle arrest by long-term downregulation of DEK.
A, Total cell numbers estimated in SK-MEL-28 (left panel) and SK-MEL-94 (right panel) after infection with control shRNA or shDEK-2 (shDEK)-expressing lentiviruses. Error bars represent SEM. B, melanoma cell lines were transduced as in A, and fifteen days after infection, cell cycle profile distribution was analyzed by flow cytometry (upper panels), and senescence features were determined by SA-β-Gal activity (blue signal in bottom panels). C, Cells were treated as in A, and whole cell lysates were processed for detection of the indicated proteins by immunoblotting. NT, no treatment
The reduction of cells in S-phase of the cell cycle (Fig. 2B, upper panels) was accompanied by cell enlargement, flattening and vacuolization (Fig. 2B, lower panels), reminiscent of classical senescent phenotypes (41). In fact, staining for senescence-associated β-galactosidase activity was positive in DEK-depleted melanoma cells (Fig. 2B, lower panels). Interestingly, cell cycle arrest and the acquisition of senescent-like features occurred in cells expressing either wild type p53 or mutated p53 (e.g. SK-Mel-94 or SK-Mel-28, respectively; Fig. 2C). Notably, DEK shRNAs did not affect the expression of p16INK4A (Fig. 2C), a senescence-associated tumor suppressor. These results demonstrate that DEK is required for the continued proliferation of metastatic melanoma cells, and point to novel p53 and p16 INK4A-independent senescence programs.
Short-term downregulation of DEK expression bypasses melanoma chemoresistance
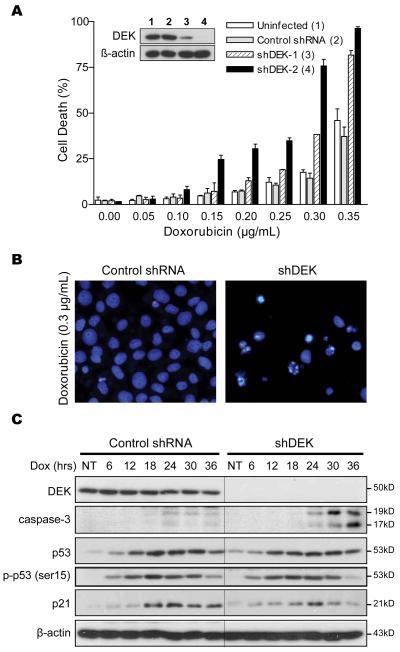
Recently, we have demonstrated that DEK can enhance resistance to DNA damaging agents by promoting efficient resolution of DNA damage foci (42). Therefore, we sought to determine whether short term downregulation of DEK (therapeutically more feasible than sustained gene inactivation) could contribute to the ability of melanoma cells to withstand genotoxic agents. The anthracycline doxorubicin is a potent anti-neoplastic agent in other tumor types, but has only limited efficacy against metastatic melanoma (8). This drug was selected to treat cells with the highest levels of DEK as they may be more dependent on this protein for survival (see Fig. 3 for SK-MEL-19 and Fig. 4 for SK-Mel-29; results with other cell lines are discussed below). SK-Mel-19 was infected with a control shRNA lentiviral vector, a vector expressing a single DEK-specific shRNA (shDEK-1), or a vector expressing two distinct DEK-specific shRNAs (shDEK-2). Two days after lentiviral infection, cells were treated with increasing doses of doxorubicin and cell death was estimated within 24-36h after treatment. Relatively high levels of doxorubicin were required to kill melanoma cells in the presence of DEK (i.e. >0.2 μg/ml; Fig. 3A). However, DEK depletion resulted in a marked increase in doxorubicin-driven cell death, particularly upon dual-shRNA targeting (Fig. 3A). Importantly, the increased response of DEK-depleted melanoma cells to doxorubicin was associated with classical apoptotic features such as chromatin condensation (Fig. 3B) and caspase processing (Fig. 3C). Therefore, although DEK may not be the sole mediator of melanoma chemoresistance, our results support a key role of this gene contributing to the malignant features of melanoma cells.
Figure 3. p53-independent induction of cell death by short-term inactivation of DEK.
A, SK-MEL-19 cells transduced with the indicated shRNAs were treated with doxorubicin for 30 hours, and cell death was determined by DAPI staining and counting of condensed and fragmented nuclei. B, fluorescence microscopy illustrating cell nuclear morphology after treatment with 0.3 μg/mL doxorubicin for 30 hrs in cells transduced with a control vector (left) or shDEK-2 (right). C, SK-MEL-19 cells transduced as in (A) were treated with 0.2 μg/mL doxorubicin and collected at the indicated times and analyzed by immunoblotting.
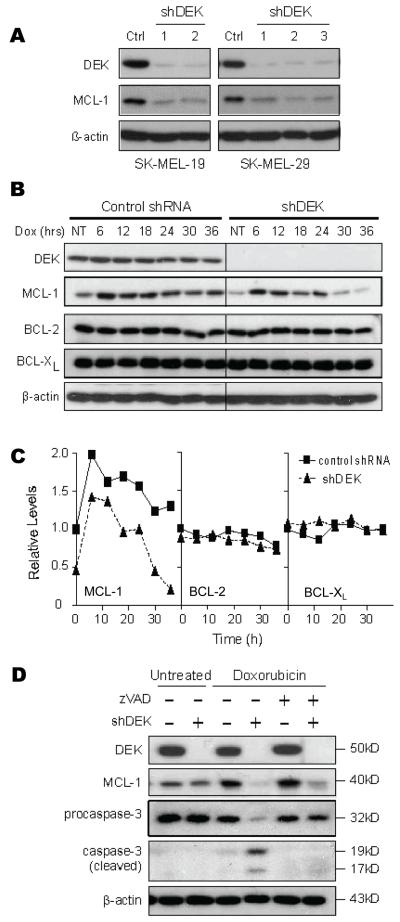
Figure 4. Caspase-independent downregulation of MCL-1 by DEK shRNA.
A, The indicated melanoma cells were infected with either a control lentivirus or one of three distinct vectors targeting DEK expression (shDEK-1, shDEK-2, shDEK-3). 96 hours after infection, cells were collected and their lysates were immunoblotted for DEK and MCL-1 expression. B, SK-MEL-19 cells transduced with the indicated shRNAs were treated with 0.2 μg/mL doxorubicin and collected for immunoblotting analyses. C, The graphs show the relative expression of MCL-1 (left panel), BCL-2 (middle panel) and BCL-xL (right panel) following doxorubicin treatment as estimated by band densitometry. Expression is shown relative to that of untreated control-transduced cells. D, Protein immunoblots in control and shDEK-infected SK-MEL-19 cells treated with 0.2 μg/mL doxorubicin alone or with doxorubicin in the presence of 50 μM of the pan-caspase inhibitor z-VAD-fmk.
DEK does not modulate p53 levels in melanoma cells
DEK has previously been shown to mediate anti-apoptotic effects through destabilization of p53 protein and inhibition of p53 activity (23, 32). However, as with the case of long-term downregulation of DEK shown above (Fig. 2C), short term expression of DEK shRNA was not found to significantly affect total p53 levels (Fig. 3C). Furthermore, the activation of p53 (detected by phosphorylation of its serine 15) and the induction of p53 targets, such as p21CIP1, were similar in DEK-expressing and deficient melanoma cells (Fig. 3C).
DEK-knockdown cells have diminished MCL-1 expression
Overexpression of anti-apoptotic BCL-2 family members has been associated with melanoma chemoresistance (3). Therefore, we tested whether DEK could control BCL-2 family members. Interestingly, DEK downregulation with two different shRNAs led to a drastic decrease in MCL-1 expression in SK-MEL-19, SK-MEL-29, SK-MEL-5, and MM576 (Fig 4A and data not shown). Notably, DEK shRNA did not alter BCL-2 or BCL-xL levels (Fig. 4B), suggesting selective effects on the apoptotic machinery.
To further assess the requirement of DEK for high MCL-1 expression, DEK depleted cells were treated with doxorubicin (which induces MCL-1 levels in melanoma cells, Fig. 4B, C). As shown in Fig. 4B DEK shRNA cells ultimately expressed minimal levels of MCL-1 after doxorubicin treatment (see quantification in Fig. 4C). Again, the effects of DEK on the apoptotic machinery were selective, as no changes in BCL-2 or BCL-xL levels were observed prior to or after doxorubicin treatment (Fig. 4B, C). Importantly, the reduction of MCL-1 expression in DEK-shRNA cells was not due to proteolytic cleavage by apoptotic caspases (43). Thus, the pan-caspase inhibitor z-VAD-fmk, although it eliminated caspase-3 cleavage, could not prevent MCL-1 downregulation in shDEK-treated cells (Fig. 4D).
Decreased MCL-1 in shDEK treated cells is not due to increased MCL-1 degradation
To determine if the decrease in MCL-1 protein in DEK-knockdown cells was due to enhanced proteasomal turnover, cells were treated with the proteasome inhibitor bortezomib. As we previously reported (39, 44), bortezomib induced a massive MCL-1 stabilization in control melanoma cells within 2-3 h of treatment (Fig. 5A). A similar rate of MCL-1 accumulation was observed in shDEK-expressing melanoma cells (Fig. 5B), although these cells have, as indicated above, intrinsically lower basal levels of MCL-1 (note that MCL-1 immunoblots in Fig. 5A were overexposed to favor the visualization of the endogenous low expression of MCL-1 in shRNA-DEK cells). Therefore, the diminished MCL-1 levels in DEK-knockdown cells were not due to increased proteosome-dependent MCL-1 degradation.
Figure 5. Transcriptional control of MCL-1 by DEK.
A. Compartative analysis of MCL-1 protein downregulation in SK-MEL-19 cells transduced with control or DEK shRNAs, and treated for the indicated times with 25 nM bortezomib (Bort). BCL-2, and β-actin are included as examples of DEK-independent proteins and for loading control, respectively. B, Quantification by densitomety of MCL-1 levels in (A) depicted with respect to non-treated control-infected cells. C-D, Control or shRNA transduced SK-MEL-19 cells were treated for 5 hours with 25 nM of the proteasome inhibitor bortezomib. Cells were then washed to remove bortezomib and incubated with cycloheximide (CHX) for the indicated times. Following treatment cells were collected and processed for immunoblotting (C) and subsequent quantification by densitomety (D). E, MCL-1 mRNA expression of control and shDEK treated SK-MEL-19 cells determined by quantitative real-time RT-PCR. F, Control and shDEK transduced SK-MEL-19 cells were transfected with either the promoterless pGL2 basic plasmid or a plasmid expressing the firefly luciferase gene under the control of the human MCL-1 promoter (-203/+10 bp). The average luciferase activity of three experiments is shown relative to that of control transduced cells transfected with the pGL2 basic plasmid. Error bars correspond to SEM. *, p < 0.05.
To assess the stability of MCL-1 protein in DEK-knockdown cells, cycloheximide, a translational inhibitor, was used to inhibit de novo MCL-1 synthesis. To better visualize the decrease in MCL-1 expression, cells were first pre-treated with bortezomib to augment MCL-1 levels. Bortezomib was subsequently removed, and cells were incubated in cycloheximide and then processed for immunoblotting analysis (see Materials and Methods for additional details). As shown in Fig. 5C,D, depletion of MCL-1 occurred with similar kinetics in both control and shDEK-treated cells.
Novel role of DEK in MCL-1 transcription
Since DEK was not affecting DEK protein stability, it could be controlling its transcription. To this end mRNA levels were estimated by quantitative RT-PCR. As shown in Fig. 5E, DEK downregulation lead to a 50% reduction in MCL-1 mRNA. To confirm the requirement of DEK for MCL-1 mRNA transcription, a reporter construct driving luciferase expression under the control of the proximal region of the human MCL-1 promoter was transduced into either control or shDEK transduced SK-MEL-19 cells (Fig. 5F). We found a 70-80% reduction in promoter activity in MCL-1 in the absence of DEK (Fig. 5F). These results demonstrate a new role for DEK in the transcriptional control of MCL-1.
Functional implication of DEK-dependent MCL-1 expression
To define the specific interplay between DEK and MCL-1 in the enhancement of melanoma chemoresistance, we generated isogenic series of melanoma cell lines with differing levels of both proteins, and we compared their response to doxorubicin. Additional isogenic cell lines were generated to alter (positively and negatively) the endogenous expression of BCL-xL. BCL-xL was chosen as an example of an antiapoptotic BCL-2 factor that is not under the control of DEK (Fig. 4B), but is also overexpressed in melanoma cells (3).
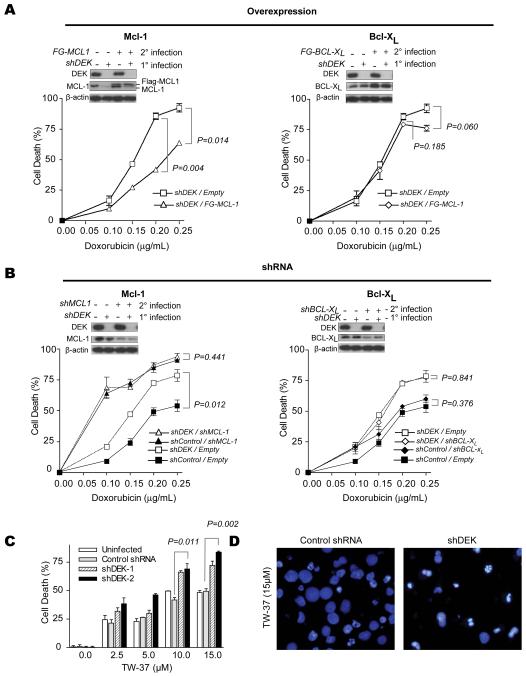
To alter MCL-1 expression we used two approaches. First, we generated lentiviral vectors to drive MCl-1 from an ectopic promoter that is not sensitive to DEK. Viral titers were selected to restore MCL-1 levels to the basal conditions (Fig. 6A, inset in left panel). In a second set of experiments, the opposite strategy was tested (Fig. 6B), namely, to abrogate MCL-1 mRNA and protein expression with shRNAs that we have previously validated (6, 39, 44). Interestingly, exogenous MCL-1 significantly protected DEK depleted melanoma cells, reducing the killing activity of doxorubicin in half (Fig 6A; P=0.014 for concentrations of doxorubicin of 0.25 μg/ml). On the other hand, when MCL-1 was depleted, DEK shRNA was not able to further enhance drug response (Fig. 6B, left panel; P=0.441).
Figure 6. Functional impact of the selective regulation of BCL-2 family members by DEK.
A, Restoration of MCL-1 levels using lentiviral vectors coding for MCL-1 under an exogenous promoter that is not sensitive to DEK shRNA downregulation (left panel). Control infections were also performed with a vector expressing BCL-xL (right panel). 72 hours after infection with MCL-1 or BCL-xL lentiviruses, cells were treated with the indicated concentration of doxorubicin for 30 hrs. Following treatment, cells were stained with DAPI and apoptotic cells were counted by fluorescence microscopy. B, inactivation of MCL-1 (left panel) or BCL-xL (right panel) with the corresponding shRNAs. Insets demonstrate shRNA-mediated knockdown of DEK and either knockdown or overexpression of BCL-xL and MCL-1 as determined by immunoblotting. C, SK-MEL-19 cells were transduced as in Fig. 3 and treated with the BH3 mimetic TW-37 for 48 hours, and then stained with trypan blue. Error bars, SEM. D, fluorescence microscopy by DAPI staining illustrating cell nuclear morphology after treatment with 15μM TW-37 in cells infected with a control shRNA expressing vector (left) or shDEK-2 (right). P values for the indicated pair-wise comparisons were determined by two-tailed Student’s t test.
In contrast to MCL-1, altering BCL-xL levels either by overexpression (Fig. 6A, right panels) or shRNAs (Fig. 6B, right panels) had no significant impact on melanoma cell viability whether in the presence or absence of DEK (see the corresponding P values in Fig. 6A, 6B).
From the data presented above, it follows that DEK shRNA may augment the therapeutic efficacy of small molecules whose anti-tumor activity depends on MCL-1 inactivation. To examine this possibility, we tested the small molecule BH3 mimetic TW-37, which we have previously shown acts primarily by inhibiting MCL-1 (6, 38). Interestingly, DEK-depleted SK-MEL-19 cells were significantly more sensitive to TW-37 treatment than their control counterparts, showing an accelerated killing (Fig. 6C; P=0.011-0.002) and induction of apoptotic like features (see chromatin condensation in Fig. 6D). These results provide proof of principle for the development of rational therapies targeting DEK/MCL-1 dependent mechanisms of cell survival in melanoma.
DISCUSSION
In this study we identified new roles for DEK in metastatic melanoma cells. Despite the high degree of heterogeneity characteristic of this tumor type, we found that the DEK protein was invariably overexpressed in all 16 metastatic cell lines investigated, independent of their genetic background. Moreover, high throughput immunohistochemical analyses revealed a marked expression of DEK in 90% of primary and metastatic melanomas studied. In the same conditions, DEK expression in normal skin and benign nevi was nearly undetectable. Importantly, this high expression of DEK was not a simple byproduct of melanoma development. Abrogation of endogenous DEK expression resulted in the disruption of characteristic malignant features of melanomas. Thus, long-term expression of DEK shRNAs led to cell cycle arrest with features of senescence. In turn, acute downregulation of DEK was sufficient to enhance the response to DNA damaging agents and BH3 mimetics (doxorubicin and TW37, respectively). Our results additionally unveiled a previously undescribed role for DEK in the regulation of a critical mediator of chemoresistance, MCL-1.
The contribution of DEK to melanoma proliferation and drug resistance provides mechanistic support for the long-suspected presence of oncogene(s) mapping to the short arm of chromosome 6 (9, 13, 14). A complication in the identification of the putative onocogene(s) in this region has been its high gene density (624 genes in the 6p21-23 interval). Moreover, this area is heavily enriched in CpG islands, and therefore, it can be subject to complex epigenetic regulation. Interestingly, a common area of copy gain at 6p22-p23 in ocular melanomas (11) had also been reported in bladder cancer and retinoblastoma (16-18). Of the seven genes in this region, only DEK was found to have a correlation between copy gain and gene expression (18). Our SKY and CGH analyses revealed duplications of chromosome 6 in melanoma cell lines expressing high levels of DEK. However, other mechanisms of regulation may favor DEK upregulation, as its expression can also be induced in cells with a normal copy number at the DEK locus.
Considering the high number of mutations and alterations in proliferative pathways that melanoma cells express, it is interesting that downregulation of a single gene (DEK) can ultimately drive them into senescence-like cell cycle arrest. In fact, melanomas are considered to be particularly efficient at overcoming premature senescence, as this is one of the early barriers to melanocyte transformation (45). Mechanisms that circumvent premature senescence in melanomas are not well characterized. In fact, a puzzling feature of this disease is that a large fraction of acquired melanomas neither mutate nor delete p53 or p16INK4a, two tumor suppressors with key roles in the induction of senescence programs (2). Therefore, DEK overexpression may represent an alternative mechanism to counteract the tumor suppressor activity of p53 and p16 in melanoma cells. The precise effects of DEK in promoting sustained proliferation will require further studies, but it is tempting to speculate that it may cooperate with other oncogenes mapping at chromosome 6p, such as c-MYC, which we have recently shown to control melanoma senescence (46).
While the requirement of sustained expression of DEK to avoid premature senescence has implications for basic aspects of melanoma, the new roles of DEK shown here in the control of the apoptotic machinery are perhaps more significant from a therapeutic perspective. Although DEK has previously been shown to participate in the regulation of gene transcription (23-25, 30, 47), promotion of MCL-1 expression is the first demonstration of a transcriptional target of DEK directly contributing to an oncogenic phenotype. The requirement of DEK for MCL-1 expression is relevant as this protein has an intrinsically short half life (5), and yet it is consistently accumulated in melanoma cells (3). In addition, MCL-1 transcription can be inhibited by p53 (48), E2F1 or E2F2 (40), all of which are expressed in melanoma cells. Therefore, DEK’s effects on MCL-1 transcription may serve to compensate for negative regulatory signals intrinsically expressed in melanoma cells. Furthermore, we and others have shown that MCL-1 is essential for long term survival of melanoma cells, and it is a main determinant of their resistance to dacarbazine, BH3 mimetics, and inhibitors of MEK or the proteasome (6, 44, 49). MCL-1 can also control melanoma cell survival under conditions of anoikis (50). Thus, enhanced MCL-1 expression caused by DEK overexpression may represent a major mechanism of melanoma cell survival in response to chemotherapeutic agents and other stress-inducing factors.
In conclusion, our results implicate overexpression of the 6p pro-oncogene DEK as an extremely frequent event in metastatic melanomas. Remarkably, this single event can have a dual effect on melanoma cell proliferation and chemoresistance (preventing senescence and inducing the anti-apoptotic MCL-1, respectively). Therefore, our results suggest that DEK may be a relevant target for therapeutic intervention in this aggressive disease.
Supplementary Material
ACKNOWLEDGEMENTS
We thank MaryBeth Riblett (Univ. Michigan) and Tonantzin G. Calvo (CNIO) for technical assistance, and M Carmen Martin and Bibiana I Ferreira (CNIO) for their help with SKY and CGH analyses. We also thank Karolyn Oetjen and Colin Duckett for the BCL-xL expression vector. M.S.K. was supported by NIH Training Grant T32-GM07863, and National Science Foundation and Rackham Predoctoral Fellowships; F.K. was supported by a William D. Robinson Fellowship from the Arthritis Foundation/Michigan Chapter and is a recipient of an Arthritis Foundation Postdoctoral Fellowship. D.S.L.K. and A.M.C. were supported by NIH Prostate SPORE P50CA69568 and NIH U01 CA111275; D.M.M. by NIH R01-AI062248 and by a Burroughs Wellcome Fund Clinical Scientist Award in Translational Research; and M.S.S by NIH R01-CA107237 and a development grant from the Spanish Association Against Cancer (AECC). E.F-R is a recipient of a post-residency award from Fundación Obra Social Caja Navarra.
REFERENCES
- 1.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–7. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 2.Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006;20:2149–82. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 3.Soengas MS, Lowe SW. Apoptosis and melanoma chemoresistance. Oncogene. 2003;22:3138–51. doi: 10.1038/sj.onc.1206454. [DOI] [PubMed] [Google Scholar]
- 4.McGill GG, Horstmann M, Widlund HR, et al. Bcl2 regulation by the melanocyte master regulator mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109:707–18. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 5.Kutuk O, Letai A. Regulation of Bcl-2 family proteins by posttranslational modifications. Curr Mol Med. 2008;8:102–18. doi: 10.2174/156652408783769599. [DOI] [PubMed] [Google Scholar]
- 6.Verhaegen M, Bauer JA, Martin de la Vega C, et al. A novel BH3 mimetic reveals a mitogen-activated protein kinase-dependent mechanism of melanoma cell death controlled by p53 and reactive oxygen species. Cancer Res. 2006;66:11348–59. doi: 10.1158/0008-5472.CAN-06-1748. [DOI] [PubMed] [Google Scholar]
- 7.Ueda Y, Richmond A. NF-kappaB activation in melanoma. Pigment Cell Res. 2006;19:112–24. doi: 10.1111/j.1600-0749.2006.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gogas HJ, Kirkwood JM, Sondak VK. Chemotherapy for metastatic melanoma: time for a change? Cancer. 2007;109:455–64. doi: 10.1002/cncr.22427. [DOI] [PubMed] [Google Scholar]
- 9.Santos GC, Zielenska M, Prasad M, Squire JA. Chromosome 6p amplification and cancer progression. J Clin Pathol. 2007;60:1–7. doi: 10.1136/jcp.2005.034389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 11.Namiki T, Yanagawa S, Izumo T, et al. Genomic alterations in primary cutaneous melanomas detected by metaphase comparative genomic hybridization with laser capture or manual microdissection: 6p gains may predict poor outcome. Cancer Genet Cytogenet. 2005;157:1–11. doi: 10.1016/j.cancergencyto.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Hoglund M, Gisselsson D, Soller M, Hansen GB, Elfving P, Mitelman F. Dissecting karyotypic patterns in malignant melanomas: temporal clustering of losses and gains in melanoma karyotypic evolution. Int J Cancer. 2004;108:57–65. doi: 10.1002/ijc.11558. [DOI] [PubMed] [Google Scholar]
- 13.Balaban G, Herlyn M, Guerry Dt, et al. Cytogenetics of human malignant melanoma and premalignant lesions. Cancer Genet Cytogenet. 1984;11:429–39. doi: 10.1016/0165-4608(84)90024-4. [DOI] [PubMed] [Google Scholar]
- 14.Pathak S, Drwinga HL, Hsu TC. Involvement of chromosome 6 in rearrangements in human malignant melanoma cell lines. Cytogenet Cell Genet. 1983;36:573–9. doi: 10.1159/000131975. [DOI] [PubMed] [Google Scholar]
- 15.Chen D, Gallie BL, Squire JA. Minimal regions of chromosomal imbalance in retinoblastoma detected by comparative genomic hybridization. Cancer Genet Cytogenet. 2001;129:57–63. doi: 10.1016/s0165-4608(01)00427-7. [DOI] [PubMed] [Google Scholar]
- 16.Evans AJ, Gallie BL, Jewett MA, et al. Defining a 0.5-mb region of genomic gain on chromosome 6p22 in bladder cancer by quantitative-multiplex polymerase chain reaction. Am J Pathol. 2004;164:285–93. doi: 10.1016/S0002-9440(10)63118-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grasemann C, Gratias S, Stephan H, et al. Gains and overexpression identify DEK and E2F3 as targets of chromosome 6p gains in retinoblastoma. Oncogene. 2005;24:6441–9. doi: 10.1038/sj.onc.1208792. [DOI] [PubMed] [Google Scholar]
- 18.Orlic M, Spencer CE, Wang L, Gallie BL. Expression analysis of 6p22 genomic gain in retinoblastoma. Genes Chromosomes Cancer. 2006;45:72–82. doi: 10.1002/gcc.20263. [DOI] [PubMed] [Google Scholar]
- 19.von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Mol Cell Biol. 1992;12:3346–55. doi: 10.1128/mcb.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waldmann T, Scholten I, Kappes F, Hu HG, Knippers R. The DEK protein--an abundant and ubiquitous constituent of mammalian chromatin. Gene. 2004;343:1–9. doi: 10.1016/j.gene.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 21.Carro MS, Spiga FM, Quarto M, et al. DEK Expression is controlled by E2F and deregulated in diverse tumor types. Cell Cycle. 2006;5:1202–7. doi: 10.4161/cc.5.11.2801. [DOI] [PubMed] [Google Scholar]
- 22.Wu Q, Li Z, Lin H, Han L, Liu S, Lin Z. DEK overexpression in uterine cervical cancers. Pathol Int. 2008;58:378–82. doi: 10.1111/j.1440-1827.2008.02239.x. [DOI] [PubMed] [Google Scholar]
- 23.Sammons M, Wan SS, Vogel NL, Mientjes EJ, Grosveld G, Ashburner BP. Negative regulation of the RelA/p65 transactivation function by the product of the DEK proto-oncogene. J Biol Chem. 2006;281:26802–12. doi: 10.1074/jbc.M600915200. [DOI] [PubMed] [Google Scholar]
- 24.Faulkner NE, Hilfinger JM, Markovitz DM. Protein phosphatase 2A activates the HIV-2 promoter through enhancer elements that include the pets site. J Biol Chem. 2001;276:25804–12. doi: 10.1074/jbc.M006454200. [DOI] [PubMed] [Google Scholar]
- 25.Gamble MJ, Fisher RP. SET and PARP1 remove DEK from chromatin to permit access by the transcription machinery. Nat Struct Mol Biol. 2007;14:548–55. doi: 10.1038/nsmb1248. [DOI] [PubMed] [Google Scholar]
- 26.Kappes F, Scholten I, Richter N, Gruss C, Waldmann T. Functional domains of the ubiquitous chromatin protein DEK. Mol Cell Biol. 2004;24:6000–10. doi: 10.1128/MCB.24.13.6000-6010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kappes F, Fahrer J, Khodadoust MS, et al. DEK is a poly(ADP-ribose) acceptor in apoptosis and mediates resistance to genotoxic stress. Mol Cell Biol. 2008;28:3245–57. doi: 10.1128/MCB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wise-Draper TM, Morreale RJ, Morris TA, et al. DEK proto-oncogene expression interferes with the normal epithelial differentiation program. Am J Pathol. 2009;174:71–81. doi: 10.2353/ajpath.2009.080330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wise-Draper TM, Allen HV, Thobe MN, et al. The human DEK proto-oncogene is a senescence inhibitor and an upregulated target of high-risk human papillomavirus E7. J Virol. 2005;79:14309–17. doi: 10.1128/JVI.79.22.14309-14317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim DW, Chae JI, Kim JY, et al. Proteomic analysis of apoptosis related proteins regulated by proto-oncogene protein DEK. J Cell Biochem. 2009 doi: 10.1002/jcb.22083. [DOI] [PubMed] [Google Scholar]
- 31.Sitwala KV, Mor-Vaknin N, Markovitz DM. Minireview: DEK and gene regulation, oncogenesis and AIDS. Anticancer Res. 2003;23:2155–8. [PubMed] [Google Scholar]
- 32.Wise-Draper TM, Allen HV, Jones EE, Habash KB, Matsuo H, Wells SI. Apoptosis inhibition by the human DEK oncoprotein involves interference with p53 functions. Mol Cell Biol. 2006;26:7506–19. doi: 10.1128/MCB.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Devany M, Kappes F, Chen KM, Markovitz DM, Matsuo H. Solution NMR structure of the N-terminal domain of the human DEK protein. Protein Sci. 2008;17:205–15. doi: 10.1110/ps.073244108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cleary J, Sitwala KV, Khodadoust MS, et al. p300/CBP-associated factor drives DEK into interchromatin granule clusters. J Biol Chem. 2005;280:31760–7. doi: 10.1074/jbc.M500884200. [DOI] [PubMed] [Google Scholar]
- 35.Grottke C, Mantwill K, Dietel M, Schadendorf D, Lage H. Identification of differentially expressed genes in human melanoma cells with acquired resistance to various antineoplastic drugs. Int J Cancer. 2000;88:535–46. doi: 10.1002/1097-0215(20001115)88:4<535::aid-ijc4>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 36.Kappes F, Damoc C, Knippers R, Przybylski M, Pinna LA, Gruss C. Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK. Mol Cell Biol. 2004;24:6011–20. doi: 10.1128/MCB.24.13.6011-6020.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denoyelle C, Abou-Rjaily G, Bezrookove V, et al. Anti-oncogenic role of the endoplasmic reticulum differentially activated by mutations in the MAPK pathway. Nat Cell Biol. 2006;8:1053–63. doi: 10.1038/ncb1471. [DOI] [PubMed] [Google Scholar]
- 38.Wang G, Nikolovska-Coleska Z, Yang CY, et al. Structure-based design of potent small-molecule inhibitors of anti-apoptotic Bcl-2 proteins. J Med Chem. 2006;49:6139–42. doi: 10.1021/jm060460o. [DOI] [PubMed] [Google Scholar]
- 39.Fernandez Y, Verhaegen M, Miller TP, et al. Differential regulation of noxa in normal melanocytes and melanoma cells by proteasome inhibition: therapeutic implications. Cancer Res. 2005;65:6294–304. doi: 10.1158/0008-5472.CAN-05-0686. [DOI] [PubMed] [Google Scholar]
- 40.Croxton R, Ma Y, Song L, Haura EB, Cress WD. Direct repression of the Mcl-1 promoter by E2F1. Oncogene. 2002;21:1359–69. doi: 10.1038/sj.onc.1205157. [DOI] [PubMed] [Google Scholar]
- 41.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–33. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Kappes F, Fahrer J, Khodadoust MS, et al. DEK is a poly(ADP-ribose)-acceptor in apoptosis and mediates resistance to genotoxic stress. Mol Cell Biol. 2008 doi: 10.1128/MCB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han J, Goldstein LA, Gastman BR, Froelich CJ, Yin XM, Rabinowich H. Degradation of Mcl-1 by granzyme B: implications for Bim-mediated mitochondrial apoptotic events. J Biol Chem. 2004;279:22020–9. doi: 10.1074/jbc.M313234200. [DOI] [PubMed] [Google Scholar]
- 44.Wolter KG, Verhaegen M, Fernandez Y, et al. Therapeutic window for melanoma treatment provided by selective effects of the proteasome on Bcl-2 proteins. Cell Death Differ. 2007;14:1605–16. doi: 10.1038/sj.cdd.4402163. [DOI] [PubMed] [Google Scholar]
- 45.Prieur A, Peeper DS. Cellular senescence in vivo: a barrier to tumorigenesis. Curr Opin Cell Biol. 2008;20:150–5. doi: 10.1016/j.ceb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 46.Zhuang D, Mannava S, Grachtchouk V, et al. C-MYC overexpression is required for continuous suppression of oncogene-induced senescence in melanoma cells. Oncogene. 2008;27:6623–34. doi: 10.1038/onc.2008.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ko SI, Lee IS, Kim JY, et al. Regulation of histone acetyltransferase activity of p300 and PCAF by proto-oncogene protein DEK. FEBS Lett. 2006;580:3217–22. doi: 10.1016/j.febslet.2006.04.081. [DOI] [PubMed] [Google Scholar]
- 48.Pietrzak M, Puzianowska-Kuznicka M. p53-dependent repression of the human MCL-1 gene encoding an anti-apoptotic member of the BCL-2 family: the role of Sp1 and of basic transcription factor binding sites in the MCL-1 promoter. Biol Chem. 2008;389:383–93. doi: 10.1515/BC.2008.039. [DOI] [PubMed] [Google Scholar]
- 49.Qin JZ, Xin H, Sitailo LA, Denning MF, Nickoloff BJ. Enhanced killing of melanoma cells by simultaneously targeting Mcl-1 and NOXA. Cancer Res. 2006;66:9636–45. doi: 10.1158/0008-5472.CAN-06-0747. [DOI] [PubMed] [Google Scholar]
- 50.Boisvert-Adamo K, Longmate W, Abel EV, Aplin AE. Mcl-1 is required for melanoma cell resistance to anoikis. Mol Cancer Res. 2009;7:549–56. doi: 10.1158/1541-7786.MCR-08-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.