Abstract
Purpose
Tumors from 50% of epidermal growth factor receptor (EGFR) mutant non - small cell lung cancer patients that develop resistance to gefitinib or erlotinib will contain a secondary EGFR T790M mutation. As most patients do not undergo repeated tumor biopsies we evaluated whether EGFR T790M could be detected using plasma DNA.
Experimental Design
DNA from plasma of 54 patients with known clinical response to gefitinib or erlotinib was extracted and used to detect both EGFR-activating and EGFR T790M mutations. Forty-three (80%) of patients had tumor EGFR sequencing (EGFR mutant/wild type: 30/13) and seven patients also had EGFR T790M gefitinib/erlotinib-resistant tumors. EGFR mutations were detected using two methods, the Scorpion Amplification Refractory Mutation System and the WAVE/Surveyor, combined with whole genome amplification.
Results
Both EGFR-activating and EGFR T790M were identified in 70% of patients with known tumor EGFR-activating (21 of 30) or T790M (5 of 7) mutations. EGFR T790M was identified from plasma DNA in 54% (15 of 28) of patients with prior clinical response to gefitinib/erlotinib, 29% (4 of 14) with prior stable disease, and in 0% (0 of 12) that had primary progressive disease or were untreated with gefitinib/erlotinib.
Conclusions
EGFR T790M can be detected using plasma DNA from gefitinib- or erlotinib-resistant patients.This noninvasive method may aid in monitoring drug resistance and in directing the course of subsequent therapy.
Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKI) are effective therapies for non-small cell lung cancer (NSCLC) patients with activating EGFR mutations. Several prospective clinical trials treating chemotherapy-naBve patients with EGFR mutations with gefitinib or erlotinib have been reported to date (1-6). Cumulatively, these studies have prospectively identified and treated over 200 patients with EGFR mutations. Together they show radiographic response rates ranging from 60% to 82% and median times to progression of 9.4 to 13.3 months in the patients treated with gefitinib and erlotinib. These outcomes are 3- to 4-folder greater than that observed with platin-based chemotherapy (20-30% and 3-4 months, respectively) for advanced NSCLC (7).
Unfortunately despite these benefits in EGFR-mutant NSCLC, all patients will ultimately develop progressive tumor growth while receiving gefitinib or erlotinib treatment. Two different mechanisms of acquired resistance in EGFR-mutant NSCLC patients have thus far been identified. These include a secondary mutation in EGFR (EGFR T790M) found in ~50% of those with acquired resistance and MET amplification in ~20% of patients (8-11). The therapeutic strategies for patients with these resistance mechanisms are also different. Irreversible EGFR inhibitors are effective in preclinical models at inhibiting the growth of EGFR T790M containing tumors in vitro and in vivo (12, 13). Several clinical trials involving irreversible EGFR inhibitors have now been initiated. However, whether these agents are effective clinically in gefitinib- and erlotinib-resistant NSCLC patients remains to be determined. Furthermore, if these agents are clinically effective, it will be important to determine the relationship to the presence/ absence of EGFR T790M mutation. Unfortunately very few patients undergo repeated tumor biopsies at the time when resistance develops to help guide appropriate therapeutic choices. Thus, there is a need to develop noninvasive methods to identify these resistance mechanisms.
A limited number of prior studies have evaluated the ability to detect EGFR-activating mutations from serum DNA of NSCLC patients treated with gefitinib (14, 15). The largest of these to date examined 42 NSCLC patients treated with gefitinib. EGFR-activating mutations were detected in 8 tumor specimens and 6 of the 8 mutations were correctly identified from serum DNA (15). None of the studies to date have specifically examined for EGFR T790M. This may be even harder to detect than an EGFR-activating mutation as EGFR T790M can sometimes represent a minor allele which may be missed by direct DNA sequencing-based methods (16).
In this study we examined the ability to detect EGFR T790M from plasma DNA from NSCLC patients that had clinically developed acquired resistance to gefitinib or erlotinib. We examine different methods of mutation detection and evaluate the benefits of whole genome amplification as a method to increase detection sensitivity.
Materials and Methods
Patients
From October 2006 to April 2008 patients with advanced NSCLC were identified using an institutional review board-approved protocol from the Thoracic Oncology clinic at the Dana Farber Cancer Institute. Only patients that had previously received single-agent gefitinib or erlotinib therapy and were at the time of the study off therapy were included in the study. In addition, patients were included if their clinical response, as defined by Response Evaluation Criteria in Solid Tumors, to gefitinib and erlotinib was known; they were willing to donate blood on one or more occasions; and they were receiving their treatment at Dana Farber Cancer Institute (17). Patients with known EGFR tumor genotype (mutant or wild type) were included only if they met the other criteria. Using these criteria we identified 50 patients previously treated with gefitinib (n = 17) or erlotinib (n = 33); 28 had a prior clinical partial response, 14 had prior stable disease, and 8 had primary progressive disease. In addition we included four randomly selected advanced NSCLC patients as negative controls who fit the inclusion criteria but had not received any therapy with either gefitinib or erlotinib or with any other EGFR-directed agent. Thirty patients had known tumor EGFR-activating mutations. All patients provided written informed consent and the studies were approved by the Dana Farber Cancer Institute Institutional Review Board.
Tumor mutation detection
Pretreatment tumor specimens were analyzed for an EGFR mutation using either direct DNA sequencing (n = 43) or our previously described DNA endonuclease-based method (18). Seven patients had gefitinib or erlotinib posttreatment specimens that contained an EGFR T790M mutation and all were detected by direct sequencing. All detected mutations were independently confirmed.
Blood sample collection and DNA extraction
Blood samples (average 5 mL each) were collected in BD Vacutainer CPT Cell Preparation Tube with Sodium Heparin (BD). Plasma was isolated according to the manufacturer's specifications and stored at -80°C until use. Plasma DNA was extracted using QIAamp DNA Micro Kit (Qiagen). DNA was eluted in 100 μL of Qiagen Buffer AE. In the DNA extraction optimization experiments two additional methods [Promega Wizard, (Promega) and NucleoSpin Plasma XS (Macherey-Nagel)] were also evaluated and used according to the manufacturer's recommended specifications.
Whole genome amplification
For whole genome amplification plasma DNA was processed either by a blunt-end ligation method described previously (19) or by an alternative method that favors the amplification of small, tumor-derived DNA (20). Whole genome amplification was carried out using GenomiPhi V2 DNA Amplification Kit (GE Healthcare).
Plasma DNA quantification using Alu qPCR
Primer sequences for Alu 115bp and Alu 247 bp fragments were previously described (21). Standard curve was constructed using serial diluted female genomic DNA (Promega; 0.01-100 pg DNA). Male genomic DNA (Promega) was used as a calibrator in the assay. The cycling conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 30 sec, 64°C for 30 sec, and 72°C for 30 sec. Reactions were run on an ABI 7500Fast real-time PCR instrument.
EGFR mutation analysis by Scorpion Amplification Refractory Mutation system Real-time PCR
EGFR mutation detection of the common EGFR-activating mutations (del E746_A750 and L858R) or the EGFR T790M resistance mutation were done using the EGFR Scorpion Amplification Refractory Mutation system (SARMS) technology (DxS Ltd.) as previously described (15). One microliter of plasma-derived or whole genome amplified DNA was added to 24 μL of master mix prepared according to manufacturer's instructions. The real-time PCR reactions were run on an ABI 7500Fast System and according to the manufacturer's recommended conditions. Comparative threshold values were calculated using 7500Fast System SDS Software. Positive samples fell into the window between the comparative threshold of the control assay, and the background comparative threshold and cutoff values were determined according to the manufacturer's instructions.
EGFR mutation analysis by WAVE/SURVEYOR
EGFR exons 18 through 21 were PCR-amplified using primers that flank the exonic regions. For the detection of insertion/deletion mutations, PCR products were loaded on to the WAVE system (Transgenomic Inc.) and resolved at 50°C. For the detection of point mutations, PCR products were subjected to enzymatic digestion using the SURVEYOR enzyme at 42°c, and the resulting products resolved on the WAVE at 50°C. Detailed protocols for exon-specific PCR and WAVE analysis were described previously (18).
Statistical analysis
Fisher's exact test was used to compare the effect of whole genome amplification on detection of EGFR mutations and to assess the association between EGFR mutation status and clinical response. Data were analyzed on a per patient basis. The Wilcoxon rank-sum test was used to compare the differences in time between the development of resistance and collection of plasma DNA in patients with and without EGFR T790M. All the exact P values were based on a two-sided hypothesis test and were computed using StatXact verson 6.1 (Cytel Software Corp.).
Results
Alu real-time PCR and optimization of plasma DNA extraction
We first established a DNA extraction procedure that resulted in the greatest yield of tumor-derived circulating DNA. Prior studies suggest that most malignant tumor-derived DNA varies in size (median, 544 bp; range, 185-926 bp) whereas apoptotic DNA from normal cells is more uniformly sized as 185 to 200 bp fragments (22). We thus adopted a previously developed real-time PCR assay to determine the ratio between Alu sequences of 115 bp (Alu115) and 247 bp (Alu247), and used it as the indication of DNA integrity (21). Alu115 represents both short, apoptotic DNA fragments and tumor-derived fragments (total circulating DNA), whereas Alu247 represents tumor-derived DNA alone. The ratio of Alu247/Alu115 was used to calculate the percentage of tumor DNA in total circulating DNA.
For these initial studies we evaluated plasma DNA from seven patients. These seven samples were not included in the larger study, nor were they subjected to whole genome amplification. Plasma DNA was extracted in parallel from each sample using three independent protocols: Qiagen, Promega Wizard, and NucleoSpin Plasma XS. The Alu 247/Alu115 ratio was determined for each sample prior to EGFR mutation analysis, and DNA input for mutational analyses was normalized to total circulating DNA (Alu115). The median total circulating DNA (Alu115) yields of the Qiagen, Promega Wizard, and NucleoSpin methods were 0.064 ng/μL, 0.021 ng/μL, and 0.086 ng/μL, respectively. The median Alu247/Alu115 ratio obtained using Qiagen, Promega Wizard, and NucleoSpin methods were 50.9%, 59.4%, and 10.9%, respectively. The DNA derived using the Qiagen extraction method was successfully amplified 100% of the time using both the SARMS and the WAVE/Surveyor methods. In contrast, DNA derived using the Promega Wizard or Nucleospin methods successfully amplified in only 75% or 67% of the reactions, respectively. Based on the high Alu247/Alu115 ratio and the ability to successfully amplify the DNA we used the Qiagen DNA extraction method for all subsequent studies.
Patient characteristics
Fifty-four patients were enrolled in this study (Table 1). Fifty of the 54 patients (93%) had received prior treatment with either gefitinib (n = 17; 31.4%) or erlotinib (n = 33; 61.1%) and all had developed disease progression at the time blood specimens were obtained. Four patients (7.5%) were never treated with either gefitinib or erlotinib and served as negative controls. The best response to prior therapy was partial response with 28 patients (56 %), followed by stable disease with 14 patients (28 %) and progressive disease with 8 patients (16 %). Tumor EGFR mutation status, obtained from baseline pre-gefitinib or -erlotinib treatment specimens, was available in 43 of 54 (80%) of patients (Table 1).
Table 1.
Patient characteristics
| No. of patients | n = 54 |
|---|---|
| Gender | |
| Male | 10 (18.5%) |
| Female | 44 (81.5%) |
| EGFR TKI treatment | |
| Gefitinib | 17 (31.4%) |
| Erlotinib | 33 (61.1%) |
| None | 4 (7.5%) |
| Response to prior EGFR TKI treatment | |
| Partial response | 28 (56%) |
| Stable disease | 14 (28%) |
| Progressive disease | 8 (16%) |
| Not treated | 4 (7.5%) |
| Tumor EGFR mutation | |
| Exon 19 deletion | 20 (37.0%) |
| L858R | 7 (12.9%) |
| L861Q | 1 (1.9%) |
| Exon 20 insertion | 2 (3.7%) |
| Wild type | 13 (24.1%) |
| Unknown | 11 (20.4%) |
Seventy-six plasma specimens were obtained from the 54 patients (median number per patient, 1; range, 1-5) and were used for DNA extraction. All DNA specimens were subjected to whole genome amplification, with the DNA quantified before and after whole genome amplification. The median concentrations were 0.252 ng/μL (range, 0.023-100.1 ng/μL) for unamplified plasma DNA samples and 52.3 ng/μL (range, 9.9-162.7 ng/μL) for whole genome-amplified DNA samples. Both unamplified plasma DNA and whole genome-amplified DNA specimens were used for subsequent genotyping studies.
EGFR mutation detection
We used two different methods, SARMS and WAVE/Surveyor, to detect EGFR activation and resistance mutations from plasma DNA. Both SARMS and WAVE/Surveyor technologies are PCR-based methods for mutation detection. SARMS uses a Scorpions primer/probe in a real-time PCR setting. Short probes allow greater allelic specificity and a lower background. The WAVE/Surveyor method combines standard PCR followed by an endonuclease digestion (Surveyor) that targets wild-type/mutant heteroduplexes. The resulting products are resolved on the WAVE HS system (18).
We first tested the sensitivity and specificity of detecting EGFR T790M with the SARMS assay using NSCLC cell lines with known EGFR T790M mutation status (H1975, H820, and H3255 GR, all known to contain an EGFR T790M mutation, and A549 that does not contain an EGFR T790M mutation). Using this assay, we determined the EGFR T790M allele frequencies for each of the cell lines: H1975 at 55%, H820 at 7%, H3255 GR at 2%, and A549 at 0%. These results were consistent with our own previous genotyping results using WAVE/Surveyor and published data (16, 23).
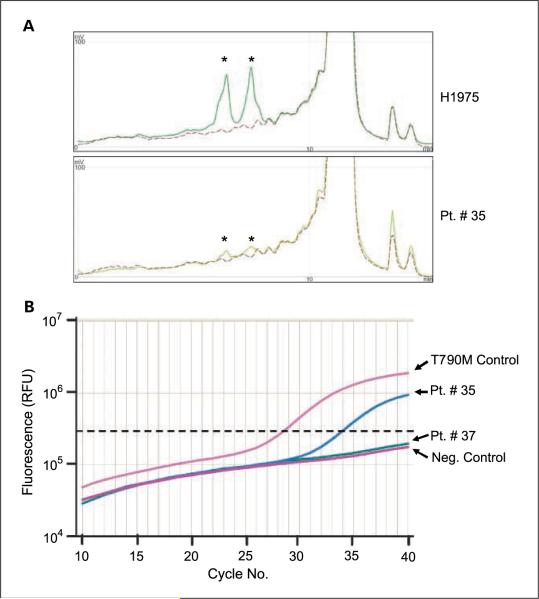
Next, we determined whether we could detect EGFR-activating mutations and the T790M resistance mutation in patient-derived plasma DNA. Based on previous reports (14, 15) and our determination of median patient plasma DNA concentration (0.252 ng/μL, which is equivalent to a median of 43 genome copies) in our sample cohort, we used 1 μL of patient plasma DNA in both the SARMS and WAVE/ Surveyor assays. Figure 1 depicts the detection of the EGFR T790M mutation in a representative patient plasma DNA sample using both the WAVE/Surveyor and the SARMS methods. Using the SARMS assay we detected 12 patients with EGFR del E746_A750, 7 patients with L858R, and 8 patients with EGFR T790M mutations. All plasma DNA samples were also independently PCR-amplified and screened for EGFR exon 19 to 21 mutations using WAVE/Surveyor as previously described (18). At the time of the study, the Scorpions assays were only available to detect two EGFR-activating mutations (del E746_A750 and L858R) and the EGFR T790M resistance mutation. Thus, we used the WAVE/Surveyor method to evaluate for the remaining EGFR mutations and also as a complementary approach to the SARMS assays. Using the WAVE/Surveyor method we detected EGFR exon 19 deletion mutations in 25 patients, no exon 20 insertion mutations, EGFR L858R mutations in 2 patients, and EGFR T790M mutations in 4 patients with. Of the 25 patients with EGFR exon 19 deletion mutations detected by WAVE/Surveyor, 11 were exon 19 deletions other than the del E746_A750 mutation. Such deletions were not a part of the SARMS assay. We compared the findings between these two mutation detection methods. The SARMS and WAVE/Surveyor detected EGFR del E746_A750 in a combined 15 patients, L858R in a combined 7 patients, and T790M in a combined 9 patients, with concordance rates of 73% (11 of 15), 28% (2 of 7), and 33% (3 of 9), respectively (Table 2).
Figure 1.
Detection of EGFR T790M using WAVE/Surveyor and SARMS. A, detection of EGFR T790M from the H1975 (EGFR L858R/T790M) cell line (top) and plasma DNA from patient 35 (bottom). Exon 20 of EGFR was amplified by PCR, the resulting product digested with Surveyor and analyzed using the WAVE-HSsystem (Materials and Methods). In the presence of EGFR T790M, two fragments (asterisk)are generated by Surveyor digestion (solid lines)fromthe positive control (H1975) and patient 35. The wild-type control (A549; dashed line)isuncut. B,SARMS analysis of EGFR T790M. Included are positive and negative control DNA samples and plasma DNA from patients 35 and 37. The horizontal dotted line represents the threshold. DNA from the negative control and patient 37 do not amplify above the threshold whereas DNA from the positive control and patient 35 both cross the threshold in the linear portion of the assay. Fluorescence was measured quantitatively in relative fluorescence units.
Table 2.
Comparison of SARMS and WAVE/ Surveyor methods in detecting EGFR exon 19 (del E746_A750), L858R, and T790M mutations from gefitinib/erlotinib-treated NSCLC patients
| EGFR mutation | Del E746_A750 | L858R | T790M |
|---|---|---|---|
| Plasma DNA alone | |||
| Total positive patients | 15 | 7 | 9 |
| SARMS-positive | 12 | 7 | 8 |
| WAVE/Surveyor-positive | 14 | 2 | 4 |
| Concordance | 11/15 | 2/7 | 3/9 |
| Plasma DNA and whole genome amplified | |||
| Total positive patients | 18 | 8 | 19 |
| SARMS-positive | 14 | 8 | 18 |
| WAVE/Surveyor-positive | 16 | 3 | 7 |
| Concordance | 12/18 | 3/8 | 6/19 |
NOTE: The data are displayed on a per patient basis.
Impact of whole genome amplification on plasma DNA-based mutation detection
We further investigated whether whole genome amplification facilitated the detection of additional EGFR mutations from plasma DNA. Whole genomeamplified DNA samples were screened for mutations in EGFR exons 19, 20, and 21 in an identical fashion to non-whole genome-amplified samples using both SARMS and WAVE/Surveyor assays. Using the SARMS assay we detected 13 additional EGFR mutations: 2 patients with EGFR del E746_A750, 1 with L858R, and 10 patients with EGFR T790M not detected from the plasma DNA. The WAVE/Surveyor method detected 7 additional patients with EGFR mutations: 3 with EGFR exon 19 deletions, 1 patient with an EGFR L858R mutation, and 3 patients EGFR T790M mutations not detected from the plasma DNA (Table 2). One of the additional EGFR exon 19 deletion mutations detected by the WAVE/Surveyor method one was a non -exon 19 del E746_A750 mutation, thus not assayed by SARMS method. Nine of 10 (90%) of the patients in which we detected an EGFR T790M using the SARMS assay also contained a concurrent EGFR-activating mutation whereas this occurred in 67% (2 of 3) of EGFR T790M containing patient specimens using the WAVE/Surveyor method.
We next combined the results obtained from non-whole genome-amplified and whole genome -amplified samples and compared the findings between SARMS and WAVE/Surveyor detection methods. The SARMS and WAVE/Surveyor detected EGFR del E746_A750 in a combined 18 patients, L858R in a combined 8 patients, and T790M in a combined 19 patients, with concordance rates of 67% (12 of 18), 38% (3 of 8), and 32% (6 of 19), respectively (Table 2).
Overall the effect of whole genome amplification seemed to have the greatest effect on the detection of EGFR T790M (Table 2). For EGFR del E746_A750 and L858R, whole genome amplification identified only 4 additional patients with mutations whereas for EGFR T790M whole genome amplification resulted in the identification of 10 additional patients (P = 0.011).
Concordance of primary tumor sequencing and clinical response with detection of plasma EGFR mutations
We compared the EGFR-activating mutation detected in plasma DNA with the tumor EGFR-activating mutation. For these studies we combined the findings from the SARMS and WAVE/Surveyor methods and included findings from the whole genome-amplified specimens. Tumor EGFR mutation status was known in 43 of 54 (80%) and not available in 11 of 54 (20%) of patients. Thirteen (13) of the 43 patients were EGFR wild-type (30%) whereas 30 (70%) had an EGFR-activating mutation in exons 19 to 21 (Table 1). Collectively the plasma-based detection methods identified 29 of 54 (54%) of patients as having an EGFR-activating mutation whereas 25 of 54 (46%) were EGFR wild-type. In the 43 patients whose tumor EGFR mutation status was known, we identified EGFR mutations from plasma DNA in 21 of 30 patients (70%). The overall concordance of tumor EGFR mutation with plasma EGFR mutation was 74% (32 of 43; Table 3). We also examined concordance as a function of the specific type of mutation (exon 19 deletion versus L858R). In the patients with a known tumor exon 19 deletion mutation there was an 85% (17 of 20) concordance with the plasma EGFR mutation whereas in those with a tumor L858R mutation the concordance rate was only 29% (2 of 7) with the plasma EGFR mutation (P = 0.011).
Table 3.
Summary of detecting EGFR-activating mutations from plasma DNA
| Plasma DNA |
|||
|---|---|---|---|
| Mutation | No Mutation | ||
| Tumor tissue | |||
| Mutation | 30 | 21 | 9 |
| No mutation | 13 | 2 | 11 |
| Not available | 11 | 6 | 5 |
| Response to prior EGFR TKI therapy | |||
| Partial response | 28 | 23 | 5 |
| Stable disease | 14 | 4 | 10 |
| Progressive disease | 8 | 2 | 6 |
| Untreated | 4 | 0 | 4 |
NOTE: NSCLC patients broken down based on known tumor EGFR mutations and based on clinical outcome with prior EGFR TKI therapy.
We also analyzed the findings based on prior response to therapy (Tables 1 and 3). EGFR-activating mutations were detected in plasma DNA from 23 of 28 (82%) patients with a complete response (CR)/partial response, 4 of 14 (28.5%) patients with stable disease, and 2 of 8 (25%) patients with progressive disease. EGFR-activating mutations detected in plasma DNA are associated strongly with a clinical response among the patients treated with gefitinib or erlotinib (P < 0.001).
Correlation of EGFR T790M detected in plasma DNA with prior drug response and tumor EGFR T790M
We evaluated the relationship with prior clinical response to gefitinib or erlotinib in patients in which EGFR T790M was detected in plasma DNA. Prior tumor-based studies suggest that EGFR T790M can be detected in 50% of NSCLC patients with a prior response (CR or partial response) to gefitinib or erlotinib therapy (10, 11). EGFR T790M was detected from plasma DNA in 35% (19 of 54) patients in this study. In the 28 patients that had a prior partial response to either gefitinib or erlotinib EGFR T790M was detected in the plasma DNA in 15 of 28 (54%) patients (Table 4). EGFR T790M was detected in 5 of 7 patients (71%) for whom posttreatment biopsy specimens were available and had been confirmed to contain an EGFR T790M by direct sequencing (Table 4). EGFR T790M was also detected in 4 of 14 (29%) of patients with stable disease. One of the four patients had a concurrent EGFR-activating mutation detected from plasma DNA. EGFR T790M was detected in none (0 of 8; 0%) of the patients with progressive disease to gefitinib or erlotinib or in patients who had never been treated with these agents (0 of 4; 0%). Collectively, these findings show that the EGFR T790M mutation detected in plasma DNA is associated strongly with a prior clinical response to gefitinib or erlotinib (P = 0.004). We further evaluated the time between the clinical development of resistance and plasma collection and the presence or absence of EGFR T790M in patients with a prior clinical response to gefitinib or erlotinib. The time between the development of clinical resistance and plasma collection was numerically longer but not significantly different (P = 0.829; Wilcoxon rank-sum test) in patients in whom we did not identify an EGFR T790M (median, 68 days; range, 1-940 days) compared with those in which an EGFR T790M was identified from plasma DNA (median, 38 days; range, 1-817 days).
Table 4.
Comparison of NSCLC patient clinical response to prior EGFR TKI therapy and known EGFR T790M-containing tumors with detection of EGFR T790M using plasma DNA
| Plasma EGFR T790M |
|||
|---|---|---|---|
| Yes | No | ||
| Response to prior EGFR TKI Therapy | |||
| Partial response | 28 | 15 | 13 |
| Stable disease | 14 | 4 | 10 |
| Progressive disease | 8 | 0 | 8 |
| Untreated | 4 | 0 | 4 |
| Tumor EGFR T790M | 7 | 5 | 2 |
Discussion
EGFR inhibitors are effective therapies against EGFR-mutant NSCLC (1-6). Given that only 10% to 15% of Caucasian NSCLC patients harbor EGFR-activating mutations, it is important to identify this subgroup of lung cancer patients (24). However, not all lung cancer diagnostic specimens are amenable to genotyping because they often contain only a limited number of cancer cells. Potential solutions to this barrier include developing more sensitive diagnostic methods and/or developing noninvasive diagnostic methods. The latter can also potentially be used to study secondary resistance alterations, such as EGFR T790M, that occur during the course of treatment.
Using both SARMS and WAVE/Surveyor we detected both the EGFR-activating and the EGFR T790M resistance mutation from plasma DNA in 70% of patients in which these mutations were known to occur in the patients' tumor specimens (Tables 3 and 4). These findings are similar to two smaller studies of plasma-based DNA analyses of EGFR-activating mutations and show a 70% concordance with the primary tumor mutation (14, 15). However, they differ from a more recent larger study which used the SARMS technique which detected only 7 (39%) EGFR mutations in plasma DNA from 18 known EGFR-mutant patients (25). The higher plasma EGFR mutation detection rate in the current study may be a result of using two mutation detection methods (SARMS and WAVE/Surveyor) and combining these with whole genome amplification. In fact, in our study SARMS alone detected only 14 (47 %) plasma EGFR mutations from 30 patients with tumor EGFR mutations (data not shown). Improvements in the sensitivity of genotyping, such as by using single-molecule sequencing or digital PCR, may further improve the ability to detect EGFR-activating mutations from plasma DNA using just a single technique as opposed to a combination of methods as in the current study.
Our study also included a large number of known EGFR-mutant patients (30) compared with prior studies(14, 15, 25). Given that, we are able to compare the ability to detect the common EGFR mutations (exon 19 deletion and L858R) from plasma DNA. Intriguingly, our techniques were significantly more likely to detect an exon 19 deletion mutation (85%; 17 of 20) than L858R (29%; 2 of 7) from plasma DNA. Although we cannot completely exclude the possibility, based on our prior studies suggesting a similar sensitivity for detecting EGFR exon 19 deletions and L858R using DNA derived from tumor tissue, this observation is unlikely due to differences in the sensitivity assays in detecting exon 19 deletion and L858R mutations (18). It is possible that this reflects a biological difference between these two different EGFR mutant cancers and needs to be evaluated in future noninvasive DNA-based studies.
In two patients EGFR-activating mutations were identified from plasma DNA but were not detected in the corresponding tumor specimen (Table 3). Although these findings may represent false positive results of our current technique, a further look into the details of these patients may suggest an alternative hypothesis. In one of the patients, the EGFR wild-type tumor specimen was obtained from the time of surgery following chemotherapy and radiation. Following tumor relapse, the patient was subsequently treated with gefitinib and after >24 months developed disease progression with new brain metastases, at which time the plasma sample was obtained. Analysis of DNA from the plasma sample showed both EGFR del E746_A750 and T790M mutations. EGFR sequencing of the brain metastasis also showed both EGFR del E746_A750 and T790M mutations. These findings suggest either clonal evolution of the patient's NSCLC or a false negative result for EGFR delE746_A750 from the original tumor sequencing. The second patient with an EGFR wild-type pretreatment tumor specimen was treated with erlotinib for 22 months with stable disease prior to developing disease progression, at which time an L858R mutation alone was detected from plasma DNA. In the pathologist's description of her tumor it is noted that her tumor specimen contains a significant portion of inflammatory cells that could interfere with genotyping by direct sequencing. Thus, it is possible that the tumor EGFR sequencing represents a false negative. Prolonged stable disease with erlotinib treatment has previously been observed in patients with EGFR-activating mutations (26).
Our studies detected EGFR T790M from plasma DNA from 54% of patients with a prior clinical response to gefitinib or erlotinib (Table 4), which is similar to the expected frequency based on prior tumor sequencing studies (10, 11). Furthermore, we identified EGFR T790M in 71% (5 of 7) using plasma DNA in patients whose tumors were known to contain an EGFR T790M mutation. Whole genome amplification had the most impact in the detection of EGFR T790M and resulted in a significantly (P = 0.011) greater number of patients in whom T790M could be identified by SARMS (Table 2). This finding may reflect the biology of EGFR T790M which can often exist as a rare allele and thus may go undetected in the absence of whole genome amplification (16). Our findings would suggest that future noninvasive studies aimed at detecting EGFR T790M should incorporate whole genome amplification to increase the likelihood of detecting this resistance mutation. Noninvasive genotyping methods, such as those described in this and other studies, may be helpful in clinically identifying patients with EGFR T790M and should be incorporated into future clinical trials of irreversible EGFR inhibitors (25). At the current time, noninvasive genotyping should not replace a repeat tumor biopsy, which remains the gold standard. However, given the paucity of posttreatment tumor biopsies from gefitinib/erlotinib-resistant NSCLC patients, these methods, although not 100% sensitive, may aid in determining whether irreversible EGFR inhibitors have a differential clinical effect in NSCLC patients with EGFR T790M. Furthermore noninvasive methods may be used to serially monitor the evolution of EGFR T790M during the course of drug treatment.
We also detected EGFR T790M in four patients that had stable disease as their primary clinical response to gefitinib or erlotinib therapy (Table 4). The mechanism(s) of acquired resistance in NSCLC patients that develop stable disease to gefitinib or erlotinib therapy, especially in cancers that are EGFR wild-type, is completely unknown. It possible that, at least in some patients, EGFR T790M also contributes to the development of acquired resistance. Noninvasive mutation detection methods, especially when used in the context of prospective clinical trials, may be well suited to begin to explore this important clinical question.
There are potential limitations to all plasma-based tumor DNA genotyping methods. The first is the ability to determine whether the isolated DNA is truly tumor-derived. For this reason we examined the Alu247/115 ratios and used this as a way to select the most effective DNA isolation method that would result in the greatest likelihood and yield of tumor-derived plasma DNA. A positive result (such as identification of EGFR T790M) is more meaningful because this is not a normal variant but tumor-derived. However, a negative result is more challenging as it is hard to determine whether such a finding represents a true negative or the lack of tumor-derived DNA. One potential method to overcome this limitation is to specifically isolate circulating tumor cells and use these for the genotyping studies (25). Currently this technique requires specialized equipment, and blood specimens need to be processed within hours after being drawn from the patient. An advantage of the plasma-based DNA method is that it does not require specialized equipment for DNA isolation. Furthermore, once the plasma has been separated, the specimen can be frozen and DNA isolated at a future date, thus allowing collection of samples from multi-institutional clinical studies. A second limitation is whether the plasma-derived DNA changes are truly reflective of the genomic changes of the bulk of the cancer. It will be important to correlate the plasma-based studies with tumor-based studies. This is particularly challenging in patients that develop gefitinib/erlotinib resistance as very few on them undergo repeated tumor biopsies. Clinical correlation provides an alternative surrogate to tumor-based studies as was used in the current study. Importantly we did not detect EGFR T790M in patients that developed progressive disease to gefitinib/erlotinib or in those that were never treated with these agents. In these patient subsets, we would not expect to detect EGFR T790M as it is thought to arise as a result of drug exposure.
An additional factor that needs to be considered in the noninvasive evaluation of EGFR T790M is the time between the development of resistance and the collection of the plasma DNA specimen for evaluation. It is possible that the ability to detect EGFR T790M will decrease over time, due to lack of selection pressure, once a patient has developed clinical progression and stopped gefitinib or erlotinib treatment. Although our studies did not show a significant time difference between resistance and plasma collection in patients with and without EGFR T790M, these observations will need to be further validated in a prospective clinical trial.
An additional limitation of the current and other noninvasive genotyping studies is the ability to only detect one mechanism of gefitinib/erlotinib resistance. MET amplification occurs in ~20% of NSCLC that develop acquired resistance to gefitinib/erlotinib and can occur both independent of and concurrently with EGFR T790M (8, 9). The amplified MET sequences are wild type and thus likely impossible to detect using a plasma DNA-based assay. MET amplification may be more effectively identified by examining copy number changes by fluorescent in situ hybridization specifically on individual circulating tumor cells. The recent advances in isolating CTC suggest that this may be possible in the near future and can be combined with genotyping studies to examine both mechanisms of geftinib/erlotinib resistance simultaneously.
Translational Relevance.
The epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKI) gefitinib and erlotinib are effective therapies for patients with non - small cell lung cancer (NSCLC) that harbor activating mutations in EGFR. However, all patients ultimately develop progressive disease (acquired resistance) while receiving treatment with gefitinib or erlotinib. The cause of acquired resistance in 50% of patients is a secondary EGFR mutation (EGFR T790M). Second-generation EGFR TKIs are now entering clinical development that can inhibit the growth of cancers with EGFR T790M and may be clinically effective. Very few patients, however, undergo repeated tumor biopsies at the time of developing acquired resistance. In this study we identify both EGFR-activating and the EGFR T790M resistance mutation from plasma DNA derived from patients that have clinically developed resistance to gefitinib or erlotinib. This noninvasive method may help identify NSCLC patients who may benefit from second-generation EGFR kinase inhibitors.
Acknowledgments
Grant support: NIH RO1CA114465-03 (P.A. Jaänne, B.Y. Yeap), National Cancer Institute Lung SPORE P50CA090578 (P.A. Jaänne, B.Y. Yeap), the Hazel and Samuel Bellin research fund (P.A.Jaänne), and Department of Defense W81 XOSHO6-1-0303 (P.A. Jaänne). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Disclosure of Potential Conflicts of Interest P. Janne, royalties from intellectual property licensed to Genzyme; Transgenomic, AstraZeneca, Roche, consultant.
References
- 1.Inoue A, Suzuki T, Fukuhara T, et al. Prospective phase II study of gefitinib for chemotherapy-naive patients with advanced non-small-cell lung cancer with epidermal growth factor receptor gene mutations. J Clin Oncol. 2006;24:3340–6. doi: 10.1200/JCO.2005.05.4692. [DOI] [PubMed] [Google Scholar]
- 2.Paz-Ares L, Sanchez JM, GarcIía-Velasco A, et al. A prospective phase II trial of erlotinib in advanced non-small cell lung cancer (NSCLC) patients (p) with mutations in the tyrosine kinase (TK) domain of the epidermal growth factor receptor (EGFR) J Clin Oncol. 2006;24 Abstract 7020. [Google Scholar]
- 3.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–9. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 4.Tamura K, Okamoto I, Kashii T, et al. Multicentre prospective phase II trial of gefitinib for advanced non-small cell lung cancer with epidermal growth factor receptor mutations: results of the West Japan Thoracic Oncology Group trial (WJTOG0403) Br J Cancer. 2008;98:907–14. doi: 10.1038/sj.bjc.6604249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asahina H, Yamazaki K, Kinoshita I, et al. A phase II trial of gefitinib as first-line therapy for advanced non-small cell lung cancer with epidermal growth factor receptor mutations. Br J Cancer. 2006;95:998–1004. doi: 10.1038/sj.bjc.6603393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sutani A, Nagai Y, Udagawa K, et al. Gefitinibfornon-small-cell lung cancer patients with epidermal growth factor receptor gene mutations screened by peptide nucleic acid-locked nucleic acid PCR clamp. Br J Cancer. 2006;95:1483–9. doi: 10.1038/sj.bjc.6603466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–8. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 8.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 9.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinibor erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–7. doi: 10.1073/pnas.0710370104. Epub 2007 Dec 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balak MN, Gong Y, Riely GJ, et al. Novel D761Yand common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12:6494–501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 11.Kosaka T, Yatabe Y, Endoh H, et al. Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clin Cancer Res. 2006;12:5764–9. doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]
- 12.Engelman JA, Zejnullahu K, Gale CM, et al. PF00299804, an irreversible pan-ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Res. 2007;67:11924–32. doi: 10.1158/0008-5472.CAN-07-1885. [DOI] [PubMed] [Google Scholar]
- 13.Li D, Ambrogio L, Shimamura T, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008;27:4702–11. doi: 10.1038/onc.2008.109. Epub 2008 Apr 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura H, Kasahara K, Kawaishi M, et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res. 2006;12:3915–21. doi: 10.1158/1078-0432.CCR-05-2324. [DOI] [PubMed] [Google Scholar]
- 15.Kimura H, Suminoe M, Kasahara K, et al. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA) Br J Cancer. 2007;97:778–84. doi: 10.1038/sj.bjc.6603949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engelman JA, Mukohara T, Zejnullahu K, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116:2695–706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–16. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 18.Janne PA, Borras AM, Kuang Y, et al. A rapid and sensitive enzymatic method for epidermal growth factor receptor mutation screening. Clin Cancer Res. 2006;12:751–8. doi: 10.1158/1078-0432.CCR-05-2047. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Harris L, Mamon H, et al. Whole genome amplification of plasma-circulating DNA enables expanded screening for allelic imbalance in plasma. J Mol Diagn. 2006;8:22–30. doi: 10.2353/jmoldx.2006.050074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mamon H, Hader C, Li J, et al. Preferential amplification of apoptotic DNA from plasma: potential for enhancing detection of minor DNA alterations in circulating DNA. Clin Chem. 2008;54:1582–4. doi: 10.1373/clinchem.2008.104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Umetani N, Kim J, Hiramatsu S, et al. Increased integrityof free circulating DNA in seraof patientswith colorectal or periampullary cancer: direct quantitative PCR for ALU repeats. Clin Chem. 2006;52:1062–9. doi: 10.1373/clinchem.2006.068577. [DOI] [PubMed] [Google Scholar]
- 22.Giacona MB, Ruben GC, Iczkowski KA, Roos TB, Porter DM, Sorensen GD. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas. 1998;17:89–97. doi: 10.1097/00006676-199807000-00012. [DOI] [PubMed] [Google Scholar]
- 23.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoSMed. 2005;2:1–11. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–46. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 25.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–77. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackman DM, Yeap BY, Lindeman NI, et al. Phase II clinical trial of chemotherapy-naive patients > or = 70 years of age treated with erlotinib for advanced non-small-cell lung cancer. J Clin Oncol. 2007;25:760–6. doi: 10.1200/JCO.2006.07.5754. [DOI] [PubMed] [Google Scholar]