Summary
DNA binding proteins play key roles in many cellular processes, including transcriptional regulation and replication. Microarray-based technologies permit high-throughput identification of binding sites and functional roles of these proteins. In particular, microarray readout either of chromatin immunoprecipitation (‘ChIP-chip’) or of DNA adenine methyltransferase fusion proteins (‘DamID’) enables the identification of in vivo genomic target sites of proteins. A complementary approach, in vitro binding of proteins directly to double-stranded DNA microarrays (‘protein binding microarrays’), permits rapid characterization of their DNA binding site sequence specificities. Recent advances in DNA microarray synthesis technologies have permitted the definition of proteins’ DNA binding sites at much higher resolution and coverage, and further advances in these and emerging technologies will further increase the efficiencies of these exciting new approaches.
Introduction
DNA binding proteins perform a variety of important functions in cells, including transcriptional regulation, chromosome maintenance, replication, and DNA repair. The interactions between transcription factors (TFs) and their DNA binding sites are of particular interest, since these interactions control critical steps in development and responses to environmental stresses, and in humans their dysfunction can contribute to the progression of various diseases. Also of significant interest are the locations of histones and their post-translational modifications, as they too contribute significantly to gene regulation. The labor involved in traditional techniques for examining the DNA binding sites of proteins limits them to analysis of a fairly small number of DNA sequences.
DNA microarrays [1,2] together with whole-genome sequences have revolutionized mRNA expression analysis, and, more recently, they have facilitated biochemical and functional genomics studies of DNA binding proteins. Chromatin immunoprecipitation of a protein of interest followed by microarray-based detection of enriched DNA fragments, referred to as ‘ChIP-chip’ or ‘genome-wide location analysis’, is currently the most widely used method for identifying in vivo TF binding sites in a high-throughput manner [3–6]. An alternative microarray-based approach for genome-scale identification of in vivo binding sites utilizes a DNA binding protein fused to DNA adenine methyltransferase (Dam), which marks DNA near the protein’s target sites [7]. The protein binding microarray (PBM) technology permits rapid, high-throughput characterization of the in vitro DNA binding specificities of DNA binding proteins by assaying their binding to double-stranded DNA microarrays [8,9, ··10].
Advances in DNA microarray synthesis technology have increased feature (spot) density, which has allowed higher resolution definition of in vivo target sites and greater coverage of genomic sequence space. Such advances also have permitted a larger fraction of sequence space to be assayed in vitro by PBMs. Because the ChIP-chip and DamID technologies and early studies using those techniques have been described in depth in a number of recent reviews [11–13], this review will focus primarily on recent studies that included technological improvements. This review also will focus on the protein binding microarray (PBM) technology, which previously has not been reviewed.
ChIP-chip
In ChIP-chip, cells are treated with a reagent, typically formaldehyde, which creates covalent crosslinks between protein and DNA. An antibody specific for a protein of interest is used to immunoprecipitate protein-bound DNA fragments, which then are labeled in an amplification reaction and hybridized to DNA microarrays to identify the protein-bound fragments. Initial ChIP-chip studies were performed on Saccharomyces cerevisiae regulatory TFs [3–6], and replication origin recognition proteins [14], using microarrays spotted with PCR amplicons covering essentially all intergenic regions in yeast. Studies of yeast TFs in multiple cellular states [15] or environmental conditions [·16] highlighted that binding of TFs can be condition-dependent. ChIP-chip also has been used in yeast to examine TATA binding protein (TBP) [17], RNA Pol II transcription initiation and elongation apparatuses [18], RNA Pol III transcription apparatus [19], and a centromere-related factor [20].
Recent ChIP-chip studies have focused on two main areas: (1) histone modifications, histone-modifying proteins, and chromatin remodeling; and (2) combinatorial and condition-specific regulation by TFs. Many recent studies have utilized arrays of oligonucleotides designed to tile a portion of the genome (‘tiling arrays’), permitting high-resolution definition of the genomic binding sites of a given protein.
A recent study that provided high-resolution mapping of nucleosome positions in yeast was accomplished by the purification of mononucleosomal DNA resulting from a digestion of genomic DNA with micrococcal nuclease, and the use of microarrays printed with 50-mer oligonucleotides tiled every 20 bp across almost all of yeast chromosome III and ∼1 kb of 230 additional promoters. Interestingly, Rando and colleagues observed nucleosome-free regions of ∼150 bp located ∼200 bp upstream of many annotated coding regions, possibly to permit accessibility of the intergenic DNA to regulatory TFs [··21]; this finding is consistent with a previous study by Lieb and colleagues that found decreased nucleosome occupancy at active promoters [22]. In contrast to other studies whose data from PCR amplicon arrays suggested the existence of a discrete, complex, combinatorial “histone code”, Rando and colleagues observed a continuous pattern of modifications across nucleosomes, suggesting a simpler, redundant “code” [··23]. Interestingly, the histone variant H2A.Z was found to occur at the transcription start sites of most genes, including those at undetectable transcription levels, suggesting a mechanism for marking the 5′ ends of both active and inactive genes [24]. In another study using such arrays, the nuclear pore associated protein Mlp1 was found to associate with alpha-factor-induced genes in an RNA-dependent manner, suggesting a mechanism for chromosome conformational changes in response to the alpha factor mating pheromone [·25]. A separate study used an Agilent DNA microarray containing ∼44,000 60-mer oligonucleotides covering most of the yeast genome at an average probe density of 266 bp to map histone acetylation and methylation at high resolution [26]. In another study, histone methylations in mouse embryonic stem cells were mapped using Affymetrix arrays custom-designed to tile noncoding regions that are highly conserved over mammalian genomes and may be important for gene regulation during development [27]. ChIP-chip studies of chromatin and its epigenetic modifications are described in a recent review [12], and so are not discussed in detail here.
The construction of microarrays for genome-scale ChIP-chip studies in metazoans has been a significant challenge due to the increased sizes of those genomes. Early ChIP-chip studies in mammalian genomes utilized various types of PCR amplicon arrays, including arrays tiling a specific genomic region of interest [28], CpG island arrays [29], and promoter arrays [30]. Promoter arrays covering roughly −750 bp to +250 bp relative to transcription start sites have been used to analyze binding of HNF1α, HNF4α, and HNF6 in post-mortem human liver and pancreas [31] and muscle regulatory TFs MyoD, myogenin, and MEF2 in differentiating murine C2C12 skeletal muscle cells [32].
Definition of in vivo binding sites at higher resolution has been due largely to the switch to oligonucleotide tiling arrays. In one study, a 10-slide set of Agilent 60-mer oligonucleotide arrays, covering −8 kb to +2 kb relative to the transcript start sites for nearly 18,000 human genes, was used to identify binding sites in human embryonic stem (ES) cells of the TFs OCT4, SOX2, and NANOG, which are important in ES cell renewal and maintenance of pluripotency [··33]. Even though important regulatory interactions can occur in promoters, a study using Affymetrix arrays representing essentially all nonrepetitive sequences on human chromosomes 21 and 22 found that most binding sites of the TFs Sp1, cMyc, and p53 were located far from transcription start sites of known protein-coding genes [··34].
ChIP-chip using anti-RNA Polymerase II and anti-TBP-associated factor 1 antibodies to isolate pre-initiation complexes from four human cell lines has enabled improved promoter mapping [35]. In a follow-up study, Ren and colleagues mapped promoters in the entire human genome using a set of 38 NimbleGen arrays containing roughly 14.5 million 50-mer oligonucleotides, designed to represent all non-repetitive DNA throughout the human genome at 100 bp resolution. Interestingly, the authors found that a large number of genes contained two or more active promoters, and also defined a set of 1,239 putative promoters that correspond to previously unannotated transcription units [··36].
A major limitation in applying ChIP-chip in other model organisms has been the availability of suitable microarrays. Using a PCR amplicon array tiling Drosophila chromosome arm 2L, Bell and colleagues found that sites of active transcription correlated with binding by the origin recognition complex and early replicating origins [·37]. Such arrays also have been used to examine heat shock factor binding in Drosophila embryos [38]. NimbleGen 60-mer arrays tiling over 36 Mb of the Drosophila genome at 100 bp resolution also have been used [39]. Binding by Escherichia coli TFs has been examined using off-the-shelf Affymetrix E. coli antisense arrays [40], while NimbleGen tiling arrays have been used to map E. coli RNA Pol binding sites [41].
Changes in the crosslinking procedure have also led to improvements in ChIP-chip. To improve the efficiency of crosslinking when formaldehyde treatment alone did not result in significant enrichment in ChIP of the histone deacetylase and repressor Rpd3, cells were treated first with dimethyl adipimidate, a protein-protein crosslinking reagent, and then with formaldehyde to create protein-DNA crosslinks [42].
Despite all the benefits of identifying in vivo binding locations, ChIP has some inherent caveats that can make the identification of DNA binding sites difficult [43]. In particular, both antibody limitations and condition-specific protein binding can result in ChIP experiments that do not provide significant enrichment of bound fragments in the immunoprecipitated sample [·16,43].
DamID
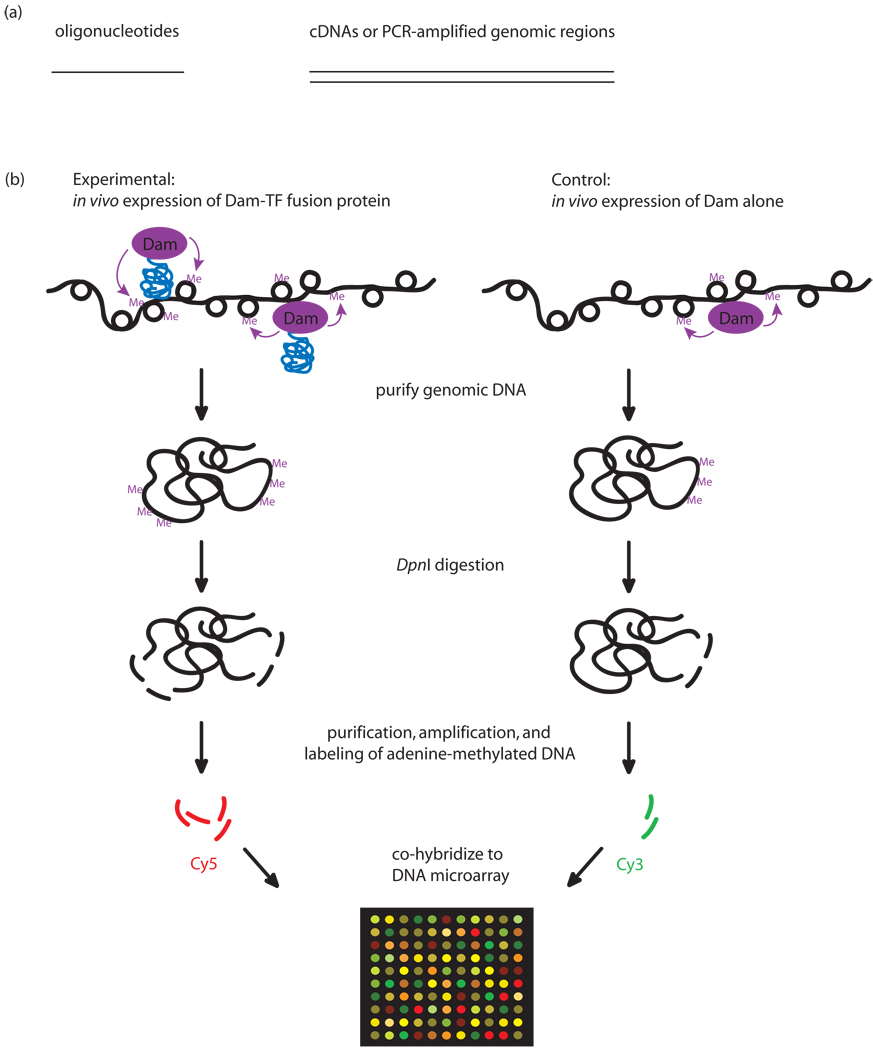
In the DamID approach, the protein of interest is overexpressed in vivo from a plasmid as a fusion to Dam. Wherever the protein binds DNA, Dam will methylate adenines within GATC sites in the vicinity of the binding sites. The methylated sites in the experimental versus control (Dam alone) samples are detected by digestion with a methyl-specific restriction enzyme, amplification, labeling, and hybridization to a microarray. DamID has been used to identify in vivo binding sites in Drosophila [7] and Arabidopsis [44], of sequence-specific TFs [45], DNA methyltransferase [44], chromatin [7] and chromatin-associated proteins using cDNA or PCR amplicon arrays, and more recently, NimbleGen 60-mer tiling arrays [·46].
Comparison of ChIP-chip and DamID indicates that these two methods for mapping in vivo TF binding sites can yield very similar results [47]. DamID has the advantage that it does not require a TF-specific antibody; however, DamID is performed using slight over-expression of a tagged-TF from a plasmid, raising concern that even this slight increase in TF concentration might result in artifactual binding at non-native binding sites. In addition, DamID is not suitable for detection of post-translational modifications. Finally, DamID does not permit high resolution mapping of binding sites, because methylation by the tethered Dam can extend over a few kb from the TF binding site [7]. Advantages and disadvantages of DamID and papers using DamID are discussed in-depth in a recent review [13].
Protein Binding Microarrays (PBMs)
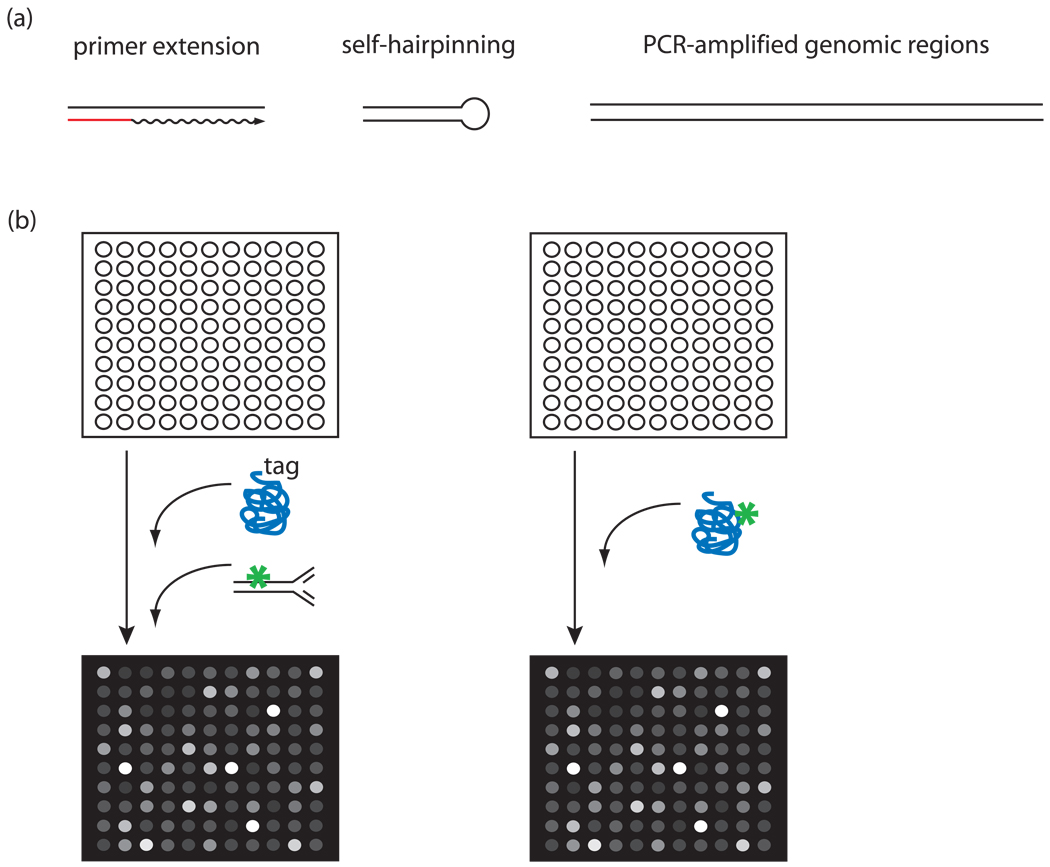
The PBM technology permits high-throughput characterization of the sequence specificities of DNA-protein interactions [8,9, ··10] (see Figure 1). Briefly, a purified, epitope-tagged protein is allowed to bind directly to a double-stranded DNA microarray. The protein-bound microarray is stained with a fluorophore-conjugated antibody; alternatively, a directly labeled protein can be used. The protein’s DNA binding specificity is determined from the significantly bound spots. Although it does not assay binding in vivo, PBMs offer a number of advantages. First, an in vitro approach does not require prior knowledge of the conditions in which a TF binds its genomic sites. PBMs can provide extensive binding preference data for each DNA sequence variant. Finally, the PBM technology is rapid, allowing the determination of the DNA binding specificities of a purified protein in a single day.
Figure 1. ChIP-chip.
(a) Types of DNAs used in ChIP-chip. Oligonucleotide tiling genomic regions (left); PCR amplification of genomic regions (right). (b) ChIP-chip experimental design. A large variety of protein-DNA and protein-protein crosslinks are created nonspecifically, due to the nonspecific nature of formaldehyde crosslinking. An antibody (orange) either specific for the protein of interest (blue) or specific for an epitope tag fused to the protein of interest is used in immunoprecipitation (IP) in the experimental sample. This IP will enrich for the target protein, including protein directly bound to genomic DNA binding sites, and also protein indirectly associated with DNA via protein-protein interactions. A control (“mock”) IP is performed using either no antibody, an irrelevant antibody, or pre-immune IgG antibodies. This mock IP is not expected to enrich for the target protein of interest.
In a proof-of-principle study, Bulyk and colleagues biochemically converted Affymetrix oligonucleotide arrays to double-stranded DNA (dsDNA) arrays by extension of a universal primer complementary to a universal primer sequence synthesized as part of each of the oligonucleotides on the arrays. Interestingly, the authors observed that the methylation-sensitivity of restriction enzymes could be detected by the use of dsDNA arrays treated with Dam methylase [8].
In the first study to assay TF binding to dsDNA arrays, Bulyk et al. used a DNA microarray spotted with short synthetic double-stranded DNAs designed to examine a phage display library of wildtype and mutant Cys2His2 zinc finger DNA binding domains of Zif268 (Egr1). Importantly, Bulyk et al. observed that spots with higher signal intensities contained higher affinity binding sites [9]. In a similar study, Udalova and colleagues examined the DNA binding specificities of Oct-1 and the NF-kB p52 homodimer. Similarly to the Bulyk et al. (2001) study, the correlation of the PBM signal intensities with binding affinity data allowed the authors to approximate the relative binding affinities for other binding site variants [48].
More recently, Mukherjee et al. examined GST-tagged yeast TFs using whole-genome yeast intergenic microarrays spotted with PCR amplicons. The PBM-derived binding site motifs for Abf1 and Rap1 were highly similar to motifs derived from ChIP-chip data [43]. Moreover, analysis of the Mig1 PBM data resulted in the Mig1 motif [··10], while analysis of the ChIPchip data [43] did not. Interestingly, many of the newly identified sites of these TFs are highly conserved across five sequenced sensu stricto yeast species and thus are potentially regulatory [··10].
The use of PCR amplicon microarrays has the advantages of covering much sequence space with relatively few spots, and representing TF binding sites in the context of their native genomic flanking sequences, including potential cofactor binding sites. In a proof-of-principle study, Doi et al. showed that Jun with its protein partner Fos bound to microarrays upon formation of the Jun/Fos heterodimer [49]. However, inherent in the use of intergenic arrays are two key limitations. First, the relative binding preferences for each of potentially multiple sites in a given intergenic region are not readily distinguished. Second, it can be difficult to separate the specific versus nonspecific contributions to the signal intensities for spotted DNAs of variable lengths.
In contrast, microarrays created with short synthetic dsDNAs representing all k-mers [8,9, ··50] permit the relative preferences for variant binding site sequences to be extracted more readily. In one recent study, Warren et al. in situ synthesized arrays of 34-mer oligonucleotides containing a 14 bp double-stranded hairpin region, with each possible 8-mer binding site sequence variant synthesized at a distinct feature on the array, to examine binding by fluorophore-conjugated engineered polyamides and the Drosophila TF Extradenticle, Cy3-labeled at a unique cysteine [··50]. However, as k (i.e., the DNA binding site length to be probed) increases, the number of possible k-mers can far exceed the number of spots that currently can be manufactured on a single microarray. To overcome this limitation, one could sample binding site space. Alternatively, instead of devoting a unique spot to each k-mer, one can instead employ a compact universal design, whereby distinct k-mers within a given dsDNA longer than k are allowed to overlap (MF Berger, AA Philippakis, et al., unpublished).
Conclusions
Most TFs in human and various model organisms have undetermined DNA binding specificities and their regulatory functions are not well understood on a genomic scale. Significant challenges include characterizing their DNA binding specificities, and determining the differential usage of their binding sites in various cell types, through development and response to cellular and environmental conditions, and in normal and disease states. Thus, the TFs’ target genes and potential combinatorial modes of transcriptional regulatory control will be discovered. Continued improvements in the synthesis of high density DNA microarrays will allow even greater coverage of DNA binding site space. As more genomes are sequenced and comprehensive data on TF-DNA binding are generated, it will become feasible to examine the co-evolution of TF protein sequence and their corresponding DNA binding sites.
Figure 2. DamID.
(a) Types of DNAs used in DamID microarray hybridizations. Oligonucleotide tiling genomic regions (left); cDNAs or PCR amplification of genomic regions (right). (b) DamID experimental design. The protein of interest (blue) is overexpressed in vivo from a plasmid as a fusion to Dam (purple). Wherever the protein binds DNA, Dam will methylate adenines within GATC sites in the vicinity of the binding sites. The methylated sites are digested with the methyl-specific restriction enzyme DpnI, which cuts only at methylated GATC sites. The smaller DpnI digestion fragments, corresponding to the methylated regions, are either purified by sucrose gradient centrifugation or specifically amplified using a methylation-specific PCR protocol. (Panel (b) was adapted from [7] and [51] with permission from Nature Publishing Group.)
Figure 3. Protein binding microarrays (PBMs).
(a) Types of DNAs used in PBMs. Short double-stranded oligonucleotides created by primer extension using a primer (red), complementary to a sequence present on spots either on an oligonucleotide array [8] or in solution [9, ··10] (left); regions of short double-stranded DNA created by self-hairpinning oligonucleotides on an oligonucleotide array [··50] (center); longer double-stranded DNAs resulting from PCR amplification of genomic regions [··10] (right). (b) PBM experimental design. Double-stranded DNA microarrays can be either bound by an epitope-tagged TF and labeled by a fluorophore-conjugated (green) antibody specific for the tag [··10] (left), or bound by a directly fluorophore-labeled (green) TF [··50] (right). (Panel (b) was adapted from [··10] with permission from Nature Publishing Group.)
Table 1.
Comparison of ChIP-chip, DamID, and PBM technologies.
 Technology Technology |
Advantages | Disadvantages |
|---|---|---|
| ChIP-chip |
|
|
| DamID |
|
|
| PBM |
|
|
Acknowledgements
I apologize to colleagues for not being able to cite their work because of space limitations. I thank Pete Estep, Rachel Patton McCord, and Mike Berger for comments on the manuscript. This work was supported in part by National Institutes of Health grants from the National Human Genome Research Institute to M.L.B. (R01 HG002966 and R01 HG003420).
References and recommended reading
Papers of particular interest, published approximately within the last two years, have been highlighted as:
· of special interest
·· of outstanding interest
- 1.Pease AC, Solas D, Sullivan EJ, Cronin MT, Holmes CP, Fodor SP. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci USA. 1994;91:5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 3.Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 4.Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 5.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 6.Reid JL, Iyer VR, Brown PO, Struhl K. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell. 2000;6:1297–1307. doi: 10.1016/s1097-2765(00)00128-3. [DOI] [PubMed] [Google Scholar]
- 7.van Steensel B, Delrow J, Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat Genet. 2001;27:304–308. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- 8.Bulyk ML, Gentalen E, Lockhart DJ, Church GM. Quantifying DNA-protein interactions by double-stranded DNA arrays. Nat Biotechnol. 1999;17:573. doi: 10.1038/9878. [DOI] [PubMed] [Google Scholar]
- 9.Bulyk ML, Huang X, Choo Y, Church GM. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc Natl Acad Sci USA. 2001;98:7158–7163. doi: 10.1073/pnas.111163698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mukherjee S, Berger MF, Jona G, Wang XS, Muzzey D, Snyder M, Young RA, Bulyk ML. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. This is the first genome-scale PBM study. This paper describes the use of whole-genome yeast intergenic PCR amplicon microarrays for the analysis of the binding specificities of epitope-tagged yeast TFs Rap1, Abf1, and Mig1. PBM-derived binding site motifs matched known DNA binding specificities of the TFs, and many new candidate regulatory sites and target genes of these TFs were identified.
- 11.Sikder D, Kodadek T. Genomic studies of transcription factor-DNA interactions. Curr Opin Chem Biol. 2005;9:38–45. doi: 10.1016/j.cbpa.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Barrera LO, Ren B. The transcriptional regulatory code of eukaryotic cells – insights from genome-wide analysis of chromatin organization and transcription factor binding. Curr Opin Cell Biol. 2006;18:1–8. doi: 10.1016/j.ceb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Orian A. Chromatin profiling, DamID and the emerging landscape of gene expression. Curr Opin Genet Dev. 2006;16:157–164. doi: 10.1016/j.gde.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, Bell SP, Aparicio OM. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science. 2001;294:2357–2360. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]
- 15.Simon I, Barnett J, Hannett N, Harbison C, Rinaldi N, Volkert T, Wyrick J, Zeitlinger J, Gifford D, Jaakkola T, et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/s0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- 16. Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. In this ChIP-chip study of 203 yeast TFs using PCR amplicon arrays, 84 TFs were examined in multiple environmental conditions. The authors observed that TF binding could be categorized as condition-invariant, condition-enabled, condition-expanded, or condition-altered. In addition, ChIP-chip data for roughly one-third of these 203 yeast TFs resulted in high quality DNA binding site motifs.
- 17.Kim J, Iyer VR. Global role of TATA box-binding protein recruitment to promoters in mediating gene expression profiles. Mol Cell Biol. 2004;24:8104–8112. doi: 10.1128/MCB.24.18.8104-8112.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 19.Moqtaderi Z, Struhl K. Genome-wide occupancy profile of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol Cell Biol. 2004;24:4118–4127. doi: 10.1128/MCB.24.10.4118-4127.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiburz BM, Reynolds DB, Megee PC, Marston AL, Lee BH, Lee TI, Levine SS, Young RA, Amon A. The core centromere and Sgo1 establish a 50-kb cohesin-protected domain around centromeres during meiosis I. Genes Dev. 2005;19:3017–3030. doi: 10.1101/gad.1373005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. The authors printed microarrays with 50-mer oligonucleotides tiled every 20 bp across all of yeast chromosome III and ∼1 kb of ∼230 additional promoters. They used these microarrays to examine mononucleosomal DNA resulting from a digestion of genomic DNA with micrococcal nuclease, in order to generate a high resolution map of nucleosome positions in yeast. This study found nucleosome-free regions of ∼150 bp located ∼200 bp upstream of many annotated coding regions, possibly to permit accessibility of the intergenic DNA for binding by regulatory TFs.
- 22.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 23. Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. The authors of this study used the oligonucleotide tiling arrays described in Yuan et al. (2005) [··21] in ChIP-chip studies using antibodies against 12 histone modifications. In contrast to other studies whose data from PCR amplicon arrays supported a complex, combinatorial “histone code”, this study found a continuous pattern of modifications across nucleosomes.
- 24.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 2005;19:1188–1198. doi: 10.1101/gad.1307205. Using the oligonucleotide tiling arrays described in Yuan et al. (2005) [··21], the authors found by ChIP-chip that the yeast nuclear pore associated protein Mlp1 associates with alpha-factor-induced genes in an RNA-dependent manner. This suggests a mechanism for changing chromosome conformation in response to the alpha factor mating pheromone.
- 26.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 27.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 28.Horak CE, Mahajan MC, Luscombe NM, Gerstein M, Weissman SM, Snyder M. GATA-1 binding sites mapped in the beta-globin locus by using mammalian chIp-chip analysis. Proc Natl Acad Sci USA. 2002;99:2924–2929. doi: 10.1073/pnas.052706999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weinmann AS, Yan PS, Oberley MJ, Huang TH, Farnham PJ. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD. An initial blueprint for myogenic differentiation. Genes Dev. 2005;19:553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. The authors of this study custom-designed a 10-slide set of Agilent 60-mer oligonucleotide arrays to cover −8 kb to +2 kb relative to the transcript start sites for nearly 18,000 human genes. They used these arrays in ChIP-chip studies to identify binding sites in human ES cells of the TFs OCT4, SOX2, and NANOG, which are important in ES cell renewal and maintenance of pluripotency. The results suggest that these TFs co-regulate subsets of their target genes and repress various transcriptional regulators important in differentiation.
- 34. Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. Using Affymetrix oligonucleotide arrays representing essentially all nonrepetitive sequences on human chromosomes 21 and 22, the authors found that most binding sites of the TFs Sp1, cMyc, and p53 were located far from transcription start sites of known protein-coding genes.
- 35.Kim TH, Barrera LO, Qu C, Van Calcar S, Trinklein ND, Cooper SJ, Luna RM, Glass CK, Rosenfeld MG, Myers RM, et al. Direct isolation and identification of promoters in the human genome. Genome Res. 2005;15:830–839. doi: 10.1101/gr.3430605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. The authors designed a set of 38 NimbleGen arrays containing roughly 14.5 million 50-mer oligonucleotides, representing all non-repetitive DNA throughout the human genome at 100 bp resolution. They utilized these arrays in ChIP-chip studies using anti-RNA Polymerase II and anti-TAF1 antibodies in order to map promoters in the entire human genome. Interestingly, they found that a large number of genes contained two or more active promoters, and also defined a set of 1,239 putative promoters that correspond to previously unannotated transcription units.
- 37. MacAlpine DM, Rodriguez HK, Bell SP. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. The authors of this study constructed a PCR amplicon array tiling Drosophila chromosome arm 2L. They used this array in ChIP-chip studies to identify binding sites of the origin recognition complex (ORC) in the Kc cell line. Interestingly, they found that sites of active transcription correlated with ORC binding and early replicating origins.
- 38.Birch-Machin I, Gao S, Huen D, McGirr R, White RA, Russell S. Genomic analysis of heat-shock factor targets in Drosophila. Genome Biol. 2005;6:R63. doi: 10.1186/gb-2005-6-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 40.Grainger DC, Overton TW, Reppas N, Wade JT, Tamai E, Hobman JL, Constantinidou C, Struhl K, Church G, Busby SJ. Genomic studies with Escherichia coli MelR protein: applications of chromatin immunoprecipitation and microarrays. J Bacteriol. 2004;186:6938–6943. doi: 10.1128/JB.186.20.6938-6943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herring CD, Raffaelle M, Allen TE, Kanin EI, Landick R, Ansari AZ, Palsson BO. Immobilization of Escherichia coli RNA polymerase and location of binding sites by use of chromatin immunoprecipitation and microarrays. J Bacteriol. 2005;187:6166–6174. doi: 10.1128/JB.187.17.6166-6174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurdistani SK, Robyr D, Tavazoie S, Grunstein M. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat Genet. 2002;31:248–254. doi: 10.1038/ng907. [DOI] [PubMed] [Google Scholar]
- 43.Lee T, Rinaldi N, Robert R, Odom D, Bar-Joseph Z, Gerber G, Hannett N, Harbison C, Thompson C, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 44.Tompa R, McCallum C, Delrow J, Henikoff J, van Steensel B, Henikoff S. Genome-wide profiling of DNA methylation reveals transposon targets of CHROMOMETHYLASE3. Curr Biol. 2002;12:65–68. doi: 10.1016/s0960-9822(01)00622-4. [DOI] [PubMed] [Google Scholar]
- 45.Orian A, van Steensel B, Delrow J, Bussemaker HJ, Li L, Sawado T, Williams E, Loo LW, Cowley SM, Yost C, et al. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. de Wit E, Greil F, van Steensel B. Genome-wide HP1 binding in Drosophila: developmental plasticity and genomic targeting signals. Genome Res. 2005;15:1265–1273. doi: 10.1101/gr.3198905. The authors used custom-designed NimbleGen 60-mer arrays in DamID studies of HP1, a major component of heterochromatin in Drosophila.
- 47.van Steensel B. Mapping of genetic and epigenetic regulatory networks using microarrays. Nat Genet. 2005;37 Suppl:S18–S24. doi: 10.1038/ng1559. [DOI] [PubMed] [Google Scholar]
- 48.Linnell J, Mott R, Field S, Kwiatkowski DP, Ragoussis J, Udalova IA. Quantitative high-throughput analysis of transcription factor binding specificities. Nucleic Acids Res. 2004;32:e44. doi: 10.1093/nar/gnh042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doi N, Takashima H, Kinjo M, Sakata K, Kawahashi Y, Oishi Y, Oyama R, Miyamoto-Sato E, Sawasaki T, Endo Y, et al. Novel fluorescence labeling and high-throughput assay technologies for in vitro analysis of protein interactions. Genome Res. 2002;12:487–492. doi: 10.1101/gr.218802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Warren CL, Kratochvil NC, Hauschild KE, Foister S, Brezinski ML, Dervan PB, Phillips GN, Jr, Ansari AZ. Defining the sequence-recognition profile of DNA-binding molecules. Proc Natl Acad Sci USA. 2006 doi: 10.1073/pnas.0509843102. The authors used a NimbleGen maskless array synthesizer to create arrays of 34-mer oligonucleotides containing a 14 bp double-stranded hairpin region, with each possible 8-mer binding site sequence variant synthesized at a distinct feature on the array. They used these ‘all 8-mer’ arrays in PBMs to examine binding by fluorophore-conjugated engineered polyamides and the Drosophila TF Extradenticle, Cy3-labeled at a unique cysteine.
- 51.van Steensel B. Mapping of genetic and epigenetic regulatory networks using microarrays. Nat Genet. 2005;37 Suppl:S18–S24. doi: 10.1038/ng1559. [DOI] [PubMed] [Google Scholar]