Abstract
Micromixing is a crucial step for biochemical reactions in microfluidic networks. A critical challenge is that the system containing micromixers needs numerous pumps, chambers, and channels not only for the micromixing but also for the biochemical reactions and detections. Thus, a simple and compatible design of the micromixer element for the system is essential. Here, we propose a simple, yet effective, scheme that enables micromixing and a biochemical reaction in a single microfluidic chamber without using any pumps. We accomplish this process by using natural convection in conjunction with alternating heating of two heaters for efficient micromixing, and by regulating capillarity for sample transport. As a model application, we demonstrate micromixing and subsequent polymerase chain reaction (PCR) for an influenza viral DNA fragment. This process is achieved in a platform of a microfluidic cartridge and a microfabricated heating-instrument with a fast thermal response. Our results will significantly simplify micromixing and a subsequent biochemical reaction that involves reagent heating in microfluidic networks.
Introduction
A compelling advantage of microfluidics comes from the parallel networking of highly sensitive, fast microfluidic components, especially for biochemical reactions, separations, and detections.1-2 In microfluidic networks,3-6 sufficient mixing of heterogeneous solutions is an initial, yet essential, step that provides the basis of biochemical reactions. However, mixing is difficult in the microfluidic regime because of two reasons: laminar flow and slow diffusion. In a smooth-walled microchannel, multiple streams usually flow in a parallel manner; viscous force is typically much more dominant than inertial force (i.e. low Reynolds number). So the transverse motion that enhances mixing is repressed, thus making micromixing process purely diffusive. However, diffusion time for homogeneous streams is excessively long compared with the time for subsequent biochemical reactions, due to the relative large size of biomolecules.7
Desirable micromixing processes in microfluidic networks need to be simple and compatible with system integration, while maintaining the ability to homogenize different solutions in a proper time scale prior to biochemical reactions. Numerous studies,8-12 however, focuses solely on the performance of micromixing and have paid relatively little attention to achieving simplicity for the topological structure, fabrication process, and off-chip setup of their micromixers. Many of the micromixing processes in these studies were based on a passive mechanism where inherent stationary structures of microchannels were used. For example, a microchannel with groove-patterns were used to enhance efficient micromixing.12-13 Due to a 3-D shape, the construction of the micromixer needs relatively laborious two-step microfabrication for the microchannel. In contrast, a curved microchannel that has a greatly simple 2-D shape harnessed micromixing.14 Although the mixing scheme was elegant, micromixing was only effective at a relatively high Reynolds number of ~10. Moreover, these passive methods, including even those using simple micromixers,15 could not function without external pumps for the micromixing, often resulting in enormous system volume when the micromixers are integrated in the microfluidic networks. Also, they often require separate chambers for subsequent biochemical reactions, thus adding more complexity to microfluidic networks.
Here, we present how temperature-programmed natural convection leads to significant mixing in the microfluidic regime. Most important, this study enables micromixing and a subsequent biochemical reaction in a single microfluidic chamber, within a simple pumpless platform (Figure 1). As a model application, we sequentially show micromixing and PCR in a single chamber, while maintaining a fast thermal response. Our system consists of two separate subunits with distinct functions: (a) a microfluidic cartridge and (b) a microfabricated heating-instrument. The microfluidic cartridge has simple 2-D microchannels and a chamber. In contrast to previous methods, our microfluidic cartridge uses capillarity16 to control the merging of two streams and their filling of the chamber; this step is an autonomous process without any external pumps. Also, solutions can be injected into inlets simply by pipettes, thus removing interfacing-tubes for external pumps. The microfabricated heating-instrument incorporates mixing and reaction heaters (Figure 1C). The mixing heaters, composed of single and dual heaters, induce natural convection for homogenizing the different solutions within the microfluidic chamber. Afterwards, the reaction heaters are used to implement the PCR process. By programming the heaters with a time sequence of applied voltages, the sample transport, micromixing, and PCR are all autonomously performed in a seamless sequence.
Figure 1.
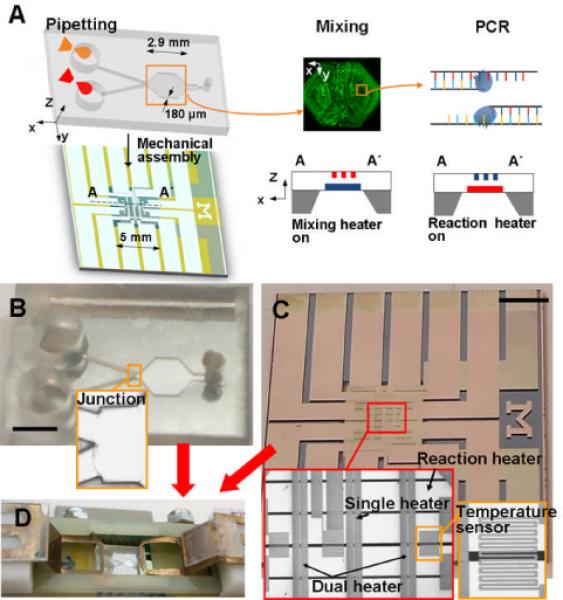
Single-chamber micromixing and PCR. (A) Sequential process of pipetting, micromixing, and PCR. After injecting solutions, their streams fill the chamber by capillarity. Upper- and lower-level heaters embedded in a membrane are used for micromixing and PCR, respectively. Natural convection induces micromixing. (B) Microfluidic cartridge. The microfluidic cartridge is a disposable component, which has simple 2-D shape microchannels and a chamber where the height is 180 μm. Sample transport is driven by capillarity in the microfluidic cartridge. The PDMS cartridge slab is coated with parylene for preventing evaporation of solutions. A stainless-steel film of 8 μm thickness is used as the sealing membrane at the bottom of the chamber. (C) Microfabricated heating-instrument. The microfabricated heating-instrument is a component for repeated use that incorporates temperature sensors and heaters within the membrane. The single and dual heaters are for micromixing, and the reaction heater is for PCR. (D) Assembly of heating instrument and microfluidic cartridge components. Scale bars, 3 mm.
Experimental Section
Microfabrication of the Microfluidic Cartridge
The microfluidic cartridge (Supporting information, Figure S3) is composed of a polydimethylsiloxane (PDMS, Sylgard 184, Ellswoth Adhesive) slab and an 8 μm-thick stainless-steel film (SUS304, Nilaco). The microfluidic channels and the chamber in the PDMS were fabricated using soft lithography.17 The mold for the soft lithography was a 4-inch Si wafer, which was patterned by deep reactive ion etch, and the surface of the mold was hydrophobically treated by SF6 gas. The precursor of PDMS was cured on the mold at room temperature and then was released from the mold. Then inlet and outlet holes were punched by using biopsy punches. To prevent vapor evaporation during the micromixing and PCR processes, a 3 μm-thick parylene layer18 was deposited on the replica-molded PDMS slab. A SU-8 (SU-8 5, Microchem) glue layer was used for the bonding of the PDMS slab and the stainless-steel film as follows: first, a 3 μm-thick film of SU-8 was spin-coated onto a silicon wafer and transferred to the PDMS slab by a contact printing method. Then the PDMS slab and the stainless-steel film were bonded together at a mild pressure followed by UV exposure and curing at 80°C for 2 min. This method is similar to the adhesive bonding process by Wu et al.19
Microfabrication of the Heating Instrument
The heating instrument was built by a conventional microfabrication process (Supporting information, Figure S4). The starting substrate was a 4-inch silicon wafer of 450-μm thickness. We thermally grew a 1-μm oxide layer on the both sides of the wafer. Then the backside of the wafer was photolithographically patterned. The substrate was dipped in a buffered hydrofluoric acid solution; simultaneously, the oxide layer on the front side of the wafer was removed for anodic bonding, and the one on the backside of the wafer was patterned as an etch mask of silicon. A glass wafer (Pyrex 7740, Sensors Prep Services, 100 μm in thickness) was anodically bonded to the silicon wafer. Ti/Pt reaction heaters (20 nm/100 nm in thickness) were patterned on the glass layer by a lift-off process using an acetone solution. Then, a 4 μm-thick oxynitride layer was deposited by a PECVD process for electrical passivation. Next, mixing heaters and temperature sensors, also made with Ti/Pt (20 nm/100 nm in thickness) layers, were formed by a lift-off process. A 1 μm thick oxynitride layer was deposited again on top of these heaters and the temperature sensors for electrical passivation. The silicon under the glass was removed by an ethylene-diamine-pyrocatechol solution. Finally, the wafer was diced into individual chips, followed by the etching of the glass layer in a hydrofluoric acid solution (49% concentration). The total thickness of the membrane combining the glass and oxynitride layers together was approximately 10 μm.
Assembly of the Microfluidic Cartridge and the Microfabricated Heating-Instrument
When the microfluidic cartridge was placed on the heating instrument, we used mechanical clamps to secure close contact between the two subunits. A mild pressure was applied on the top surface of the microfluidic cartridge to prevent the buckling of the stainless-steel membrane. A thermoelectric cooler (MI1021T, Marlow industries) under the heating instrument was used for fast cooling of the microfluidic cartridge. To effectively cool the chamber while maintaining fast heating at low power, we made a 60 μm air-gap (Supporting information, Figure S5) between the thermoelectric cooler and the membrane at the heating instrument.
Temperature Control
The calibration of the resistive-type temperature-sensors was performed with the microfabricated heating-instrument incubated in a convection oven. The resistances of the sensors were measured using a four-point probe technique at varying temperature (Supporting information, Figure S6), and the data were recorded using a computer program (LabVIEW 7.1, National instruments). The slope and intercept from a linear fit were used to obtain the temperature-resistance correlation for the temperature sensing. For micromixing, open-loop temperature-control was used. In the single heating (SH) and dual heating (DH) modes, a constant voltage was applied to the heaters using a DC power supply (E3646A, Agilent), whereas, in the alternating heating (AH) mode, square pulses were applied using a function generator (33250A, Agilent) and an op amp (OPA544T, Texas instrument). In the PCR process, temperature was controlled by varying the voltage of a DC power supply (Model 1760, BK precision) with a LabVIEW program based on a proportional-integral algorithm.20, 21 The temperature of the thermoelectric cooler was maintained at 17±1°C.
Measurement of Flow Speed and Micromixing
In the measurement of the flow speed at steady-state, we used fluorescent microbeads of 8 μm diameter (35-3, Duke scientific) as flow tracers. We obtained the value of the local flow speed from the length of a particle pathline captured by image analysis software (ImageJ, National Institutes of Health) and its corresponding flight time. The average and the maximum flow speeds were measured for microbeads flowing on the mid plane of the chamber (i.e., the half height of the chamber). Although microbeads on other planes could be measured due to the depth of field of the microscope (±25 μm), the measurement error was estimated to be 6.3% assuming a parabolic velocity profile across the chamber height.
Standard deviation, (σ = ⟨(I - ⟨I⟩)2⟩½), was used to quantify the degree of mixing, where I is the normalized intensity of each pixel (Supporting information, Figure S7). The grayscale value of each pixel's intensity was processed by using computer software (MATLB, Mathworks). To provide optical access to the vertically placed microfluidic chamber for a stereo microscope (SZX16, Olympus) equipped with a CCD camera (MP3.3, Qimaging), we used a 45° inclined mirror (Supporting information, Figure S8). We used a fluorescein (46980, Sigma-aldrich) with low temperature sensitivity22 to visualize the micromixing process. The solution used in the measurement of flow speed and the micromixing-degree commonly contained sucrose (S1888, Sigma-aldrich) of 10% (w/w) and sodium dodesyl sulfate (L4390, Sigma-aldrich) of 1.8% (w/w). The sucrose was added to increase the solution density up to 1.05 g/cm3, which allows neutral buoyancy of the suspended fluorescent microbeads at room temperature; the calculated viscosity of the solution resulting from adding sucrose was 1.35 mPa/s at room temperature. The sodium dodesyl sulfate served as a surfactant to help the channel filling and merging processes at the junction of the microchannels.
PCR
A DNA fragment of 690 bp from the influenza viral strain A/LA/1/87 was amplified using the on-chip PCR.20, 21 For obtaining the DNA sample, the hemaggultinin gene (HA1) region of influenza viral RNA was reverse transcribed, amplified, ligated into the pGEM-T vector, and cloned into E. coli. The cloned plasmid was used to synthesize RNA in vitro with T7 RNA polymerase; then the RNA samples were subject to reverse transcription to produce the final DNA samples that were used for the on-chip PCR. For the micromixing-PCR process, two different solutions were injected in the two inlets, respectively. A 2 ng/μL DNA template in a TE buffer with Tween-20 was injected in one inlet. A reaction mixture of 0.2 mM dNTP, 60 mM Tris-HCl, 15 mM NH4SO4, 1.5 mM MgCl2, 0.3 mM primers, and Taq DNA polymerase at 50 units/mL (Invitrogen) was injected in the other inlet. Tween-20 was used to match the surface tension of the two solutions. Mineral oil was used in the two inlets and the outlet for preventing the evaporation of solutions. The forward and reverse primers used here are 5′-GTTTGTTTCTCTGGTACATTCCGC-3′ and 5′-CAACTGTTACCCTTATGATGTGCC-3′, respectively. The thermocycling protocol consisted of a predenaturation process at 94°C for 5 s, then an amplification process following a temperature cycle of 94°C for 5 s, 55 °C for 10 s, and 72 °C for 20 s, and a final elongation process at 72°C for 30 s. We varied the total cycle numbers as 10, 20, and 30. The PCR product in each total cycle was collected in a tube from three PCR runs to compensate for the sample loss during the collecting and dispensing processes. Finally, the reaction sample in each tube was dispensed with a 1.5 μL volume in a single lane of a 1% agarose gel electrophoresis prestained with ethidium bromide.
Results and Discussion
Natural Convection-driven Flows in a Chamber
In microfluidic devices, typical chamber height is in the range of a few tens or hundreds of micrometer. Due to this relatively small size, it has been commonly accepted that natural convection is weak or even negligible in the microfluidic regime. In fact, the pathlines (Figure 2A), shown by the trajectories of individual fluorescent particles, indicate that the convective flow is weak and probably of no use for micromixing, where the microfluidic chamber has a height as small as 180 μm, and gravity acts normal to the chamber-plane (x-y plane in Figure 2A).
Figure 2.
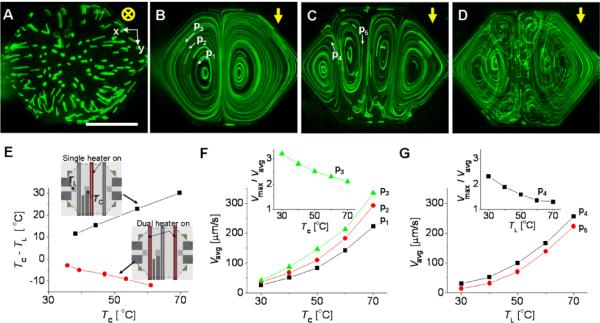
Natural convection-driven flows in the chamber. (A to D) Flow trajectories of fluorescent microparticles taken for 35 s. Measured maximum temperatures, Tmax, in A to D are 52, 51, 46, and 50°C, respectively. The yellow arrows indicate the gravity direction, and the white arrows depict the flow direction of the individual fluorescent particles of 8 μm diameter. The single heater is turned on in A and B and the dual heater is turned on in C. The single and dual heaters are alternatively turned on and off in D. To see the vertical image of B to D, we used a 45° inclined mirror (Supporting information, Figure S8) (E) Temperature gradient in the chamber. TC and TL are the fluid temperatures measured at the temperature sensors of the center and the left of the chamber-region, respectively. (F and G) Flow speed of the fluorescent microparticles in B and C. The single heater is turned on in F and the dual heater is turned on in G. Vavg is the flow speed averaged over the individual microparticles in a single loop, whereas Vmax is the maximum flow speed for the same loop. The height of the chamber is 180 μm. Scale bar, 1 mm.
Natural convection is generated only when buoyancy force overcomes the resistance imposed by viscous force. Buoyancy force (Fb) is induced by density gradient and gravity, and strongly dependant on the characteristic length (L) of the chamber as Fb ~ L3. When a heater is on the chamber-plane, L is the height of the chamber if the chamber-plane is perpendicular to gravity whereas L is the width of the chamber if the chamber-plane is parallel to gravity. Although the typical height of microfluidic chamber is in the micrometer scale, the width of the chamber can be more than one millimeter. Following the above simple principle, we can enhance natural convection in the microfluidic device by simply flipping the chamber 90° to make the chamber-plane parallel to gravity. This configuration greatly increases L from 180 μm to ~1.4 mm, thus enhancing Fb almost a thousand orders. As shown in Figure 2B, natural convection is significantly enhanced. In this case, the Rayleigh number, which is the ratio of buoyant flow-driving forces to diffusive flow-resisting forces, is calculated to be 2.6×103, thus indicating that the process is primarily driven by buoyant force.
The flow pattern in Figure 2B was obtained with the single heater (Figure 1B) turned on, namely the single heating (SH) mode. The SH mode generated one pair of counter-rotating vortices symmetrically positioned along the chamber center where the single heater was located. The number of vortices increased when we used the dual heater, namely the dual heating (DH) mode. The DH mode generated two pairs of counter-rotating vortices (Figure 2C), which were also symmetric along the chamber center. As shown in Fig 2B and 2C, the pathlines formed nearly closed loops, where the starting and ending points of each loop are only slightly different. When the single and dual heaters were alternatively turned on and off, we could obtain new flow patterns (Figure 2D); we refer to this mode as the alternating heating (AH) mode.
The temperature of the chamber was measured using two resistive-type temperature-sensors integrated in the microfabricated heating-instrument. The temperature data reasonably represent the temperature distribution according to our theoretical simulation (Supporting information, Figure S1), which indicates that the spatial temperature gradient primarily exists in the lateral direction and is symmetric about the chamber center. In the SH mode the temperature is high at the center of the chamber, whereas in the DH mode the temperature is low at the same position (Figure 2E). The flow patterns of the SH and DH modes (Figure 2B and 2C) are affected by the temperature gradient: the fluid moves up in the hot temperature regions near the activated heaters, and moves down in the cold regions. Although we found that thermal expansion deflected the membrane at the bottom of the microfluidic cartridge during the device operation, (Supporting information, Figure S2), this deflection did not change the flow patterns in the SH and DH modes and enabled close contact between the microfluidic cartridge and the microfabricated heating-instrument.
The average flow speeds (Figure 2F and 2G), which were measured for each single loop (Figure 2B and 2C), polynomially grow with the temperature. At a given geometry the flow speed (V) is influenced by thermal expansion coefficient (β), temperature gradient (ΔT), and kinematic viscosity (ν) of the solution as V ∝ βΔT/ν.23 Increasing the temperature of the chamber causes both ΔT and β to increase but ν to decrease. Thus, the net result of heating the solution is the rapid increase of the flow speed. In addition, the flow is accelerated near heaters, yielding maximum speed near the heating region. The maximum speed at the single loop is several times (1.3 - 3.1) faster than the average speed at the single loop (Figure 2F and 2G Insets).
Both the SH and DH modes can generate vortex flow-patterns for micromixing. However, with these modes alone, the vortices in each closed loop do not overlap and the flow is relatively fast only in a small region of the chamber near each activated heater. In contrast, micromixing can be significantly enhanced when we use the AH mode (Figure 2D): the device operation in the AH mode can overlap the pathlines of different closed loops and increase the fast flow regions by the continuous alternation of the SH and DH modes. In previous studies of flow-generation by natural convection, the flow patterns had only one24-27 or two28 vortices, which would be insufficient for micromixing according to results in our following study on micromixing.
Micromixing
We characterized the micromixing process in the microfluidic chamber using a fluorescent dye. Two solutions were first injected into the two inlets using pipettes, and then their streams filled each channel, merged at the junction, and finally reached the microfluidic chamber (1 μL, 180 μm-heights) by capillarity (Supporting information, Movie S1). The essential part of the capillarity-based stream control is the merging process at the junction; we previously showed that merging process can be passively controlled by setting an appropriate aspect-ratio of the microchannels.16
When the streams reached the outlet, heat was applied for the micromixing. The micrographs in Figure 3A show the time evolution of micromixing process in each heating mode. When the heaters were turned on, the flows in the SH and DH modes started to move up along the heaters. The flow motion outside the heating region was, however, relatively slow, and individual flows rotated only along specified paths. With this flow behavior, the mixing process was not completed in 36 s. In contrast, micrographs in the AH mode (Figure 3A) show that the two distinct solutions were greatly homogenized in the same amount of time.
Figure 3.
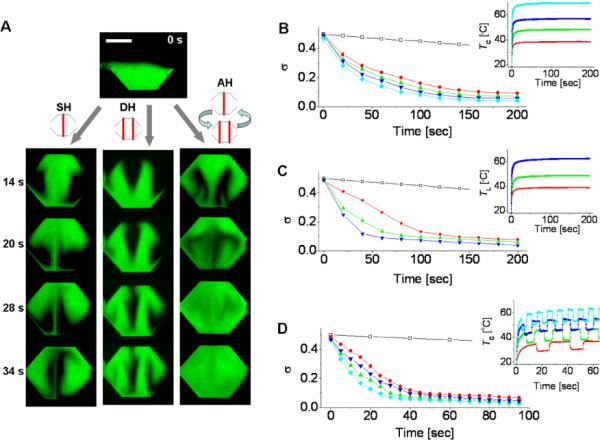
Micromixing performance in different heating modes. (A) Time evolution of micromixing in the single heating (SH), dual heating (DH), and alternating heating (AH) modes. The values of Tmax are 57, 48, and 56°C in the SH, DH, and AH modes, respectively. (B to D) Time variation of the standard deviation, σ, from the measurement of fluorescent intensity. Shown are data for the SH mode in B, the DH mode in C, and the AH mode in D. The unmixed and perfectly mixed cases are set by σ = 0.5 and σ = 0, respectively. The insets show the fluid temperatures measured using the sensors in Figure 2E (TL or TC). Their line colors correspond to those in the σ vs time plots. For comparison, data for micromixing purely due to diffusion are shown with the points (□). Scale bar, 1 mm.
We quantified the degree of mixing as a function of time using the standard deviation, σ. The decrease in the values of σ in Figures 3B to 3D with time indicates the progression of the micromixing processes. In the SH and DH modes, we applied constant voltages with a step profile; thus the temperatures gradually increased and reached final maximum values (Insets in Figures 3B and 3C). In the AH mode, we alternatively applied constant voltages to the single and dual heaters. The alternating period of the AH mode was selected based on the flow time for which each fluorescent solution arrives at the top wall of the chamber after the initial stage of heating. As shown in Figure 4, the mixing time became faster at higher temperature. However, we found that the mixing using the AH mode at 37°C was even faster than any other cases observed in the SH and DH modes, thus clearly verifying the efficiency of the AH mode-based operation for micromixing.
Figure 4.
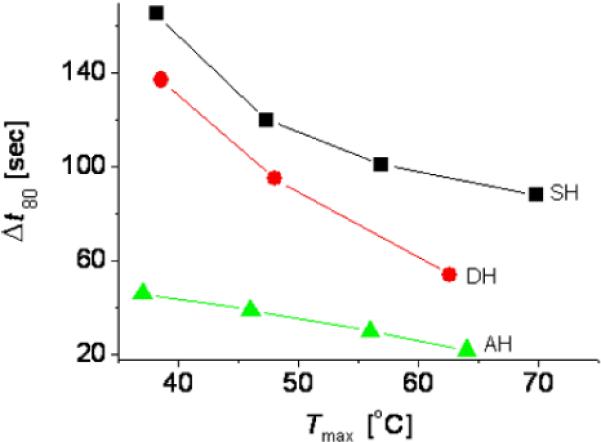
Quantitative comparison of micromixing performance in the SH, DH, and AH modes. Δt80 is the time at which the micromixing process is 80% completed. It is determined from the data points at σ = 0.1 in Figure 3B-D. Tmax is the maximum fluid temperature measured by the temperature sensors in Figure 2E.
In the AH mode, a sufficient degree of mixing can be achieved within a few tens of seconds per one microliter volume of solutions. This mixing performance is comparable to the highest performance reported in previous studies (see the Table 1 of reference 9), which took one to several hundreds of seconds per one microliter of solution.29 It should be noted that these studies, however, used complex channel geometries or active components that are likely difficult to integrate in microfluidic networks. Even a few studies demonstrated integration of micromixers in microfluidic networks, these micromixers typically faced limited performance. For instance, a fully-integrated system for the DNA purification30 used a rotary micromixer incorporating three peristaltic valves, but the mixing time was several minutes even for one nanoliter volume. Another example is a cell-culture system31 that used versatile Braille pins for micromixing, pumping, and valving; although mixing rate was not explicitly reported, the maximum pumping rate of a solution was approximately 0.6 μL/min, meaning that mixing performance is > 100 s/μL. Even when a passive mixer with 3-D groove-patterns12 was used in a DNA hybridization system,13 the mixing performance was more than 8 min per 5 μL (i.e. > 96 s/μL) because the integrated peristaltic pumps did not provide a sufficient pumping-rate.
PCR
To prove the single-chamber micromixing and reaction concept, we finally demonstrated autonomous PCR of a DNA fragment from the influenza viral strain A/LA/1/87, following the sequence of sample transport and micromixing. We started with injecting two solutions using a pipette (Figure 5A). To ensure the autonomous operation, we pre-programmed the heating instrument with a computer prior to the experiment. After mixing the solutions in the AH mode, the PCR was performed for three different total cycle numbers of 10, 20, and 30 (Figure 5B). The heating area for the PCR was 3.7 times larger than the contact area of the chamber, thus making the temperature-distribution uniform such that TC - TL < 2°C even at 94°C. Despite the relatively large heating surface, our system still maintained fast thermal response (Figure 5B inset). Owing to the use of the stainless-steel membrane (thermal conductivity: 13 W/Km, thickness: 8 μm) as the bottom cover of the microfluidic cartridge, heat conduction is confined only at the chamber region. Also, the thin membrane consisting of a 10 μm-thick Pyrex-oxynitride composite in the microfabricated heating-instrument enables spatially confined heat conduction to the chamber in the microfluidic cartridge, thus enabling fast heating and cooling rates. After the whole PCR process was finished, the PCR products were extracted and then detected using agarose gel-electrophoresis outside the cartridge. Figure 5C shows that the influence of PCR cycles on the signal enhancement of PCR products. Figure 5D demonstrates the necessity of our microfluidic mixer for the subsequent PCR process; without a microfluidic mixing process, the signal from the amplified PCR products was much weaker. Both figure 5C and 5D verify that DNA sample was successfully amplified by our micromixing and PCR process.
Figure 5.
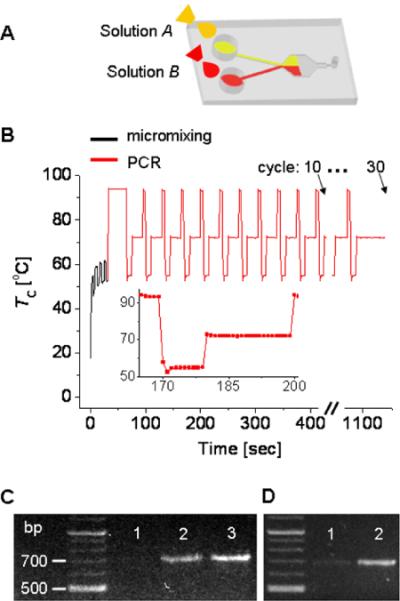
Natural convection-driven micromixing and PCR in a single microfluidic chamber. (A) Pumpless sample transport by capillarity. Solution A is a DNA template and solution B is a reaction mixture of primer, enzyme, and dNTPs. (B) Time sequence of heating used for the AH mode micromixing and the PCR process. The inset shows a single heat cycle of the PCR process with a time interval of 1 sec between the adjacent data points. PCR-based amplification of a DNA fragment from the influenza viral strain A/LA/1/87 is performed for 10, 20, and 30 cycles. (C) Influence of PCR cycles. Lanes 1, 2, and 3 correspond to the amplified PCR products after the microfluidic mixing and the subsequent 10, 20, and 30 PCR cycles, respectively. (D) Improvement of PCR by microfluidic mixing. Both lane 1 and lane 2 are the amplified PCR products after the 20 cycle. However, lane 1 is a control without a microfluidic mixing process at AH mode.
Technological Advantages of the System
Our results prove remarkable advantages for practical use. Our system can achieve both micromixing and PCR in a single chamber without adding any other microfluidic components. The single-chamber micromixing approach is broadly applicable in a wide range of biochemical reactions that require heating processes at 40°C - 135°C, which include cell lysis,32 DNA hybridization,13, 33 protein digestion,34 and biosynthesis5 as well as PCR.4 In conventional microfluidic systems, achieving biochemical reactions normally requires additional heating chambers besides micromixing channels, thereby adding more complexity to the system. Without sacrificing fast thermal response, the heaters and temperature sensors are designed as modular components originally separated from the microfluidic cartridge and integrated on the surface of the microfabricated heating-instrument. Although this type of system enables repeated use of the heating instrument and makes a microfluidic cartridge truly disposable, a previous study35 employing this approach showed relatively slow thermal response due to the large thermal mass of the system, which resulted in heating and cooling rates of 13.4°C/s and 6.4°C/s at 280 nL reaction-volume, respectively. In contrast, our system yields a heating rate of 40°C/s and a cooling rate of 35°C/s at 1 μL reaction-volume, thus greatly saving the transient time. The sample transport in our device is purely induced by capillarity, which eliminates the necessity of pumps for micromixing. In comparison, even the simple passive mixers developed in many of previous studies use pumps, rendering lab-on-a-chip system-integration difficult with the added complexity of the instrument setting.
Conclusions
We have demonstrated the utility of natural convection - a phenomenon that has been widely believed to be weak or even negligible in the microfluidic regime - for micromixing and a subsequent biochemical reaction in a single microfluidic chamber. The process is achieved by alternatively activating micro integrated heaters of different modes in a proper gravitational direction. Our microfluidic system has three technological advantages over conventional ones: (a) the fast thermal response, (b) the small number of microfluidic device components, and (c) the pumpless operation, which all together lead to significantly less idle time, device fabrication, and manual operation. Our study shows that natural convection can induce micromixing at a rate as high as a few tens of seconds per microliter even at a low fluidic temperature of ~ 37°C. With computer-programmed operation of the heaters, we have achieved autonomous on-chip PCR for a DNA sample from the influenza viral strain A/LA/1/87. Currently, the sample delivery by using capillary pumping is limited to solutions that have similar surface tension and viscosity. However, this pumping scheme can be applied to solutions with different fluidic properties, by modifying the geometry and hydrophilicity of the channel.16, 36 As such, our approach holds great promise for a wide variety of microfludic applications involving thermally induced chemical and biochemical reactions. Our single-chamber scheme would be suited for system integration of microfluidic components involving all of sample preparation, reaction, and detection processes without adding complexity.
Supplementary Material
Acknowledgement
This work was supported by National Science Foundation grant No. ECCS-0601237 (K.K.), National Institutes of Health grant No. P01-HG001984 and 1-R01-EB006789-01A2 (M.A.B.) and the University of Michigan Mechanical Engineering Graduate Fellowship (S.-J.K.). The authors would like to thank the staff and members of the Lurie Nanofabrication Facility for their assistance in device fabrication.
Footnotes
Supporting Information Available Additional information as noted in text. These materials are available free of charge via the Internet at http://pubs.acs.org
References
- (1).Dittrich PS, Manz A. Nat. Rev. Drug Discov. 2006;5:210–218. doi: 10.1038/nrd1985. [DOI] [PubMed] [Google Scholar]
- (2).Janasek D, Franzke J, Manz A. Nature. 2006;442:374–380. doi: 10.1038/nature05059. [DOI] [PubMed] [Google Scholar]
- (3).Sawetzki T, Rahmouni S, Bechinger C, Marr DW. Proc. Natl. Acad. Sci. USA. 2008;105:20141–20145. doi: 10.1073/pnas.0808808105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Burns MA, Johnson BN, Brahmasandra SN, Handique K, Webster JR, Krishnan M, Sammarco TS, Man PM, Jones D, Heldsinger D, Mastrangelo CH, Burke DT. Science. 1998;282:484–487. doi: 10.1126/science.282.5388.484. [DOI] [PubMed] [Google Scholar]
- (5).Lee CC, Sui G, Elizarov A, Shu CJ, Shin YS, Dooley AN, Huang J, Daridon A, Wyatt P, Stout D, Kolb HC, Witte ON, Satyamurthy N, Heath JR, Phelps ME, Quake SR, Tseng HR. Science. 2005;310:1793–1796. doi: 10.1126/science.1118919. [DOI] [PubMed] [Google Scholar]
- (6).Weigl BH, Yager P. Science. 1999;283:346–347. [Google Scholar]
- (7).Diffusion time of a globular protein (D = 8×10-5 mm2/s) across the chamber of width w = 1 mm is calculated as t = w2/(2D) = 104 min and diffusion time of a 4kbp DNA (D = 2×10-4 mm2/s) is 83 min. In contrast, a biochemical reaction such as PCR is usually less than one hour in microfluidic devices
- (8).Nguyen NT, Wu ZG. J. Micromech. Microeng. 2005;15:R1–R16. [Google Scholar]
- (9).Toonder J, Bos F, Broer D, Filippini L, Gillies M, de Goede J, Mol T, Reijme M, Talen W, Wilderbeek H, Khatavkar V, Anderson P. Lab Chip. 2008;8:533–541. doi: 10.1039/b717681c. [DOI] [PubMed] [Google Scholar]
- (10).Liu RH, Stremler MA, Sharp KV, Olsen MG, Santiago JG, Adrian RJ, Aref H, Beebe D, Beebe J. J. Microelectromech. Syst. 2000;9:190–197. [Google Scholar]
- (11).Lee HY, Voldman J. Anal. Chem. 2007;79:1833–1839. doi: 10.1021/ac061647q. [DOI] [PubMed] [Google Scholar]
- (12).Stroock AD, Dertinger SK, Ajdari A, Mezic I, Stone HA, Whitesides GM. Science. 2002;295:647–651. doi: 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- (13).Liu J, Williams BA, Gwirtz RM, Wold BJ, Quake S. Angew. Chem. Int. Ed. 2006;45:3618–3623. doi: 10.1002/anie.200503830. [DOI] [PubMed] [Google Scholar]
- (14).Sudarsan AP, Ugaz VM. Proc. Natl. Acad. Sci. USA. 2006;103:7228–7233. doi: 10.1073/pnas.0507976103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Seong GH, Crooks RM. J. Am. Chem. Soc. 2002;124:13360–13361. doi: 10.1021/ja020932y. [DOI] [PubMed] [Google Scholar]
- (16).Kim S-J, Lim YT, Yang H, Shin YB, Kim K, Lee DS, Park SH, Kim YT. Anal. Chem. 2005;77:6494–6499. doi: 10.1021/ac0504417. [DOI] [PubMed] [Google Scholar]
- (17).Duffy DC, McDonald JC, Schueller OJA, Whitesides GM. Anal. Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- (18).Shin YS, Cho K, Lim SH, Chung S, Park SJ, Chung C, Han DC, Chang JK. J. Micromech. Microeng. 2003;13:768–774. [Google Scholar]
- (19).Wu HK, Huang B, Zare RN. Lab Chip. 2005;5:1393–1398. doi: 10.1039/b510494g. [DOI] [PubMed] [Google Scholar]
- (20).Pal R, Yang M, Lin R, Johnson BN, Srivastava N, Razzacki SZ, Chomistek KJ, Heldsinger DC, Haque RM, Ugaz VM, Thwar PK, Chen Z, Alfano K, Yim MB, Krishnan M, Fuller AO, Larson RG, Burke DT, Burns MA. Lab Chip. 2005;5:1024–1032. doi: 10.1039/b505994a. [DOI] [PubMed] [Google Scholar]
- (21).Wang F, Yang M, Burns MA. Lab Chip. 2008;8:88–97. doi: 10.1039/b711770a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Vreeland WN, Locascio LE. Anal. Chem. 2003;75:6906–6911. doi: 10.1021/ac034850j. [DOI] [PubMed] [Google Scholar]
- (23).Imberger J. J. Fluid Mech. 1974;65:247–260. [Google Scholar]
- (24).Krishnan M, Ugaz VM, Burns MA. Science. 2002;298:793. doi: 10.1126/science.298.5594.793. [DOI] [PubMed] [Google Scholar]
- (25).Wheeler EK, Benett W, Stratton P, Richards J, Chen A, Christian A, Ness KD, Ortega J, Li LG, Weisgraber TH, Goodson K, Milanovich F. Anal. Chem. 2004;76:4011–4016. doi: 10.1021/ac034941g. [DOI] [PubMed] [Google Scholar]
- (26).Krishnan M, Agrawal N, Burns MA, Ugaz VM. Anal. Chem. 2004;76:6254–6265. doi: 10.1021/ac049323u. [DOI] [PubMed] [Google Scholar]
- (27).Agrawal N, Hassan YA, Ugaz VM. Angew. Chem. Int. Ed. 2007;46:4316–4319. doi: 10.1002/anie.200700306. [DOI] [PubMed] [Google Scholar]
- (28).Braun D, Goddard NL, Libchaber A. Phys. Rev. Lett. 2003;91:158103. doi: 10.1103/PhysRevLett.91.158103. [DOI] [PubMed] [Google Scholar]
- (29).In some applications such as protein-folding kinetics (Hertzog, et al. Anal. Chem. 2008;78:4299–4306. doi: 10.1021/ac051903j.), fast mixing using an ultra-small volume (i.e. picoliter range) is essential. However, in microfludic networks numerous biochemical reactions such as PCR and DNA hybridization are performed in a chamber, where the volume is more than hundreds of nanoliter, and the micromixing is a pre-process for the reactions. Thus we believe the use of the parameter (s/μL), which shows the required time to mix solutions at a given chamber-volume, is justified especially for the micromixers in microfluidic networks
- (30).Hong JW, Studer V, Hang G, Anderson WF, Quake SR. Nat. Biotechnol. 2004;22:435–439. doi: 10.1038/nbt951. [DOI] [PubMed] [Google Scholar]
- (31).Gu W, Zhu XY, Futai N, Cho BS, Takayama S. Proc. Natl. Acad. Sci. USA. 2004;101:15861–15866. doi: 10.1073/pnas.0404353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Alonso A, Saxena M, Williams S, Mustelin T. J. Biol. Chem. 2001;276:4766–4771. doi: 10.1074/jbc.M006497200. [DOI] [PubMed] [Google Scholar]
- (33).Kim K, Yang H, Park SH, Lee DS, Kim SJ, Lim YT, Kim YT. Chem. Commun. 2004:1466–1467. doi: 10.1039/b402914c. [DOI] [PubMed] [Google Scholar]
- (34).Turapov OA, Mukamolova GV, Bottrill AR, Pangburn MK. Anal. Chem. 2008;80:6093–6099. doi: 10.1021/ac702527b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Easley CJ, Karlinsey JM, Landers JP. Lab Chip. 2006;6:601–610. doi: 10.1039/b600039h. [DOI] [PubMed] [Google Scholar]
- (36).Kim S-J, Lim YT, Yang H, Kim K, Kim YT. unpublished results.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


