Abstract
Although it is well established that the ventral telencephalon is the primary source of GABAergic cortical interneurons in rodents, little is known about the specification of specific interneuron subtypes. It is also unclear whether the potential to achieve a given fate is established at their place of origin or by signals received during their migration to or during their maturation within the cerebral cortex. Using both in vivo and in vitro transplantation techniques, we find that two major interneuron subgroups have largely distinct origins within the MGE. Somatostatin (SST)-expressing interneurons are primarily generated within the dorsal MGE, while parvalbumin (PV)-expressing interneurons primarily originate from the ventral MGE. In addition, we show that significant heterogeneity exists between gene expression patterns in the dorsal and ventral MGE. These results suggest that, like the spinal cord, neuronal fate determination in the ventral telencephalon is largely the result of spatially segregated, molecularly distinct microdomains arranged on the dorsal-ventral axis.
Keywords: MGE, somatostatin, parvalbumin, microarray, interneuron subtypes
Introduction
Proper functioning of the cerebral cortex requires a network of excitatory and inhibitory stimuli primarily produced by projection neurons (using glutamate as a neurotransmitter) and interneurons (using GABA as a neurotransmitter), respectively. Interneurons, while comprising a minority of cortical neurons, play vital roles in modulating cortical output and plasticity (Wang et al., 2004a; Whittington and Traub, 2003) and have also been implicated in several developmental processes, including the regulation of neuronal proliferation and migration during corticogenesis and the development of cortical circuitry (Hensch, 2005; Owens and Kriegstein, 2002).
Fate-mapping experiments both in vivo and in vitro have demonstrated that the ventral forebrain is the primary source of cortical interneurons in rodents [for recent reviews see (Corbin et al., 2001; Marin and Rubenstein, 2001; Wonders and Anderson, 2006)]. Different interneuron subgroups, as defined by their expression of largely non-overlapping chemical markers (Gonchar and Burkhalter, 1997; Kawaguchi and Kubota, 1997), show regional specificity for their generation. Culture experiments suggested that Calretinin-expressing interneurons with bipolar or bitufted dendritic trees originate in the dorsal, non-Nkx2.1 expressing region of the caudal ganglionic eminence (CGE), whereas both the SST and the PV-expressing subgroups originate in mainly in the MGE (Xu et al., 2004). Homotopic transplants of MGE or CGE progenitors in vivo found similar results, with the important addition of physiological parameters to subgroup-defining characteristics to (Butt et al., 2005).
Success in the identification of distinct origins of interneuron subgroups has promoted efforts to determine factors that specify subgroup fate. In embryos just prior to birth, loss of Nkx2.1 expression results in loss of neurons expressing SST as well as NPY (Anderson et al., 2001), a neuropeptide expressed in interneuron populations that partially overlaps with SST-expressing cortical interneurons. In addition, studies using both primary cultures of Nkx2.1 cortex, and in vitro transplants of cells from the Nkx2.1-/- MGE-like region onto cortical feeder cells, suggest that Nkx2.1 is required for the initial specification of the MGE-derived interneuron subgroups (Xu et al., 2004). Upstream of Nkx2.1, sonic hedgehog expression is required during the age range of neurogenesis to maintain Nkx2.1 expression and the generation of Nkx2.1-dependent interneuron fates (Xu et al., 2005). Downstream of Nkx2.1, the transcription factor Lhx6 also appears to be required for the interneurons to achieve the SST or PV expressing phenotypes (V. Pachnis, personal communication; Du and Anderson, submitted).
Despite these advances little is known about the differential specification of SST versus PV expressing interneurons. Is their fate determined in the MGE or by factors they encounter during or after migration to the cerebral cortex? In this paper, we first demonstrate that cortical interneuron neurochemical subgroup identity is primarily specified within a progenitor's place of origin and does not appear to require cues received during migration to the cortex. By assessing the fates of different subregions of the MGE along the dorsal/ventral axis, we show a strong fate bias exists for SST+ and NPY+ interneurons being generated by progenitors in the dorsal MGE (dMGE) and PV+ interneurons from the ventral MGE (vMGE). Finally, an RNA microarray-based analysis reveals that a high level of molecular heterogeneity is present in the dorsal or ventral MGE that may account for the specification of these interneuron subgroups. As similar findings have recently been reported by another group (Flames et al., 2007), evidence is accruing that, like the ventral spinal cord, the cell fate in the ventral telencephalon is partially determined by spatially distinct progenitor domains.
Materials and methods
In vivo transplantation
E13.5 dams were sacrificed and the GFP+ embryos placed in ice-cold Hank's buffer. The brains were removed and placed in NB/B27 medium, and the ventral forebrain exposed by removing the dorsal and lateral regions of the cortex (see Figure 1A). At this age, the sulcus between MGE and LGE can easily be visualized. Using fine forceps, this sulcus was slightly extended caudally, and a cut made towards the midline at the rostral-most level of the thalamus. The MGE was then gently freed from the underlying tissue and dissociated by trituration with a fired glass pipette. In the case of dorsal and ventral MGE donor tissue, cells were collected from 250μm thick coronal sections as shown in Figure 2A. Samples obtained from four separate slices were combined to maximize the cell density. Each combined sample (four slices) was considered an individual experiment for statistical purposes (n=3×4 slices = 12 slices total).
Figure 1. Cortical interneuron subgroup identity is determined within the MGE.
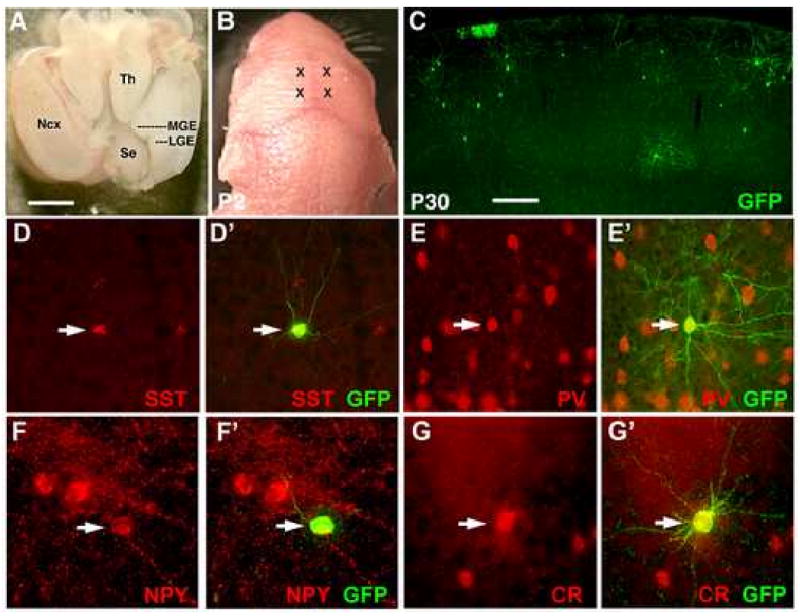
(A) The brain of an E13.5 embryo as viewed from above. Rostral is to the panel bottom. To isolate the MGE, the neocortex (Ncx) was dissected away as shown for the right hemisphere. The sulcus between the MGE and LGE is extended slightly, and a cut made along the medial-lateral axis at the level of the thalamus (Th). The MGE can then be separated free from the remainder of the brain. (B) Two injection sites per hemisphere were made into each neonate (marked by X's). (C) By P30, transplanted GFP+ cells are found throughout the cortex, of which the majority has adopted cortical interneuron morphologies. (D-G) Transplanted cells co-express interneuron subgroup markers including somatostatin (D-D′), parvalbumin (E-E′), neuropeptide Y (F-F′), and calretinin (G-G′). Scale bar in (A) is 1mm, in (C) is 200μm.
Figure 2. A dorsal/ventral fate potential bias exists for MGE-derived progenitors transplanted both in vivo and in vitro.
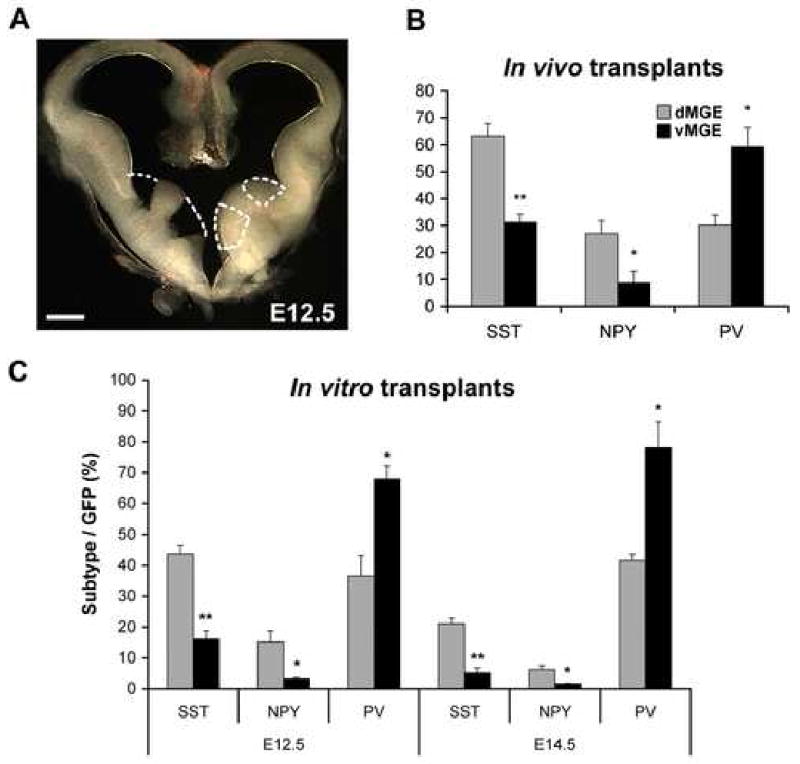
(A) The MGE of an E12.5 embryo dissected into dorsal and ventral domains as shown. Dissected domains were limited to the lighter-colored, periventricular region that is produced in these dark-field images by the higher cell packing density of the proliferative zone. Dissections of E13.5 and E14.5 MGE are performed similarly. (B) When E13.5 dorsal (d) and ventral (v) MGE progenitors are transplanted in vivo, both SST- and NPY-expressing interneurons are preferentially generated from dMGE transplants, while PV-expressing interneurons are preferentially generated from vMGE transplants. (C) At both E12.5 and E14.5, dMGE transplants also show a bias towards generating SST- and NPY-expressing interneurons, while the vMGE transplants preferentially give rise to PV-expressing interneurons when transplanted in vitro onto cortical feeders. Unpaired t-test: * p<0.05; ** p<0.005. Scale bar in (A) is 200μm.
Donor cells were resuspended in 15-30μl of NB/B27 medium to a cell concentration of roughly 40 cells/nl. The cells were then suction-filled into a pulled glass micropipette (0.5mm I.D., 1mm O.D.) fitted to an oocyte nanoinjector (Nanoinject II, Drummond). The use of this injector allowed for the minimization of tissue damage by injecting small, well-defined volumes at a relatively slow rate (23nl/s). Each pup received ten injections of 69nl each into each of four injection sites placed 1mm lateral to the midline, and either 1mm caudal to bregma or 1mm rostral to the interaural line (see Figure 1B). The micropipette tip was placed 1mm deep to the pial surface, allowing for the injection of cells mainly into layers 3, 4 and 5. (Figure 1C).
Co-cultures of MGE-derived cells on cortical feeders
Primary cortical feeder cultures (100,000 cells per 36 mm2 well of 16-well chamber slides, Lab-Tek) were prepared from the dissociated cortices of neonatal pups as previously described (Xu et al., 2004). Dorsal and ventral MGE donor cells were obtained as described for the in vivo transplants, except that each telencephalic slice was treated as a separate experiment. Two thousand cells per well were added to cortical feeder cultures prepared 1 day previously, and the cultures were maintained as previously described (Xu et al., 2004).
Tissue collection and imunohistochemistry
Neonates receiving cell injections were sacrificed at P30. The mice were perfused intracardially with 0.9% NaCl followed by 4% PFA and postfixed in fresh 4% PFA for 4 hours. The brains were then removed and sectioned in the coronal plane at 40μm on a vibratome. In vitro cultures were fixed for 20 minutes in 4% PFA except for those being processed for GABA immunolabeling, which were fixed in 4% PFA + 0.5% glutaraldehyde. Cell profile counts were made in somatosensory cortex, between the genu of the corpus callosum and the hippocampal commissure.
Primary antibodies used for immunofluorescence labeling included anti-BrdU (mouse, Chemicon, 1:400; rat, Serotec, 1:200), calretinin (rabbit, Chemicon, 1:2000; mouse, Swant, 1:5000), GABA (rabbit, Sigma, 1:5000), GFP (rabbit or chick, Molecular Probes, 1:2000), Nkx6.2 (guinea pig, Johan Ericson, 1:1000), NPY (rabbit, Immunostar, 1:2000), parvalbumin (mouse, Chemicon, 1:5000), and somatostatin (rat, Chemicon, 1:400). Fluorescent secondary antibodies were Alexa line (Molecular Probes, 1:500). Triple labeling of in vitro cell cultures was performed using a Cy5-conjuated secondary (Jackson Immunoresearch; Xu et al., 2004). The nuclear marker DAPI (300 nM) was applied with the secondary antibodies. Signal was detected by epifluorescence microscopy (Nikon E800) and images acquired with a cooled CCD camera (Coolsnap HQ, Roper; Metamorph software, Universal Imaging).
BrdU labeling and birthdating
For in vivo labeling of S-phase donor cells, one injection of BrdU (100mg/kg, interperitoneally) was made 1 hour prior to sacrificing the dam. Following in vitro culture on cortical feeder cells, GFP+ donor cells were processed for triple-labeling immunofluorescence as described below.
For birthdating studies, injections of BrdU (50mg/kg, i.p.) were made every two hours times five. Twenty-five days after birth, animals for each age of injection (E12.5, E14.5, E16.5) were deeply anethsthetized and perfused with 4% paraformaldehyde (PFA) in PBS. Their brains were removed and postfixed in 4% PFA for four hours. Following cryoprotection in 30% sucrose and embedding in OCT (Tissue Tek), 10μm thick coronal sections were mounted onto glass slides and stored at -80°C.
For visualization of BrdU and SST, sections were first processed for somatostatin (rat anti-somatostatin; Chemicon; 1:400) followed by secondary immunofluorescence (Alexa 488-conjugated anti-rat or rabbit; Molecular Probes; 1:500). The slides were then fixed in 4% PFA for 30 minutes and processed for BrdU immunofluorescence by incubation in 50% formamide, 2xSSC at 55°C for 10 min followed by 2N HCl in PBS at 37°C for 15 min and then 0.1 M boric acid for 2 min. Sections were then rinsed, blocked in 5% fetal calf serum, and treated overnight with mouse anti-BrdU (Chemicon; 1:400). The following day, the slides were treated with secondary antibodies for both BrdU (Alexa 594-conjugated anti-mouse; Molecular Probes; 1:500) and for SST.
GeneChip Oligonucleotide Array
Sample preparation for GeneChip arrays
RNA samples from dorsal and ventral MGE were obtained as follows. Embryos were harvested from pregnant wildtype (non-GFP) dams at E13.5. Dorsal and ventral MGE tissue samples were obtained as described above (see also Figure 1), and placed on ice in RNAlater stabilization reagent (Qiagen, Chatsworth, CA). All dorsal/ventral MGE samples from one litter were pooled and the RNA extracted and purified using the RNeasy Mini Kit (Qiagen). This pooled RNA was then considered a single sample, with three separate litters used for the three samples used for the array hybridization. Prior to use in amplification studies, the size integrity profile of all RNA samples was determined with an Agilent Bioanalyzer. All RNA preparations provided (A260/280) ratios greater than 1.8. RNA concentration was determined by Nanodrop spectrophotometry.
RNA Amplification, Labeling and GeneChip Array Hybridization
2 μg total RNA was first reverse transcribed using a T7-Oligo(dT) Promoter Primer in the first-strand cDNA synthesis reaction. Following RNase H-mediated second-strand cDNA synthesis, the double-stranded cDNA is purified and serves as a template in the subsequent in vitro transcription (IVT) reaction. The IVT reaction is carried out in the presence of T7 RNA Polymerase and a biotinylated nucleotide analog/ribonucleotide mix for complementary RNA (cRNA) amplification and biotin labeling. The biotinylated cRNA targets are then cleaned up, fragmented, and hybridized to GeneChip array mouse genome 430 2.0. Hybridization conditions, array washing, staining (with GeneChip Fluidics Station 450), and scanning (with GeneChip Scanner 3000) were carried out according to the Affymetrix Expression Analysis technical Manual (Affymetrix, Santa Clara, CA).
Data collection and analysis
Affymetrix GeneChip Operating Software (GCOS) was used as the image acquisition software for the mouse 430 2.0 arrays. The signal, which represents the intensity of each gene, was extracted from the image. The target intensity value from each chip was scaled to 500. Data normalization, log transformation, statistical analysis and pattern study were performed with GeneSpring software (Silicon Genetics, Redwood City, CA).
Array data were globally normalized by using GeneSpring software. Firstly, all of the measurements on each chip were divided by the 50th percentile value (per-chip normalization). Secondly, each gene was normalized to the median value of the samples (per-gene normalization).
Statistical comparison between the dorsal and ventral MGE samples was performed using a Welch t-test with log-transformed data, with p<.05. There are totally 1483 genes scored as differentially expressed between two groups. To further enhance the stringency for reporting differentially expressed genes in table S1, only those genes scored at “present” in all three samples in which it showed higher expression levels were included. An exception was made in a few cases where two of three samples showing higher expression levels were scored as “present”, and none of three from the lower-expression samples were “present”.
Results
Neurochemically defined cortical interneuron subgroup identity is primarily specified within the MGE
In order to investigate the mechanisms underlying cortical interneuron fate specification, we first asked whether progenitor subgroup identity is established within the progenitor's place of origin (the MGE) or whether the eventual fate is influenced by instructive cues received en route to the cortex. Supporting the former hypothesis is the finding that the spatial and temporal origins of cortical interneurons is predictive of both their neurochemical (Xu et al., 2004), and their intrinsic physiological profile (Butt et al., 2005; Miyoshi et al., 2007), as differentiated neurons. Furthermore, while sonic hedgehog signaling is required for the maintenance of Nkx2.1 expression and subsequent specification of multiple cortical interneuron subgroups that are generated from within the MGE, deletion of the hedgehog effector protein smoothened in postmitotic interneuron progenitors did not significantly affect cortical interneuron specification, migration, differentiation, or survival (Xu et al., 2005). These findings further suggest that cortical interneuron fate specification, at least with regard to hedgehog signaling, occurs in the MGE, rather than by instructive cues received during migration to the cortex.
To further explore the hypothesis that neurochemical aspects of cortical interneuron fate are specified within the MGE, we transplanted MGE progenitors taken from E13.5 GFP-expressing embryos directly into the cortex of cold-anesthetized P2 neonates (Figure 1A-B). These pups were then sacrificed at P30 and the eventual fates of the GFP-expressing transplant cells were analyzed by double immunofluorescence labeling for GFP and various interneuron subgroup markers including SST, PV, CR, and NPY, a peptide that partially overlaps with SST expression (Gonchar and Burkhalter, 1997; Kubota and Kawaguchi, 1994). By P30, GFP+ neurons can be seen scattered throughout the cortex (Figure 1C), the vast majority of which exhibit interneuron-like, non-pyramidal morphologies. Substantial percentages of double labeling (for quantification, see Table 1) were seen with GFP and SST (Figure 1D), PV (Figure 1E), and NPY (Figure 1F). A smaller percentage of GFP+ cells co-labeled for CR (Figure 1G). As recent evidence has suggested that the vertically oriented CR+ cells with bipolar or bitufted CR+ dendritic morphologies primarily originate within the caudal ganglionic eminence (Butt et al., 2005; Lopez-Bendito et al., 2004; Xu et al., 2004), the GFP+/CR+ population from the MGE are likely to be largely composed of the SST+/CR+ multipolar population (Wang et al., 2004b; Xu et al., 2006). These findings are in agreement with a recent publication, in which grafts of MGE progenitors were shown to differentiate into GABAergic interneurons upon transplantation into P3-4 neonatal cortex (Alvarez-Dolado et al., 2006).
Table 1. Interneuron subgroup specification is not affected by transplant host environment.
| E13.5 → P2 Ctx in vivo |
E13.5 → E13.5 MGE in utero1 |
E14.5 → P0 Ctx in vitro2 |
||||
|---|---|---|---|---|---|---|
| % | Total Cells | % | Total Cells | % | Total Cellsa | |
| SST | 37.7 | 435 | 34.8 | 443 | 26.3 | 1399 |
| PV | 52.9 | 359 | 57.3 | 389 | 59.1 | 473 |
| NPY | 9.8 | 193 | 6.8 | 443 | N.S. | N.S |
| Calr | 9.3 | 216 | 3.5 | 578 | 0 | 515 |
N.S. – not studied
The data presented for the in vitro cultures are expressed as an average percentage of co-labeling across multiple cell cultures (each containing roughly 100-200 GFP+ neurons).
Together, these data are strikingly similar to those previously published by Butt et al. (2005) from their isochronic in utero transplants and by Xu et al (2004) using a cortical feeder co-culture system (for summary, see Table 1). The similarity in results from homotopic MGE transplants versus transplants directly into cortical environments in vitro or in vivo suggests, at least for the vertically oriented CR+ population versus those that express PV or SST, the potential to attain these interneuron subgroup identities is established in the MGE or CGE. Furthermore, this process does not require factors encountered during their migration into the cerebral cortex.
Distinct origins of PV and SST-expressing interneurons within the MGE
In addition to the data presented above, multiple lines of evidence suggest that PV+ and SST+ cortical interneurons originate primarily within the MGE where they are dependent on the transcription factor Nkx2.1 for their specification (for review see Wonders and Anderson, 2006). The important question remains as to how these subgroups become differentially specified. Our initial approach to this question is to determine whether, as in the spinal cord (Briscoe et al., 2001; Jessell, 2000; Liem et al., 1997; Muroyama et al., 2002; Zhuang and Sockanathan, 2005), there are distinct progenitor domains along the dorsal-ventral axis of the MGE. To test this possibility, progenitors were dissected from the proliferative zones of the dorsal or ventral MGE (as shown in Figure 2A), then transplanted directly into neonatal cortex (as in Figure 1). Like whole MGE transplants, both dorsal and ventral MGE transplants generate SST+, and PV+ interneurons similar to those shown in Figure 1D-E. However, dorsal MGE (dMGE) transplants showed roughly double the percentage of co-expression with SST than ventral MGE (vMGE) transplants (n=3; dMGE 63.24±4.61%, vMGE 31.21±2.98%; p=0.007). Conversely, vMGE transplants showed a bias towards co-expression with PV (n=3; dMGE 30.23±3.78%, vMGE 59.18±7.19%; p=0.04). These findings, summarized in Figure 2B, suggest the existence of a cell-fate bias within the MGE that preferentially directs dMGE progenitors towards SST-expressing and vMGE progenitors towards PV-expressing fates.
To further investigate this difference in fate potential, similar dorsal vs. ventral (d/v) MGE transplant experiments were performed using a previously described in vitro co-culture system (Xu et al., 2004; Xu et al., 2005). This system mimics the development of MGE progenitors in vivo and has the advantage of allowing for higher throughout and more experimental control. d/vMGE progenitors (Figure 2A) from E12.5 and E14.5 GFP-expressing embryos were transplanted onto primary cortical feeders prepared the previous day from P0 neonates (Xu et al., 2004). After 2-4 weeks in vitro, the fates of the d/vMGE neurons (GFP+) were examined by co-labeling with SST, NPY, or PV. Similar to the observations made in vivo, E14.5 dMGE transplants were more likely to generate SST interneurons compared to vMGE transplants (Figure 2C; n=3; p<0.005) whereas PV interneurons are primarily generated from ventral transplants (n=3; p<0.05). This effect was also observed in transplants from E12.5 transplants for both SST (n=6; p<0.0001) and PV (n=3; p<0.05). At both ages, NPY was primarily generated from dMGE (at E12.5, n=4; p<0.05; at E14.5, n=3; p<0.05), consistent with its partial overlap with the SST+ population.
Although a significant difference in fate potential was seen between the dorsal and ventral MGE, significant heterogeneity remained within the transplants. One possible explanation for this is that postmitotic interneuron progenitors from the vMGE migrate through the dMGE en route to the cortex, resulting dilution of the “dMGE-born” sample (Figure 3A). To identify transplanted cells that were in S-phase of the cell cycle immediately prior to transplantation, BrdU (100mg/kg) was injected into E12.5 dams 1-hour prior to sacrifice. Following 14 days in vitro, there was no significant difference in the percentage of BrdU expression by GFP+ neurons from dMGE or vMGE transplants (Figure 3F; n=3; p=0.17). In contrast, there was a 3.75-fold higher co-localization of SST with BrdU/GFP in dMGE cultures compared to vMGE cultures (Figure 3E; n=3; p=0.005). As previous studies have shown that tangentially migrating interneurons are postmitotic (Polleux et al., 2002; Xu et al., 2003; Xu et al., 2005) the finding that the percent co-labeling for SST within S-phase-labeled (BrdU+/GFP+) dMGE transplants was significantly higher than for the non BrdU-labeled dMGE transplant (69.93±2.15% vs. 43.70±2.71%; p<0.001) supports the hypothesis that postmitotic progenitors from the vMGE are contributing to the dMGE transplants. In sum, these data suggest that interneuron subgroups have distinct origins within the MGE, with the dMGE primarily generating SST+ interneurons and the vMGE generating PV+ interneurons.
Figure 3. Dorsal MGE cells labeled in S-phase 1 hr prior to transplantation in vitro show a strong bias towards SST+ interneuron generation.
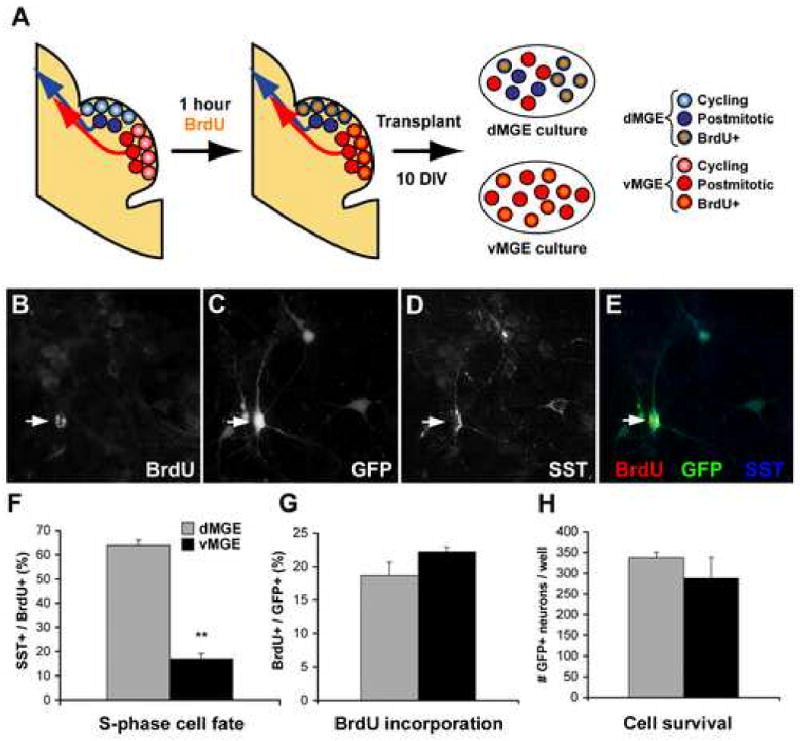
(A) Embryos are exposed to BrdU for 1 hour, allowing for the labeling of those cells in S-phase immediately prior to transplantation. This labeling makes it possible to separate “dMGE born” and “vMGE born” progenitors based on their expression of BrdU (orange). (B-E) Immunofluorescence labeling for BrdU (B), GFP (C), and SST (D) in transplant cultures grown for 10 DIV. (F) dMGE progenitors that had been in S-phase 1hr prior to transplant as labeled by an injection of BrdU (100 mg/kg) show a very pronounced bias towards SST interneuron generation compared to vMGE. (G-H) There is no significant difference in BrdU incorporation (% of cells that are BrdU+ at 10 DIV) or cell survival between dorsal and ventral MGE transplants.
Unpaired t-test: * p<0.05; ** p<0.005.
SST and PV interneurons in superficial or deeper cortical layers are born contemporaneously
While the evidence presented above suggests the existence of a spatial bias of SST and PV interneuron progenitors within the MGE, an alternative or additional mechanism for differentially specifying these subgroups is one in which SST+ and PV+ interneurons are born at different times. There is precedence for this hypothesis, as SST or calbindin and CR/vasoactive intestinal peptide (VIP)-expressing interneurons of a given layer appear to have different temporal origins (Cavanagh and Parnavelas, 1988; Cavanagh and Parnavelas, 1989; Yozu et al., 2004). However, to our knowledge no direct comparison of the birthdates of SST+ versus PV+ interneurons has been published in the mouse.
To determine the age at which SST and PV interneurons exit the cell cycle in vivo in the mouse, a cohort of proliferating cells were labeled with injections of BrdU in a paradigm designed to approximate the labeling of the entire cycling population at a given age (see methods). Injections were made at either E12.5, E14.5, or E16.5 to label the early, mid, and later phases of neurogenesis. Cryosections from these animals were then processed at P25 for immunofluorescence. Injections made at E12.5 labeled cells throughout cortical layers II-VI, with the strongest labeling being in the deeper cortical layers, presumably due to their exiting the cell cycle prior to diluting their BrdU content by subsequent divisions. Consistent with the well-established “inside-out” gradient of overall cortical neurogenesis (Angevine and Sidman, 1961; Caviness and Sidman, 1973), the E14.5 and E16.5 injections labeled increasingly superficial cells and fewer deep layer cells (data not shown; see also Xu et al., 2004).
To determine the birthdates of SST+ and PV+ interneurons, double immunofluorescence labeling was performed for BrdU and either PV (Figure 4A) or SST (Figure 4B), and the percentage of each interneuron subtype within the somatosensory cortex that incorporated BrdU at each time point was determined. As shown in Figure 4C, there was no significant difference between the percentages of either interneuron subgroup that incorporated BrdU at any of the three time points studied. This suggests that SST+ and PV+ interneuron progenitors of a given layer undergo their final S-phase at similar times during development, consistent with reports of the overall birthdates of GABAergic cortical neurons (Fairén et al., 1986; Peduzzi, 1988).
Figure 4. SST- and PV-expressing interneuron subgroups are generated contemporaneously during development.
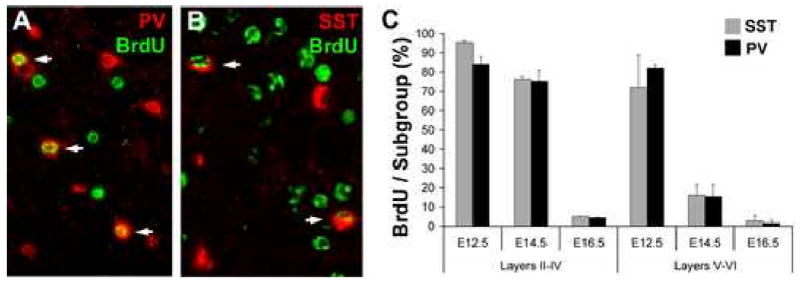
The cortex of P30 mice, exposed to BrdU at E12.5, E14.5, or E16.5 as previously published (Xu et al., 2004), was processed for immunofluorescence for BrdU and either parvalbumin (A; PV) or somatostatin (B; SST) with arrows indicating co-expression. (C) The percentages of SST/PV co-labeling with BrdU are not significantly different at any of the ages studied in either the superficial (Layers II-IV) or deep (Layers V-VI) cortex.
Differences in gene expression within the dorsal or ventral MGE
Together, these findings suggest that SST+ and PV+ interneurons are specified within different subregions of the MGE, which implies that there are differences in gene expression between the dorsal and ventral MGE. However, aside from the dependence of SST+ and PV+ interneurons on the expression of Nkx2.1 (Xu et al., 2004; Xu et al., 2005) and Lhx6 (V. Pachnis, personal communication; Du and Anderson, unpublished data), little is known about the genetic specification of those subgroups. To address this question, we analyzed the gene expression profiles of dorsal and ventral MGE progenitors using oligonucleotide microarrays. After establishing a twofold change cutoff and P value < .05, between the two populations (see methods), and we find a total of 687 probes (331 in dMGE, 296 in vMGE) that appear preferentially expressed in one population relative to the other. This number has been further refined by only excepting transcript identifications in which all three samples in the enriched source read as “present” (2 or 3 read as present were accepted in a few cases where 0/3 were present in the lower-level sample). Based on these criterion, a complete list of identified genes is presented in Table S1 (48 enriched in dMGE and 59 vMGE).
A considerable number of the identified genes in Table S1 have been confirmed or suggested (depending on quality of the coronal section and in situ signal) as differentially expressed on the dorsal-ventral axis of the MGE based on ours and previous studies. Interestingly, a number of dorsally enriched genes are either downstream effectors of (Gli1, Gli2, Hhip1), or are transcriptional targets of (Nkx6.2), the sonic hedgehog signaling pathway (Figure 5 and (Flames et al., 2007; Loulier et al., 2005; Stenman et al., 2003; Tole et al., 2000; Xu et al., 2005). In fact, Gli2, Nkx6.2, and Hhip1 are 3 of the top 5 “hits” for the dorsally enriched genes.
Figure 5. An array based approach to identify genes enriched in the dorsal or ventral MGE.
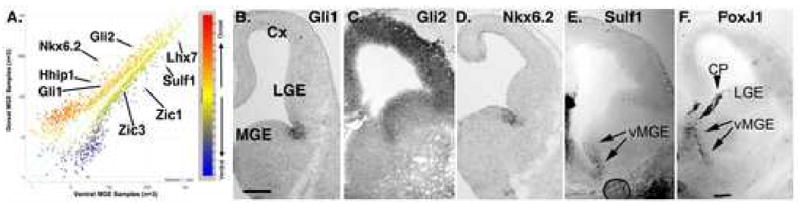
(A) The plot shows results averaged from 3 separately prepared, non-amplified RNA samples of dorsal or ventral MGE (dMGE, vMGE), run on the Affimatrix 430 2.0 array. Distance from the origin reflects strength of signal. Distance from the diagonal line is proportion to relative difference between dorsal and ventral signals for that RNA such that RNAs above the line are enriched in the dMGE, whereas RNAs below the line are enriched in the vMGE. (B-D) Several of the genes identified as enriched in the dMGE are known to be selectively expressed. Gli1 and Nkx6.2 expression in the dMGE have been shown to remain dependent on Shh signaling after initial patterning has been established (Xu et al., 2005). (E) Hedgehog Interacting Protein 1 (Hhip1) is also expressed in the dMGE along with another Shh signaling inhibitor, patched (Loulier et al., 2005). Hhip1 acts as an antagonist of Hedgehog signaling (Chuang et al., 2003), although it could conceivably also act to concentrate Hh proteins into a defined region where net Hh signaling could be enhanced (Saha and Schaffer, 2006).
vMGE-enriched genes that have been confirmed by ours or previous studies include sulfatase 1, sulfatase 2, brevican, and FoxJ1 (Figure 5), as well as Zic1 and Zic3 (Inoue et al., 2007), Lhx7/8 (Flames et al., 2007). In sum, these results suggest that a considerable level of molecular heterogeneity exists on the dorsal-ventral axis of the MGE, and correlates with the bias for generation of different cortical interneuron subgroups from these regions.
Discussion
Previous work has demonstrated that cortical interneurons expressing PV and SST primarily originate within the MGE (Butt et al., 2005; Valcanis and Tan, 2003; Wichterle et al., 2001; Xu et al., 2004). Additionally, both interneuron subgroups require the expression of Nkx2.1 for their specification (Xu et al., 2004; Xu et al., 2005). However, it has been unclear whether the potential to express PV or SST during postnatal development is imposed on MGE-derived interneurons during their genesis in the MGE, during migration to the cortex, or during post-migratory development. Here we present evidence that different progenitor domains exist within the MGE along the dorsal/ventral axis, with the dorsal MGE primarily generating SST+ interneurons and the ventral MGE primarily generating PV+ interneurons. These results suggest that aspects of interneuron fate determination are established at their cites of origin in the MGE.
A bias for the generation of interneuron subgroups within the dorsal-ventral axis of the MGE
Similar to those of recently published study (Flames et al., 2007), our results suggest that relative to eachother, vMGE progenitors are about twice as likely to become PV+ interneurons, and dMGE progenitors are about twice as likely to become SST+ interneurons (Figure 2). Moreover, when only those cells that were in s-phase shortly before transplantation are considered, thus excluding migratory vMGE cells transiting dMGE subventricular zone, the dorsal bias for generating SST+ cells grows to 4:1 (Figure 3). So why should these seemingly arbitrary neurochemical markers display tendencies for distict origins within the MGE? The calcium binding protein PV and the neuropeptide SST are expressed in non-overlapping subgroups of cortical and striatal interneurons that together comprise about 65% of cortical interneurons in mice (Gonchar and Burkhalter, 1997; Kubota and Kawaguchi, 1994; Tamamaki et al., 2003). Both subgroups are composed of a number of distinct interneuron types, where an interneuron type is defined by distinctive combinations of physiological, neurochemical, morphological, and connectivity-based characteristics. While it may be surprising that the PV versus SST neurochemical features should have largely distinct origins, these subgroups also tend to feature other differences. For example, the PV-expressing subgroup includes large basket cells, nest basket cells, and chandelier cells. Both the large basket and the chandelier cells express the potassium channel Kv3.1 that contributes to the “fast spiking” characteristic of these cells (Weiser et al., 1995). The SST-expressing subgroup neither generally expresses this channel nor generally has fast-spiking characteristics. In addition, the connectivity of the PV-expressing subgroup tends to include innervation of the cell soma or axon initial segment of pyramidal neurons (DeFelipe, 1997). In contrast, the SST-expressing subgroup tends to target distal dendrites (Katona et al., 1999)(but see also (Gonchar et al., 2002)). Thus, the seemingly arbitrary PV versus SST characteristic also generally correlates with differences in interneuron physiology and connectivity. Since the transplantation studies are suggestive that an incomplete bias, or gradient of interneuron fates are generated on the dorsal-ventral axis of the MGE, future studies will be needed to determine whether types of interneurons within each group, that either do or do not share the group's connectivity or physiological tendencies, also are biased in terms of their origins. For example, it may be that PV is a less-specific characteristic of ventral MGE interneurons than the tendency to innervate the cell soma, or the tendency to have a fast-spiking physiology.
In addition to the question of whether PV versus SST expression per se is the most parsimonious or relevant way to distinguish most MGE-derived interneurons, a caveat to the data presented on BrdU birthdating of these subgroups deserves mention. Although the data suggest that no gross difference exists in the time of generation of PV versus SST-expressing interneurons within a given cortical layer (Figure 4), the resolution of this experiment is inadequate to determine whether subtypes of SST-expressing interneurons from distinct subcortical regions have distinct temoral origins. In fact, recent evidence suggests that within these subgroups, physiologically- (and possibly connectivity-) defined subtypes are generated in a temporal sequence (Miyoshi et al., 2007).
It also should be noted that the MGE gives rise to multiple types of neurons in addition to the SST and PV interneuron subgroups of the cerebral cortex, such as the same neurochemically-defined subgroups in the striatum (Kubota and Kawaguchi, 1994; Marin et al., 2000), and PV+, GABAergic projection neurons of the globus pallidus (Xu et al., in press). Evidence that striatal and cortical interneurons can be lineage-related (Reid and Walsh, 2002) suggests that the dorsal-ventral bias for SST versus PV+ interneurons applies to the far less numerous striatal population in addition to the cortical population. It is more difficult to assess whether displaced PV+ pallidal projection neurons are strongly represented among the cortical transplants and whether these would have the same bias towards vMGE generation. However, a small minority of PV+ neurons from these transplants are heavily spiny (Welagen and Anderson, unpublished data), a prominent characteristic of PV+ pallidal projection neurons, suggesting that their survival through the transplantation procedure or in postnatal cortex may be compromised.
Microdomains of gene expression along the dorsal-ventral axis of the MGE
Although as measured by PV versus SST interneuron subgroup generation in the MGE is better described as a bias or gradient than as generation within discreet domains, the presence of this bias in generation of interneuron subgroups has important implications for the fate determination of these cells. First, it suggests that the potential to achieve the PV or SST-expressing fate is specified at the cell's origins in the MGE, rather than during later stages of migration or post-migratory differentiation. Together with the evidence that whole-MGE transplants results in similar ratios of PV, SST, and CR-expressing cells regardless of being transplanted homotopically in vivo, heterotopically and heterochronically into the neonatal cortical plate in vivo, or onto a feeder layer made of dissociated neonatal cortex, these results suggest that key decisions for interneuron fate are probably being made during the final cell cycle such that later cues are primarily permissive rather than directive of achieving a particular fate (Figure 1). This idea is further supported by our previous finding that cycling MGE progenitors lose Nkx2.1 expression and their ability to generate PV or SST-expressing interneurons in response to acute reductions of Shh signaling (Xu et al., 2005).
An important implication of this notion is that cell based therapy of seizures disorders or other neuropsychiatric conditions using MGE-derived progenitors, or embryonic stem cells directed into MGE-like progenitors, should have predictable fate potential based on the source/molecular profile at the time of transplantation. Of course, cortical factors will still be required for interneuron survival, migration, and connectivity within the cortex (Marty et al., 1997; Trinh et al., 2006).
A second key implication of this bias in interneuron subgroup origin is that molecular differences should exist in the progenitor zone that contribute to this bias. Indeed, a number of genes, including Nkx6.2, Gli1, Gli2, and Hhip1, are enriched within the sulcus region of the dorsal MGE. To further establish the presence of molecular differences between the dorsal and ventral MGE, we conducted a microarray experiment on RNA samples from E13.5 MGEs. All the of above genes were identified in this array as significantly enriched in the dorsal MGE, along with about 100 others that appeared to be enriched in either the dorsal or ventral domain (Tables S1). Of the ventrally enriched genes, the transcription factor FoxJ1, that functions in cell polarization and cilia formation (Huang et al., 2003; Zhang et al., 2004), and sulfatase 1, whose activity can alter heparin sulfate proteoglycans such that the morphogen Shh becomes concentrated (Danesin et al., 2006), have been confirmed by in situ hybridization. Other interesting “hits” in the array include Zic1 and Zic3, cell fate-influencing transcription factors that in addition to the array data have recently been shown to be highly enriched in the vMGE (Inoue et al., 2007). Although differential expression of the large majority of array-identified genes has not yet been confirmed, the fact that nearly all of the handful of transcripts known to be differentially expressed in the MGE are indeed identified by the array strongly suggests that many of the unconfirmed transcripts, that include potentially fate-regulating proteins, are correctly identified. A causal link between any of these genes and interneuron fate determination remains to be determined.
Paradoxical enhancement of Shh signaling with the dorsal-most MGE
The presence of Gli1, Gli2, and Hhip1 suggest that Shh signaling is relatively enhanced in the dorsal-most region of the MGE, and this signaling appears to be required for the maintenance of Nkx6.2 expression (Xu et al., 2005). In addition, although loss of Shh signaling during the period of neurogenesis greatly reduced the expression of Nkx2.1 in S-phase cells of the MGE, the effect is most pronounced in the dorsal MGE (Xu et al, 2005; the same study showed a more pronounced loss of SST+ than PV+ interneurons in postnatal cortex of NestinCre:Shhfl/fl mice).
However, studies of Shh mRNA expression show highest levels of detection in the proliferative zone of the preoptic region that underlies the MGE, and in the mantle region of the MGE. Thus, if any SHH protein gradient existed, it would be expected to be ventral high-dorsal low as in the spinal cord. This paradox raises the possibility that SHH concentration is substantially distorted by variable diffusion within the extracellular matrix, and that another signaling pathway, such as that of fibroblast growth factors (FgFs), might be relatively more important for ventral MGE expression of interneuron-fate determining genes (Gutin et al., 2006).
In sum, we provide evidence for genetic microdomains in the MGE that correlate with biases in the generation of two major subgroups of cortical interneurons. These data suggest that, together with the fate-related domains of the lateral ganglionic eminence, to a significant extent cell fate in ventral telencephalon is spatially regulated similar to that seen in the spinal cord. While we would expect that, as also holds for spinal cord, important aspects of interneuron fate diversity will be also generated by temporal regulation (Miyoshi et al., 2007) and by Notch-delta mediated fate sequestration (Mizuguchi et al., 2006; Mizutani et al., 2007), the spatial segregation of specified progenitors should enhance efforts to identify fate-determining factors of cortical interneuron subtypes.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez-Dolado M, Calcagnotto ME, Karkar KM, Southwell DG, Jones-Davis DM, Estrada RC, Rubenstein JL, Alvarez-Buylla A, Baraban SC. Cortical inhibition modified by embryonic neural precursors grafted into the postnatal brain. J Neurosci. 2006;26:7380–9. doi: 10.1523/JNEUROSCI.1540-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SA, Marin O, Horn C, Jennings K, Rubenstein JL. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development. 2001;128:353–63. doi: 10.1242/dev.128.3.353. [DOI] [PubMed] [Google Scholar]
- Angevine JB, Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Chen Y, Jessell TM, Struhl G. A hedgehog-insensitive form of patched provides evidence for direct long-range morphogen activity of sonic hedgehog in the neural tube. Mol Cell. 2001;7:1279–91. doi: 10.1016/s1097-2765(01)00271-4. [DOI] [PubMed] [Google Scholar]
- Butt SJ, Fuccillo M, Nery S, Noctor S, Kriegstein A, Corbin JG, Fishell G. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 2005;48:591–604. doi: 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- Cavanagh ME, Parnavelas JG. Development of somatostatin immunoreactive neurons in the rat occipital cortex: a combined immunocytochemical-autoradiographic study. J Comp Neurol. 1988;268:1–12. doi: 10.1002/cne.902680102. [DOI] [PubMed] [Google Scholar]
- Cavanagh ME, Parnavelas JG. Development of vasoactive-intestinal-polypeptide-immunoreactive neurons in the rat occipital cortex: a combined immunohistochemical-autoradiographic study. J Comp Neurol. 1989;284:637–45. doi: 10.1002/cne.902840410. [DOI] [PubMed] [Google Scholar]
- Caviness VS, Jr, Sidman RL. Time of origin or corresponding cell classes in the cerebral cortex of normal and reeler mutant mice: an autoradiographic analysis. J Comp Neurol. 1973;148:141–51. doi: 10.1002/cne.901480202. [DOI] [PubMed] [Google Scholar]
- Chuang PT, Kawcak T, McMahon AP. Feedback control of mammalian Hedgehog signaling by the Hedgehog-binding protein, Hip1, modulates Fgf signaling during branching morphogenesis of the lung. Genes Dev. 2003;17:342–7. doi: 10.1101/gad.1026303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin JG, Nery S, Fishell G. Telencephalic cells take a tangent: non-radial migration in the mammalian forebrain. Nat Neurosci. 2001;4 1:1177–82. doi: 10.1038/nn749. [DOI] [PubMed] [Google Scholar]
- Danesin C, Agius E, Escalas N, Ai X, Emerson C, Cochard P, Soula C. Ventral neural progenitors switch toward an oligodendroglial fate in response to increased Sonic hedgehog (Shh) activity: involvement of Sulfatase 1 in modulating Shh signaling in the ventral spinal cord. J Neurosci. 2006;26:5037–48. doi: 10.1523/JNEUROSCI.0715-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFelipe J. Types of neurons, synaptic connections and chemical characteristics of cells immunoreactive for calbindin-D28K, parvalbumin and calretinin in the neocortex. J Chem Neuroanat. 1997;14:1–19. doi: 10.1016/s0891-0618(97)10013-8. [DOI] [PubMed] [Google Scholar]
- Fairén A, Cobas A, Fonseca M. Times of generation of glutamic acid decarboxylase immunoreactive neurons in mouse somatosensory cortex. Journal of Comparative Neurology. 1986;251:67–83. doi: 10.1002/cne.902510105. [DOI] [PubMed] [Google Scholar]
- Flames N, Pla R, Gelman DM, Rubenstein JL, Puelles L, Marin O. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007;27:9682–95. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonchar Y, Burkhalter A. Three distinct families of GABAergic neurons in rat visual cortex. Cerebral Cortex. 1997;7:347–58. doi: 10.1093/cercor/7.4.347. [DOI] [PubMed] [Google Scholar]
- Gonchar Y, Turney S, Price JL, Burkhalter A. Axo-axonic synapses formed by somatostatin-expressing GABAergic neurons in rat and monkey visual cortex. J Comp Neurol. 2002;443:1–14. doi: 10.1002/cne.1425. [DOI] [PubMed] [Google Scholar]
- Gutin G, Fernandes M, Palazzolo L, Paek H, Yu K, Ornitz DM, McConnell SK, Hebert JM. FGF signalling generates ventral telencephalic cells independently of SHH. Development. 2006;133:2937–46. doi: 10.1242/dev.02465. [DOI] [PubMed] [Google Scholar]
- Hensch TK. Critical period plasticity in local cortical circuits. Nat Rev Neurosci. 2005;6:877–88. doi: 10.1038/nrn1787. [DOI] [PubMed] [Google Scholar]
- Huang T, You Y, Spoor MS, Richer EJ, Kudva VV, Paige RC, Seiler MP, Liebler JM, Zabner J, Plopper CG, Brody SL. Foxj1 is required for apical localization of ezrin in airway epithelial cells. J Cell Sci. 2003;116:4935–45. doi: 10.1242/jcs.00830. [DOI] [PubMed] [Google Scholar]
- Inoue T, Ota M, Ogawa M, Mikoshiba K, Aruga J. Zic1 and Zic3 regulate medial forebrain development through expansion of neuronal progenitors. J Neurosci. 2007;27:5461–73. doi: 10.1523/JNEUROSCI.4046-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–9. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Katona I, Acsady L, Freund TF. Postsynaptic targets of somatostatin-immunoreactive interneurons in the rat hippocampus. Neuroscience. 1999;88:37–55. doi: 10.1016/s0306-4522(98)00302-9. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kubota Y. GABAergic cell subtypes and their synaptic connections in rat frontal cortex. Cereb Cortex. 1997;7:476–86. doi: 10.1093/cercor/7.6.476. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Kawaguchi Y. Three classes of GABAergic interneurons in neocortex and neostriatum. Japanese Journal of Physiology. 1994;44 2:S145–8. [PubMed] [Google Scholar]
- Liem KFJ, Tremml G, Jessell TM. A role for the roof plate and its resident TGFbeta-related proteins in neuronal patterning in the dorsal spinal cord. Cell. 1997;91:127–138. doi: 10.1016/s0092-8674(01)80015-5. [DOI] [PubMed] [Google Scholar]
- Lopez-Bendito G, Sturgess K, Erdelyi F, Szabo G, Molnar Z, Paulsen O. Preferential origin and layer destination of GAD65-GFP cortical interneurons. Cereb Cortex. 2004;14:1122–33. doi: 10.1093/cercor/bhh072. [DOI] [PubMed] [Google Scholar]
- Loulier K, Ruat M, Traiffort E. Analysis of hedgehog interacting protein in the brain and its expression in nitric oxide synthase-positive cells. Neuroreport. 2005;16:1959–62. doi: 10.1097/01.wnr.0000187632.91375.81. [DOI] [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. Journal of Neuroscience. 2000;20:6063–76. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Rubenstein JL. A long, remarkable journey: Tangential migration in the telencephalon. Nature Reviews Neuroscience. 2001;2:780–790. doi: 10.1038/35097509. [DOI] [PubMed] [Google Scholar]
- Marty S, Berzaghi Mda P, Berninger B. Neurotrophins and activity-dependent plasticity of cortical interneurons. Trends in Neurosciences. 1997;20:198–202. doi: 10.1016/s0166-2236(96)01026-0. [DOI] [PubMed] [Google Scholar]
- Miyoshi G, Butt SJ, Takebayashi H, Fishell G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J Neurosci. 2007;27:7786–98. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi R, Kriks S, Cordes R, Gossler A, Ma Q, Goulding M. Ascl1 and Gsh1/2 control inhibitory and excitatory cell fate in spinal sensory interneurons. Nat Neurosci. 2006;9:770–8. doi: 10.1038/nn1706. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–5. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Muroyama Y, Fujihara M, Ikeya M, Kondoh H, Takada S. Wnt signaling pays an essential role in neuronal specification of the dorsal spinal cord. Genes Dev. 2002;16:548–553. doi: 10.1101/gad.937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens DF, Kriegstein AR. Is there more to GABA than synaptic inhibition? Nat Rev Neurosci. 2002;3:715–27. doi: 10.1038/nrn919. [DOI] [PubMed] [Google Scholar]
- Peduzzi JD. Genesis of GABA-immunoreactive neurons in the ferret visual cortex. J Neurosci. 1988;8:920–31. doi: 10.1523/JNEUROSCI.08-03-00920.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polleux F, Whitford KL, Dijkhuizen PA, Vitalis T, Ghosh A. Control of cortical interneuron migration by neurotrophins and PI3-kinase signaling. Development. 2002;129:3147–60. doi: 10.1242/dev.129.13.3147. [DOI] [PubMed] [Google Scholar]
- Reid CB, Walsh CA. Evidence of common progenitors and patterns of dispersion in rat striatum and cerebral cortex. J Neurosci. 2002;22:4002–14. doi: 10.1523/JNEUROSCI.22-10-04002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha K, Schaffer DV. Signal dynamics in Sonic hedgehog tissue patterning. Development. 2006;133:889–900. doi: 10.1242/dev.02254. [DOI] [PubMed] [Google Scholar]
- Stenman JM, Wang B, Campbell K. Tlx controls proliferation and patterning of lateral telencephalic progenitor domains. J Neurosci. 2003;23:10568–76. doi: 10.1523/JNEUROSCI.23-33-10568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamaki N, Yanagawa Y, Tomioka R, Miyazaki J, Obata K, Kaneko T. Green Fluorescent Protein Expression and Colocalization with Calretinin, Parvalbumin, and Somatostatin in the GAD67-GFP Knock-In Mouse. Journal of Comparative Neurology. 2003;467:60–79. doi: 10.1002/cne.10905. [DOI] [PubMed] [Google Scholar]
- Tole S, Ragsdale CW, Grove EA. Dorsoventral patterning of the telencephalon is disrupted in the mouse mutant extra-toes(J) Dev Biol. 2000;217:254–65. doi: 10.1006/dbio.1999.9509. [DOI] [PubMed] [Google Scholar]
- Trinh HH, Reid J, Shin E, Liapi A, Parnavelas JG, Nadarajah B. Secreted factors from ventral telencephalon induce the differentiation of GABAergic neurons in cortical cultures. Eur J Neurosci. 2006;24:2967–77. doi: 10.1111/j.1460-9568.2006.05194.x. [DOI] [PubMed] [Google Scholar]
- Valcanis H, Tan SS. Layer Specification of Transplanted Interneurons in Developing Mouse Neocortex. Journal of Neuroscience. 2003;23:5113–5122. doi: 10.1523/JNEUROSCI.23-12-05113.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Tegner J, Constantinidis C, Goldman-Rakic PS. Division of labor among distinct subtypes of inhibitory neurons in a cortical microcircuit of working memory. Proc Natl Acad Sci U S A. 2004a;101:1368–73. doi: 10.1073/pnas.0305337101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Toledo-Rodriguez M, Gupta A, Wu C, Silberberg G, Luo J, Markram H. Anatomical, physiological and molecular properties of Martinotti cells in the somatosensory cortex of the juvenile rat. J Physiol. 2004b;561:65–90. doi: 10.1113/jphysiol.2004.073353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser M, Bueno E, Sekirnjak C, Martone ME, Baker H, Hillman D, Chen S, Thornhill W, Ellisman M, Rudy B. The potassium channel subunit KV3.1b is localized to somatic and axonal membranes of specific populations of CNS neurons. J Neurosci. 1995;15:4298–314. doi: 10.1523/JNEUROSCI.15-06-04298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington MA, Traub RD. Interneuron diversity series: inhibitory interneurons and network oscillations in vitro. Trends Neurosci. 2003;26:676–82. doi: 10.1016/j.tins.2003.09.016. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Turnbull DH, Nery S, Fishell G, Alvarez-Buylla A. In utero fate mapping reveals distinct migratory pathways and fates of neurons born in the mammalian basal forebrain. Development. 2001;128:3759–71. doi: 10.1242/dev.128.19.3759. [DOI] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006 doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- Xu Q, Cobos I, De La Cruz E, Rubenstein JL, Anderson SA. Origins of cortical interneuron subtypes. J Neurosci. 2004;24:2612–22. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, De La Cruz E, Anderson SA. Cortical interneuron fate determination: diverse sources for distinct subtypes? Cereb Cortex. 2003;13:670–6. doi: 10.1093/cercor/13.6.670. [DOI] [PubMed] [Google Scholar]
- Xu Q, Tam M, Anderson SA. Fate-mapping Nkx2.1-lineage cells in the mouse telencephalon. Journal of Comparative Neurology. doi: 10.1002/cne.21529. in press. [DOI] [PubMed] [Google Scholar]
- Xu Q, Wonders CP, Anderson SA. Sonic hedgehog maintains the identity of cortical interneuron progenitors in the ventral telencephalon. Development. 2005;132:4987–98. doi: 10.1242/dev.02090. [DOI] [PubMed] [Google Scholar]
- Xu X, Roby KD, Callaway EM. Mouse cortical inhibitory neuron type that coexpresses somatostatin and calretinin. J Comp Neurol. 2006;499:144–60. doi: 10.1002/cne.21101. [DOI] [PubMed] [Google Scholar]
- Yozu M, Tabata H, Nakajima K. Birth-date dependent alignment of GABAergic neurons occurs in a different pattern from that of non-GABAergic neurons in the developing mouse visual cortex. Neurosci Res. 2004;49:395–403. doi: 10.1016/j.neures.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Zhang M, Bolfing MF, Knowles HJ, Karnes H, Hackett BP. Foxj1 regulates asymmetric gene expression during left-right axis patterning in mice. Biochem Biophys Res Commun. 2004;324:1413–20. doi: 10.1016/j.bbrc.2004.09.207. [DOI] [PubMed] [Google Scholar]
- Zhuang B, Sockanathan S. Dorsal-ventral patterning: a view from the top. Curr Opin Neurobiol. 2005;15:1–5. doi: 10.1016/j.conb.2005.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


