Abstract
If breast cancers arise independently in each breast the odds ratio (OR) for bilateral breast cancer for carriers of CHEK2*1100delC should be ~5.5, the square of the reported OR for a first primary (OR, 2.34). In the subset of bilateral cases with one or more affected relatives, the predicted carrier OR should be ~9. We have tested these predictions in a pooled set of 1,828 cases with 2 primaries and 7,030 controls from 8 studies. The second primary OR for CHEK2*1100delC carriers was 6.43 (95% confidence interval, 4.33-9.56; P < 0.0001), significantly greater than the published estimate for a first primary (P < 0.001) but consistent with its square. The predicted increase in carrier OR with increasing numbers of affected relatives was seen using bilateral cases from the UK (Ptrend = 0.0003) and Finland (Ptrend = 0.37), although not using those from the Netherlands and Russia (P = 0.001 for heterogeneity between countries). Based on a standard genetic model, we predict lifetime risks for CHEK2*1100delC carrier and noncarrier daughters of bilateral breast cancer cases of 37% and 18%, respectively. Our results imply that clinical management of the daughter of a woman with bilateral breast cancer should depend on her CHEK2*1100delC carrier status. This and other moderate penetrance breast cancer susceptibility alleles, together with family history data, will thus identify increasing numbers of women at potentially very high risk. Before such predictions are accepted by clinical geneticists, however, further population-based evidence is needed on the effect of CHEK2*1100delC and other moderate penetrance alleles in women with a family history of breast cancer.
Introduction
The average lifetime breast cancer risk in a typical Western woman is ~10%. Individual risks probably range from <2% to >50% (1), but apart from carriers of BRCA1 or BRCA2 mutations, women at very high risk cannot yet be identified by genetic testing alone. This very wide variation in genetic risk in the general population is predicted by a model in which a large number of “moderate or low penetrance” (2) alleles act in combination to confer high risks in women who carry large numbers of such alleles, and several such alleles have recently been discovered in candidate gene (3-7) and genome-wide (8-11) studies. An important implication of this polygenic model is that a single moderate-penetrance allele such as CHEK2*1100delC that doubles the risk in women with no family history is also likely to double the substantially higher risk in women with affected relatives. Predicted personal risks based only on family history rarely reach the threshold at which prophylactic treatment would usually be considered (~10% by age 50 or ~30% lifetime risk),16 but combining information on carrier status for moderate and low-penetrance alleles and family history may substantially increase the number of women seen in genetics clinics whose predicted risk reaches this level. Women with bilateral breast cancer are themselves at high genetic risk (12) and the lifetime risk among their female first-degree relatives is ~20%. We have analyzed the prevalence of CHEK2*1100delC in 1828 bilateral breast cancer cases in relation to family history to compare observed and predicted carrier odds ratios (OR). This comparison also constitutes a test of the polygenic model’s predictions of lifetime risk for carriers of CHEK2*1100delC with and without a first degree relative with bilateral breast cancer.
Materials and Methods
Full details of ascertainment of cases and controls for each of the studies have been published previously (3, 4, 13-22). A summary is given in Supplementary Table S1. All of the studies include predominantly, or exclusively, White Northern European subjects. All subjects gave written informed consent, and all studies were approved by the appropriate ethics committee or local institutional review board.
Genotyping methods in each study are described elsewhere (3, 13-16). Study-specific bilateral ORs and exact 95% confidence intervals (95% CI) were calculated using standard methods. Trends in OR for family history and age were calculated among cases, ignoring controls. The pooled OR was estimated by logistic regression with study as a stratifying covariate. Heterogeneity between studies was tested using likelihood ratio tests to compare logistic regression models with and without genotype-stratum interaction terms. Statistical analyses were carried out using Stata statistical software version 9.0 (Stata Corporation).
Lifetime breast cancer risks in the unaffected daughter of a bilateral case were derived from the Breast and Ovarian Analysis of Disease Incidence and Carrier Estimation Algorithm breast cancer model (23), which incorporates BRCA1 and BRCA2 mutations with a polygenic background, and has been calibrated against pooled population-based data on familial risks from several sources (24). We assumed that both cancers in the bilateral mother were diagnosed at age 50 y and that the status of all other female relatives was unknown. The model predictions thus represent the risk to the average 40-year-old daughter of a bilateral breast cancer case over all possible family histories (including both genetic and nongenetic familial factors), but the predictions are not strongly dependent on either the age at diagnosis of the index case or the presence of additional unaffected female relatives (Supplementary Table S2). Lifetime predicted risks in an unaffected 40-year-old daughter were calculated in relation to the daughter’s carrier status for BRCA1, BRCA2, and CHEK2*1100delC. The risk in CHEK2*1100delC carriers was calculated by multiplying predicted incidence rates at each age by 2.34, the OR estimate derived from pooled data on 10,860 breast cancers and 9,065 controls (15). Ideally, the risk for carriers of CHEK2*1100delC should be based on a model in which the polygenic variance is the residual variance after taking into account the effect of CHEK2*1100delC. The contribution of CHEK2*1100delC to the polygenic variance, however, is predicted to be <1%, and such an adjustment would not, therefore, affect the lifetime predicted risks.
Results
Eight studies from five Northern European countries (UK, Finland, the Netherlands, Germany, and Russia) contributed data to these analyses. The pooled OR estimate from these studies is 6.43 (95% CI, 4.33-9.56; P < 0.0001; heterogeneity χ2 = 10.25 (degrees of freedom, 6); P = 0.11; Fig. 1). The OR increases with each additional affected first-degree relative in the British and Finnish studies (OR per relative, 2.39; 95% CI, 1.53-3.73; Ptrend = 0.0003 for British studies; OR per relative, 1.51; 95% CI, 0.62-3.66; Ptrend = 0.37 for HEBCS; Table 1). There is, however, no consistent trend with family history for the other studies (Rotterdam, ABCS, and St. Petersburg) for which data on affected relatives were available (P = 0.001 for between-study heterogeneity). There is no significant trend with increasing age at first diagnosis in any study or overall, although the pooled OR per decade (1.00; 95% CI 0.75-1.31) is consistent with the modest reduction seen in older unselected cases (15).
Figure 1.
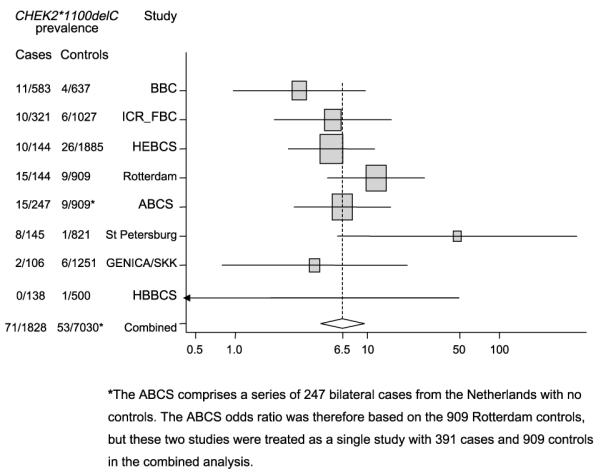
Estimated ORs with 95% CIs for the breast cancer ORs associated with CHEK2*1100delC. The area of each square is proportional to the variance of the log OR. The Hannover Breast Cancer study, where there were no carriers among cases is represented as a line with no square as the log OR and its variance cannot be calculated. An approximate OR and 95% CI were derived for Supplementary Table S1.
Table 1. Proportion of carriers of CHEK2*1100delC in women with bilateral breast cancer by number of affected first-degree relatives and by age group.
| United Kingdom |
Finland |
the Netherlands |
Russia |
Germany |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BBC |
ICR_FBC |
Pooled OR |
HEBCS |
Rotterdam |
ABCS |
Pooled OR |
St. Petersburg |
GENICA/SKK |
||||
| n/N (%) | n/N (%) | OR (95% CI) | n/N (%) | OR (95% CI) | n/N (%) | n/N (%) | OR (95% CI) | n/N (%) | OR (95% CI) | n/N (%) | OR (95% CI) | |
| Controls | 4/637 (0.63) | 6/1,027 (0.58) | 1.00 (Ref) | 26/1,885 (1.38) | 1.00 (Ref) | 9/909 (0.99) | 1.00 (Ref) | 1/821 (0.12) | 1.00 (Ref) | 6/1,251 (0.48) | 1.00 (Ref) | |
| All bilaterals | 11/583 (1.89) | 10/321 (3.12) | 3.93 (1.76-9.39) | 10/144 (6.94) | 5.34 (2.2-11.7) | 15/144 (10.42) | 15/247 (6.07) | 8.31 (3.79-20.1) | 8/145 (5.16) | 47.88 (6.30-2,126.8) | 2/106 (1.87) | 3.99 (0.4-22.6) |
| Family history | ||||||||||||
| Bilaterals 0 FDR | 4/400 (1.00) | 1/96 (1.04) | 1.68 (0.45-5.44) | 2/55 (3.64) | 2.70 (0.30-11.3) | 7/59 (11.86) | 10/109 (9.17) | 11.26 (4.63-29.1) | 8/114 (7.02) | 47.81 (9.30-464.3) | N/K | — |
| Bilaterals 1 FDR | 5/158 (3.16) | 3/153 (1.96) | 4.37 (1.48-12.4) | 7/78 (8.97) | 7.05 (2.49-17.4) | 5/58 (8.62) | 1/53 (1.89) | 5.71 (1.63-18.3) | 0/28 (0) | — | N/K | — |
| Bilaterals 2+ FDR | 2/25 (8.00) | 6/72 (8.33) | 10.91 (3.17-33.9) | 1/11 (9.09) | 7.15 (0.16-53.5) | 3/27 (11.11) | 0/19 (0) | 6.98 (1.17-29.2) | — | — | N/K | — |
| Age at first diagnosis | ||||||||||||
| Bilaterals <40 | 3/99 (3.03) | 3/68 (4.41) | 6.28 (1.85-19.3) | 0/12 (0) | 0 | 4/40 (10.00) | 5/91 (5.49) | 9.38 (3.20-27.2) | 2/25 (8.00) | 55.09 (3.8-774.5) | 0/6 (0) | 0 |
| Bilaterals 40-49 | 4/233 (1.72) | 5/124 (4.03) | 4.28 (1.52-11.8) | 4/37 (10.81) | 8.67 (2.1-27.0) | 4/59 (6.78) | 10/151 (6.62) | 7.14 (2.83-18.9) | 1/52 (1.92) | 12.42 (0.2-240.8) | 1/20 (5.00) | 10.92 (0.23-96.5) |
| Bilaterals 50+ | 4/250 (1.60) | 2/129 (1.55) | 2.66 (0.79-8.13) | 6/95 (6.32) | 4.82 (1.58-12.4) | 7/44 (15.91) | 0/5 (0) | 16.67 (4.97-52.6) | 5/63 (7.94) | 70.69 (7.63->3,000) | 1/80 (1.25) | 2.62 (0.06-22.0) |
NOTE: Data from the HBBCS are included in pooled analyses of all studies and tests of heterogeneity between studies but are not included in this table as there were no carriers among 138 cases. There were 113, 21, and 3 cases with 0, 1, and 2 affected first-degree relatives, respectively, with missing information for 1 case. There were 13, 29, 46, and 50 cases age <40, 40-49, 50-59, and 60+, respectively. There was missing information on family history ABCS (n = 66), St. Petersburg (n = 3), and missing age information on age at diagnosis BBC (n = 1), Rotterdam (n = 1), St. Petersburg (n = 5). Abbreviation: N/K, not known; FDR, first-degree relatives.
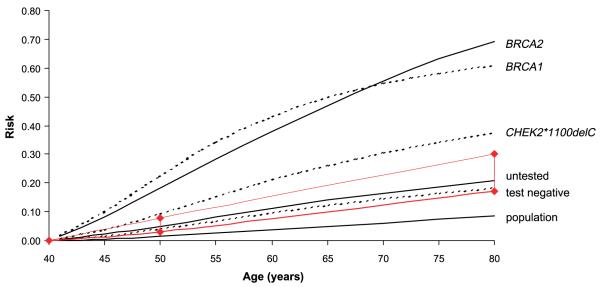
The lifetime (to age 80 years) breast cancer risk to a 40-year-old daughter of a woman with bilateral breast cancer and unknown BRCA and CHEK2 carrier status predicted by the Breast and Ovarian Analysis of Disease Incidence and Carrier Estimation Algorithm is 21% (Fig. 2), close to the observed lifetime risk of 24% in mothers and sisters of bilateral breast cancer cases in the BBC study (13). A negative result for a BRCA mutation screen reduces this only slightly to 18%. On the assumption that CHEK2*1100delC multiplies the risk caused by alleles in genes other than BRCA1 and BRCA2, our analysis suggests that this lifetime risk is doubled to 37% in carriers, more than half the risk to a carrier of BRCA1 (61%) or BRCA2 (69%). Her risk between age 40 and 50 years is 4% if she has a negative BRCA mutation screen but is increased to 9% if she is found to be a carrier of CHEK2*1100delC. If she has not been tested the daughter’s relative risk (standardized incidence ratio) compared with the general population is 3.7 at age 40 to 50 years, falling to 1.8 by age 70 to 80 years (Table 2). If she is a carrier of CHEK2*1100delC, the corresponding relative risks are 6.8 and 3.8.
Figure 2.
Lifetime predicted risks in the unaffected 40-year-old daughter of a bilateral breast cancer case in relation to the daughter’s carrier status for BRCA1, BRCA2, and CHEK2*1100delC. Risks in the unaffected daughter were derived from the Breast and Ovarian Analysis of Disease Incidence and Carrier Estimation Algorithm breast cancer model and assume that both cancers in the bilateral mother were diagnosed at age 50 y. For the purposes of this analysis, “lifetime risk” is defined as risk by age 80 y in accordance with NICE guidelines for the classification of women at risk of familial breast cancer.
Table 2. Age-specific relative risks in daughters of bilateral breast cancer cases compared with the general population.
| Age (y) | OR |
|||||
|---|---|---|---|---|---|---|
| Population | Untested | BRCA1/2 negative | CHEK2* 1100delC | BRCA1 | BRCA2 | |
| 40-49 | Ref | 3.69 | 2.91 | 6.81 | 18.68 | 14.85 |
| 50-59 | Ref | 3.10 | 2.61 | 6.12 | 14.02 | 12.45 |
| 60-69 | Ref | 2.46 | 2.13 | 4.99 | 8.95 | 13.38 |
| 70-79 | Ref | 1.83 | 1.63 | 3.83 | 6.29 | 13.00 |
Discussion
As predicted, our pooled bilateral OR estimate for CHEK2*1100delC carriers (6.43; 95% CI, 4.33-9.56) is significantly higher than the published estimate of 2.34 (95% CI, 1.72-3.20) for unselected cases (difference between ORs: Z, 3.92; P < 0.001) but consistent with its square (5.48; ref. 15). Each additional affected first degree relative should increase the carrier OR for bilaterals by a further factor of ~1.67 (an excess risk half that in unselected cases, whose reported OR is 2.34). The CHEK2*1100delC carrier OR in familial cases is thus expected to be ~9 (2.34 × 2.34 × 1.67). The British and Finnish data are consistent with a trend of this order but cases from the Netherlands and St. Petersburg show no familial trend in OR (P < 0.001 for heterogeneity between countries). This heterogeneity is not confined to studies of bilateral breast cancer cases: in studies comparing unselected first primary breast cancers with and without affected relatives, an increased prevalence of CHEK2*1100delC has been seen in familial breast cancer cases in some (3, 14) but not all studies (25, 26).
The absence of any familial effect in bilateral cases from the Netherlands and St. Petersburg seems likely to reflect a combination of systematic effects and chance variation. There is epistasis between CHEK2*1100delC and inactivating mutations in BRCA1 and BRCA2 (3), and epistatic effects with these or other risk alleles that differ in frequency between populations could have contributed to the significant heterogeneity between countries that we observed in relation to family history. Referral of familial cases from population subgroups or other regions may also have affected results in some studies. The reported carrier frequency for CHEK2*1100delC varies between 1.3% and 0.5% in Northern European Caucasians (15), and may be even lower locally. There were 6 of 651 carriers in GENICA German population controls and 0 of 600 in KORA German population controls (GENICA versus KORA, P = 0.03), and only 1 of 821 in controls from the Russian study, which gave the highest carrier OR (OR, 47.88; 95% CI, 6.30-2,126.8). The high prevalence (10.4%) of the Ashkenazi BRCA1 5382insC variant in bilateral cases from the Russian study could reflect nonrandom referral to this specialized research institute (27).
Methods for calculating a woman’s personal risk and guidelines for counseling and management in primary, secondary, or tertiary care are still evolving. Under current UK guidelines,16 a woman should be offered magnetic resonance imaging and mammographic surveillance in secondary care if her predicted breast cancer risk is between 3% and 8% from age 40 to 49 years or her lifetime risk is between 17% and 30%. This “gray area” of concern but limited intervention is shown in Fig. 2. Above this level, she should be offered tertiary care, including risk-reducing surgery.16 As the carrier OR for CHEK2*1100delC measured in unselected cases is a relative measure the implication of being a carrier in terms of absolute risk will depend on the woman’s family history of breast cancer. The model on which Fig. 2 is based assumes that CHEK2*1100delC interacts multiplicatively with other “polygenes” to increase the familial OR by roughly the same factor as it does in the overall population, and the data from the United Kingdom and Finland support this assumption. The predicted risks shown in Fig. 2 imply that the clinical management of the daughter of a woman with bilateral breast cancer should be different if she were found to carry CHEK2*1100delC. Byrnes et al. (28) have reviewed the evidence for an interaction between moderate penetrance alleles in CHEK2, ATM, BRIP1, and PALB2 and polygenes in the context of familial breast cancer cases, and they too have concluded that detection of these variants in women with a strong family history of breast cancer may be of considerable clinical consequence. Before clinical geneticists accept such predictions for CHEK2*1100delC or other moderate penetrance variants, however, they will want more consistent and more extensive population-based evidence on the effect of CHEK2*1100delC and other moderate or low-penetrance alleles in women with a family history of breast cancer.
Supplementary Material
Table I s. Characteristics of the studies and distribution of CHEK2*1100delC genotype in breast cancer cases with two primary breast cancers and controls
Table 2s. Lifetime predicted risks to the unaffected 40 year old daughter of a bilateral breast cancer case according to age at diagnosis of the mother and number of additional unaffected female relatives.
Acknowledgments
We thank Research Nurse N. Puolakka for sample and data collection; the contribution of Prof. Dr. Flora Van Leeuwen (NKI-AVL) and Prof. Dr. Jan Klijn (EUR-DDHK, Rotterdam) for patient recruitment; Yon Ko, Beate Pesch, and Thomas Brüning (GENICA) and Hans Ulrich Ulmer and Hartmut Frenzel (SKK) for recruitment and data collection; and Professor N Rahman, Professor M R Stratton and S Seal for allowing us to use data from the Institute of Cancer Research Family Breast Cancer study (ICR_FBC) and the Breast Cancer Association Consortium for discussions from which this analysis arose.
Grant support: The BBC study was funded by Cancer Research-UK and Breakthrough Breast Cancer. ICR_FBC study was funded by Cancer Research UK. The families are recruited by the Breast Cancer Susceptibility Collaboration (UK). The controls are from the British 1958 Birth Cohort DNA collection funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02. The HEBCS was supported by The Academy of Finland (project 110663), Helsinki University Central Hospital Research Funds, The Sigrid Juselius Foundation and The Finnish Cancer Society. The Rotterdam Breast Cancer study was funded by Koningin Wilhelmina Fonds Dutch Cancer Society (grant DDHK 2003-2862). The ABCS was supported by the Dutch Cancer Society (grant DCS-NKI 01-2425). The HBBCS was supported by an intramural grant of Hannover Medical School. The St Petersburg Breast Cancer study was supported by INTAS (grant 05-1000008-7870), Russian Agency for Science and Innovations (grant 02.512.11.2101), Grant for Helmholtz-Russia Joint Research Groups (grant HRJRG-006/07-04-92282-a) and Russian Federation for Basic Research (grant 07-04-00122-a) The GENICA study was supported by the German Human Genome Project and funded by the Federal Ministry of Education and Research (BMBF) Germany grants 01KW9975/5, 01KW9976/8, 01KW9977/0 and 01KW0114. Genotyping analyses were supported by the Deutsches Krebsforschungszentrum in Heidelberg.
Footnotes
Note: Supplementary data for this article are available at Cancer Epidemiology Biomarkers and Prevention Online (http://cebp.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
References
- 1.Pharoah PD, Antoniou A, Bobrow M, et al. Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet. 2002;31:33–6. doi: 10.1038/ng853. [DOI] [PubMed] [Google Scholar]
- 2.Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40:17–22. doi: 10.1038/ng.2007.53. [DOI] [PubMed] [Google Scholar]
- 3.Meijers-Heijboer H, van den Ouweland A, Klijn J, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31:55–9. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 4.Renwick A, Thompson D, Seal S, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38:873–5. doi: 10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- 5.Rahman N, Seal S, Thompson D, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–7. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox A, Dunning AM, Garcia-Closas M, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39:352–8. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 7.Erkko H, Xia B, Nikkila J, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446:316–9. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 8.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–9. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 11.Gold B, Kirchhoff T, Stefanov S, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci U S A. 2008;105:4340–5. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peto J, Mack TM. High constant incidence in twins and other relatives of women with breast cancer. Nat Genet. 2000;26:411–4. doi: 10.1038/82533. [DOI] [PubMed] [Google Scholar]
- 13.Johnson N, Fletcher O, Naceur-Lombardelli C, et al. Interaction between CHEK2*1100delC and other low-penetrance breast-cancer susceptibility genes: a familial study. Lancet. 2005;366:1554–7. doi: 10.1016/S0140-6736(05)67627-1. [DOI] [PubMed] [Google Scholar]
- 14.Vahteristo P, Bartkova J, Eerola H, et al. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am J Hum Genet. 2002;71:432–8. doi: 10.1086/341943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CHEK2 Breast Cancer Case-Control Consortium CHEK2*1100delC and susceptibility to breast cancer: a collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. Am J Hum Genet. 2004;74:1175–82. doi: 10.1086/421251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broeks A, de Witte L, Nooijen A, et al. Excess risk for contralateral breast cancer in CHEK2*1100delC germline mutation carriers. Breast Cancer Res Treat. 2004;83:91–3. doi: 10.1023/B:BREA.0000010697.49896.03. [DOI] [PubMed] [Google Scholar]
- 17.Broeks A, Braaf LM, Huseinovic A, et al. Identification of women with an increased risk of developing radiation-induced breast cancer: a case only study. Breast Cancer Res. 2007;9:R26. doi: 10.1186/bcr1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinmann D, Bremer M, Rades D, et al. Mutations of the BRCA1 and BRCA2 genes in patients with bilateral breast cancer. Br J Cancer. 2001;85:850–8. doi: 10.1054/bjoc.2001.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chekmariova EV, Sokolenko AP, Buslov KG, et al. CHEK2 1100delC mutation is frequent among Russian breast cancer patients. Breast Cancer Res Treat. 2006;100:99–102. doi: 10.1007/s10549-006-9227-7. [DOI] [PubMed] [Google Scholar]
- 20.Justenhoven C, Hamann U, Pesch B, et al. ERCC2 genotypes and a corresponding haplotype are linked with breast cancer risk in a German population. Cancer Epidemiol Biomarkers Prev. 2004;13:2059–64. [PubMed] [Google Scholar]
- 21.Pesch B, Ko Y, Brauch H, et al. Factors modifying the association between hormone-replacement therapy and breast cancer risk. Eur J Epidemiol. 2005;20:699–711. doi: 10.1007/s10654-005-0032-0. [DOI] [PubMed] [Google Scholar]
- 22.Rashid MU, Jakubowska A, Justenhoven C, et al. German populations with infrequent CHEK2*1100delC and minor associations with early-onset and familial breast cancer. Eur J Cancer. 2005;41:2896–903. doi: 10.1016/j.ejca.2005.04.049. [DOI] [PubMed] [Google Scholar]
- 23.Antoniou AC, Cunningham AP, Peto J, et al. The BOADICEA model of genetic susceptibility to breast and ovarian cancers: updates and extensions. Br J Cancer. 2008;98:1457–66. doi: 10.1038/sj.bjc.6604305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antoniou AC, Pharoah PP, Smith P, et al. The BOADICEA model of genetic susceptibility to breast and ovarian cancer. Br J Cancer. 2004;91:1580–90. doi: 10.1038/sj.bjc.6602175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernstein JL, Teraoka SN, John EM, et al. The CHEK2*1100delC allelic variant and risk of breast cancer: screening results from the Breast Cancer Family Registry. Cancer Epidemiol Biomarkers Prev. 2006;15:348–52. doi: 10.1158/1055-9965.EPI-05-0557. [DOI] [PubMed] [Google Scholar]
- 26.Weischer M, Bojesen SE, Ellervik C, et al. CHEK2*1100delC genotyping for clinical assessment of breast cancer risk: meta-analyses of 26,000 patient cases and 27,000 controls. J Clin Oncol. 2008;26:542–8. doi: 10.1200/JCO.2007.12.5922. [DOI] [PubMed] [Google Scholar]
- 27.Sokolenko AP, Mitiushkina NV, Buslov KG, et al. High frequency of BRCA1 5382insC mutation in Russian breast cancer patients. Eur J Cancer. 2006;42:1380–4. doi: 10.1016/j.ejca.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 28.Byrnes GB, Southey MC, Hopper JL. Are the so-called low penetrance breast cancer genes, ATM, BRIP1, PALB2 and CHEK2, high risk for women with strong family histories? Breast Cancer Res. 2008;10:208. doi: 10.1186/bcr2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table I s. Characteristics of the studies and distribution of CHEK2*1100delC genotype in breast cancer cases with two primary breast cancers and controls
Table 2s. Lifetime predicted risks to the unaffected 40 year old daughter of a bilateral breast cancer case according to age at diagnosis of the mother and number of additional unaffected female relatives.