Abstract
Previously, we showed that the Vpu protein from subtype C human immunodeficiency virus type 1 (HIV-1) was efficiently targeted to the cell surface, suggesting that this protein has biological properties that differ from the well-studied subtype B Vpu protein. In this study, we have further analyzed the biological properties of the subtype C Vpu protein. Flow cytometric analysis revealed that the subtype B Vpu (strain HXB2) was more efficient at down-regulating CD4 surface expression than the Vpu proteins from four subtype C clinical isolates. We constructed a simian-human immunodeficiency virus virus, designated as SHIVSCVpu, in which the subtype B vpu gene from the pathogenic SHIVKU-1bMC33 was substituted with the vpu from a clinical isolate of subtype C HIV-1 (strain C.96.BW16B01). Cell culture studies revealed that SHIVSCVpu replicated with slightly reduced kinetics when compared with the parental SHIVKU-1bMC33 and that the viral Env and Gag precursor proteins were synthesized and processed similarly compared to the parental SHIVKU-1bMC33. To determine if substitution of the subtype C Vpu protein affected the pathogenesis of the virus, three pig-tailed macaques were inoculated with SHIVKU-1bMC33 and circulating CD4+ T-cell levels and viral loads were monitored for up to 44 weeks. Our results show that SHIVSCVpu caused a more gradual decline in the rate of CD4+ T cells in pig-tailed macaques compared to those inoculated with parental subtype B SHIVKU-1bMC33. These results show for the first time that different Vpu proteins of HIV-1 can influence the rate at which CD4+ T-cell loss occurs in the SHIV/pig-tailed macaque model.
Keywords: Vpu, SHIV, Macaques, Pathogenesis, Subtype C HIV-1, CD4+ T-cell depletion
Introduction
All the naturally occurring primate lentiviruses encode for Tat, Rev, Nef, Vif and Vpr accessory proteins. In addition to these accessory proteins, human immunodeficiency virus type 1 virus (HIV-1) and a select number of simian immunodeficiency virus (SIV) isolates (SIVcpz, SIVden, SIVgsn, SIVmon, SIVmus) also encode for a Vpu protein (Barlow et al., 2003; Courgnaud et al., 2002, 2003; Dazza et al., 2005; Huet et al., 1990). The Vpu protein is a small transmembrane phosphoprotein synthesized off the same mRNA as the Env precursor and has two known functions within the cell (Schwartz et al., 1990; Maldarelli et al., 1993). Vpu is known to interact with the CD4 molecule within the rough endoplasmic reticulum (RER) and re-translocate the protein to the proteasome for degradation (Fujita et al., 1997; Schubert et al., 1998). By removing the CD4, it permits unbound gp160 to be transported to the cell surface and incorporated into the budding virion (Willey et al., 1992). Previous studies have shown that the interaction of the subtype B Vpu with the CD4 molecule is dependent on the presence of two casein kinase II (CK-II) phosphorylation sites (Paul and Jabbar, 1997). The Vpu protein also enhances virion release from infected cells (Klimkait et al., 1990). Although the mechanism of enhanced virion release is unknown, some investigators have linked this property of Vpu to the transmembrane domain and its ion channel properties (Cordes et al., 2001; Ewart et al., 1996; Grice et al., 1997; Park et al., 2003; Schubert et al., 1996a,b). Other investigators have shown that the certain ion channel-blocking compounds could inhibit the ion channel activity of Vpu (Ewart et al., 2002, 2004).
Because SIVmac strains commonly used in pathogenicity studies do not encode for the Vpu protein, we have used the pathogenic simian human immunodeficiency virus (SHIV), which has the tat, rev, vpu and env genes of HIV-1 in a genetic background of the SIVmac239, to analyze the role of Vpu in pathogenicity. Infection with these pathogenic SHIVs results in high viral loads, a rapid loss of CD4+ T cells within 1 month of infection, and severe depletion within lymphoid organs such as the thymus, lymph nodes and spleen. Using pathogenic molecular clones that express altered Vpu proteins, we showed that both the transmembrane domain and cytoplasmic domains of the Vpu protein contribute to the severe CD4+ T-cell loss in these macaques (Singh et al., 2003; Hout et al., 2004, 2005).
To date, all studies assessing the role of Vpu in macaques have been performed using the well-studied subtype B Vpu protein from a laboratory-adapted HIV-1 isolate. Recently, we reported that the subtype C Vpu proteins were efficiently transported to the cell surface, indicating that the subtype C Vpu protein may have biological properties that differ from the better studied subtype B Vpu protein (Pacyniak et al., 2005). In this study, we report that the subtype C Vpu protein is less efficient at down-regulating CD4 from the cell surface than the subtype B Vpu. Using the pathogenic molecular clone, SHIVKU-1bMC33, we report on the construction of a SHIV in which the subtype B vpu was exchanged with the vpu from a clinical isolate of subtype C HIV-1 (SHIVSCVpu). Our results show that following inoculation into macaques, SHIVSCVpu had a decreased rate of CD4+ T-cell loss compared with the parental SHIVKU-1bMC33. These results show for the first time that different Vpu proteins can influence the rate of CD4+ T-cell loss in the SHIV/macaque model.
Results
CD4 down-regulation by subtype B and C Vpu proteins
The sequence of the subtype B and C Vpu proteins analyzed in this study are shown in Fig. 1. Previously, we showed that fusion of the Vpu protein to enhanced green fluorescent protein (EGFP) still resulted in the ability to down-modulate cell surface CD4 (Hout et al., 2005, 2006; Singh et al., 2003). We analyzed the efficiency of cell surface CD4 down-regulation by the subtype B and C Vpu fusion proteins. HeLa CD4+ cells were transfected with vectors expressing either the subtype B or C Vpu fusion proteins. At 48 h post-transfection, live cells were immunostained for CD4 and analyzed by flow cytometry to measure the intensity of cell surface CD4 expression. As shown in Fig. 2, the cells transfected with the vector expressing the subtype B fusion (VpuEGFP) consistently down-regulated CD4 more efficiently (p<0.008) than cells transfected with the vector expressing the subtype C Vpu fusion protein (VpuSCEGFP1). We also analyzed the ability of three additional subtype C Vpu fusions to EGFP (IN21301, BW04.07, and BW06.H51) fusion proteins to down-regulate CD4 surface expression. As shown in Fig. 2, all three Vpu proteins were significantly less efficient (p<0.05) at preventing CD4 surface expression compared to the HXB2 Vpu protein.
Fig. 1.
Comparison of the Vpu sequences in SHIVKU-1bMC33 and SHIVSCVpu.
Fig. 2.
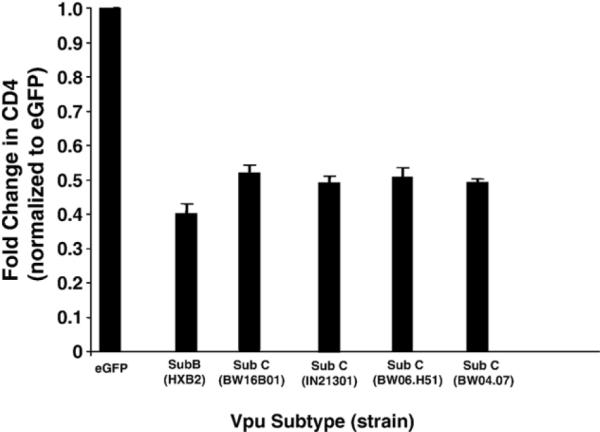
The subtype C Vpu is less efficient at down-regulating surface CD4 than the subtype B Vpu protein. HeLa CD4+ cells were transfected with plasmids pcegfp, pcvpuegfp, pcvpuSCegfp1 or pcvpuSCegfp21301, pcvpuSCegfpBW06. H51, or pcvpuSCegfpBW04.07. At 48 h, live cells were immunostained for CD4. Cells expressing EGFP or EGFP fusion proteins were assessed for CD4 down-regulation using flow cytometry.
The SHIVSCVpu virus expresses the Vpu protein within infected cells
We analyzed the expression of the Vpu protein in C8166 cultures inoculated with parental SHIVKU-1bMC33 or SHIVSCVpu. Cultures were inoculated with equivalent amounts of virus and at 5 days post-inoculation, cells were radiolabeled and Vpu proteins immunoprecipitated from cell lysates. As shown in Fig. 3, a protein with an Mr of 16,000 was immunoprecipitated from SHIVKU-1bMC33-inoculated cultures. A Vpu protein was also immunoprecipitated from SHIVSCVpu-inoculated C8166 cultures with a slightly slower mobility in SDS-PAGE, which may be due to the larger size (85 vs. 82 amino acids) of the subtype C Vpu protein (Fig. 3).
Fig. 3.
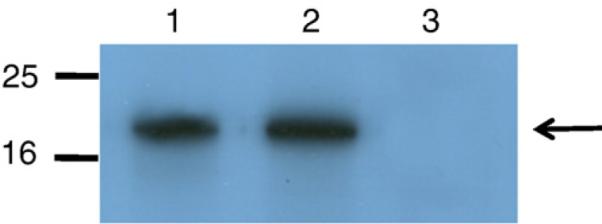
The Vpu protein is expressed in C8166 cells infected with SHIVSCVpu. C8166 cells were infected with SHIVSCVpu or SHIVKU-1bMC33 for 5 days. The cells were starved for methionine/cysteine for 2 h and then radiolabeled for 12 h with 35S-methionine/cysteine. Cell lysates were prepared as described in the text and Vpu proteins immunoprecipitated with an antisera generated against the cytoplasmic domain of the subtype B Vpu protein or the subtype C Vpu protein. (Lane 1) Vpu proteins immunoprecipitated from SHIVKU-1bMC33-inoculated cultures. (Lane 2) Vpu proteins immunoprecipitated from SHIVSCVpu-inoculated cultures. (Lane 3) Vpu proteins immunoprecipitated from uninfected cultures.
Pulse-chase analysis reveals that SHIVSCVpu precursor proteins are synthesized and processed with similar kinetics compared to parental SHIVKU-1bMC33
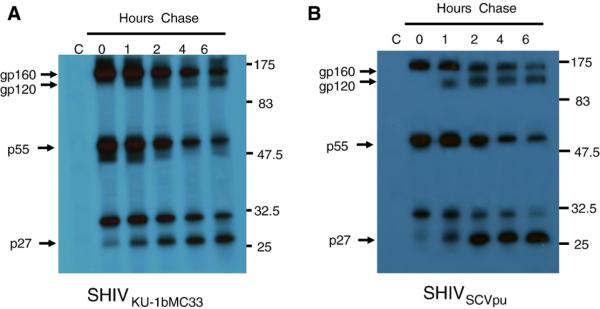
Previous studies have shown that Vpu expression results in more efficient release of virus particles from infected cells. We used pulse-chase analysis to determine if substitution of the subtype B vpu for the subtype C vpu would result in similar release of viral proteins from infected cells. These results indicate that the substitution of the subtype B vpu for the subtype C vpu did not significantly alter the processing of Gag and Env precursor proteins (Fig. 4).
Fig. 4.
Pulse-chase analysis of the SHIVSCVpu. To determine whether viral structural proteins were released with reduced efficiency in SHIVSCVpu-inoculated cultures, C8166 cells were with inoculated 103 TCID50 of either SHIVSCVpu or SHIVKU-1bMC33. At 7 days post-infection, the medium was removed and infected cells were incubated in methionine/cysteine-free medium for 2 h. The cells were radiolabeled for 30 min with 1 mCi per ml of 35S-Translabel (methionine and cysteine, ICN Biomedical, Costa Mesa, CA) and the radiolabel chased for various periods (0-6 h) in DMEM containing 100× unlabeled methionine/cysteine. SHIV proteins were immunoprecipitated from cell lysates using plasma pooled from several pig-tailed monkeys infected previously with SHIV as described in the Materials and methods section. Uninfected C8166 cells, radiolabeled and chased for 6 h, served as a negative control (lane C). All immunoprecipitates were collected on protein A Sepharose, the beads washed three times with RIPA buffer, and the samples resuspended in sample reducing buffer. Samples were boiled and the SHIV specific proteins analyzed by SDS-PAGE (10% gel). Proteins were then visualized by standard autoradiographic techniques. (A) Results of pulse-chase analysis of viral proteins immunoprecipitated from SHIVKU-1bMC33 infected cell lysates. (B) Results of pulse-chase analysis of viral proteins immunoprecipitated from SHIVSCVpu-infected cell lysates.
SHIVSCVpu replicates with delayed kinetics and is less cytopathic compared to parental SHIVKU-1bMC33
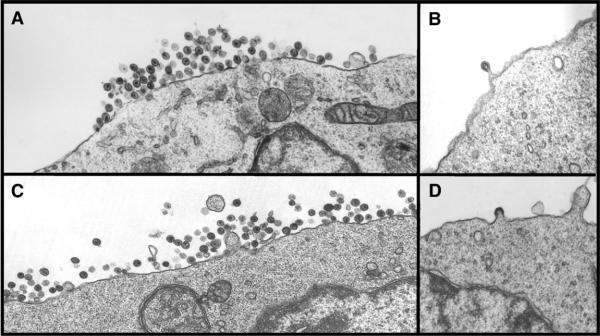
We analyzed the growth kinetics of the parental SHIVKU-1bMC33 and the SHIVSCVpu following inoculation of C8166 cell cultures. As shown in Fig. 5, SHIVSCVpu replicated with reduced kinetics as compared to the parental SHIVKU-1bMC33. While the amount of p27 released appeared to be delayed by 2 days, the rate of p27 release from 7 to 10 days was reproducibly similar. At the end of the experiment, the amount of p27 released from SHIVSCVpu-inoculated cultures was 86% of the p27 found in the parental SHIVKU-1bMC33-inoculated cultures. However, the kinetics of p27 release was different from C8166 cells inoculated with SHIVTM, which has a scrambled transmembrane domain (Hout et al., 2005), or from cultures inoculated with novpuSHIVKU-1bMC33, which lacks the vpu sequences prior to the env (Stephens et al., 2002). We also compared the pattern of virus maturation in cultures inoculated with parental SHIVKU-1bMC33 and SHIVSCVpu using electron microscopy. The results shown in Fig. 6 indicate that SHIVSCVpu matures from the cell surface similar to parental SHIVKU-1bMC33. The reduced kinetics of p27 release were also reflected in the time before the appearance of syncytial cytopathology. With the parental SHIVKU-1bMC33, cytopathology generally appears 3 to 4 days post-inoculation, whereas SHIVSCVpu-inoculated cultures displayed cytopathology starting 5 to 6 days post-inoculation. Additionally, the syncytia formation did not appear to be as extensive as SHIVKU-1bMC33, suggesting that this subtype C Vpu protein could influence the ability to form syncytia in culture (data not shown).
Fig. 5.
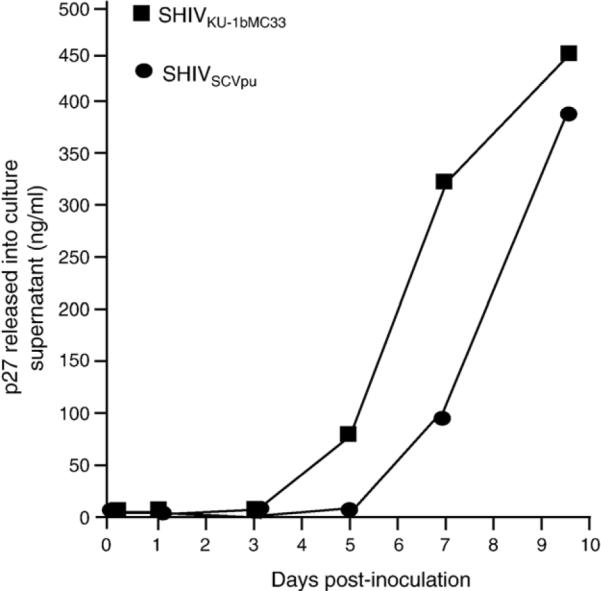
The kinetics of replication of SHIVKU-1bMC33 and SHIVSCVpu in C8166 cell cultures. Cultures of C8166 cells were inoculated with either SHIVKU-1bMC33 or SHIVSCVpu as described in the text. Aliquots of the culture medium were assayed for the presence of p27 antigen. The growth curves were performed in triplicate.
Fig. 6.
Electron microscopy examination of C8166 cells inoculated with SHIVKU-1bMC33 or SHIVSCVpu. C8166 cells were inoculated with either SHIVSCVpu or SHIVKU-1bMC33 for 7 days. Cells were washed three times with PBS and processed for electron microscopy as described in the Materials and methods section. (A) C8166 cells inoculated with SHIVSCVpu. (B) C8166 cells inoculated with parental SHIVSCVpu showing a virus particle maturing at the cell surface. (C) C8166 cells inoculated with parental SHIVKU-1bMC33. (D) C8166 cells inoculated with parental SHIVKU-1bMC33 showing a particle maturing at the cell surface.
The subtype C vpu does not influence the incorporation of envelope glycoprotein into viral particles
We determined if substitution of the subtype B vpu with the subtype C vpu would influence the incorporation of Env into viral particles. SHIVKU-1bMC33 and SHIVSCVpu were used to infect C8166 cells and radiolabeled at 6 days post-inoculation. The virus containing culture medium was harvested and virus pelleted through a 20% sucrose cushion. The pellet was lysed and the SHIV proteins immunoprecipitated using an anti-SHIV serum. The immunoprecipitated proteins were analyzed by SDS-PAGE and densitometry to determine the ratio of Gag p27 protein to Env gp120. As shown in Fig. 7, the ratio of immunoprecipitated p27 to gp120 was approximately equal for the two viruses SHIVKU-1bMC33 (ratio 4.31) than for SHIVSCVpu (ratio 4.89), suggesting that exchanging the subtype B vpu for the subtype C vpu did not influence the level of Env (gp120) associated with particles.
Fig. 7.
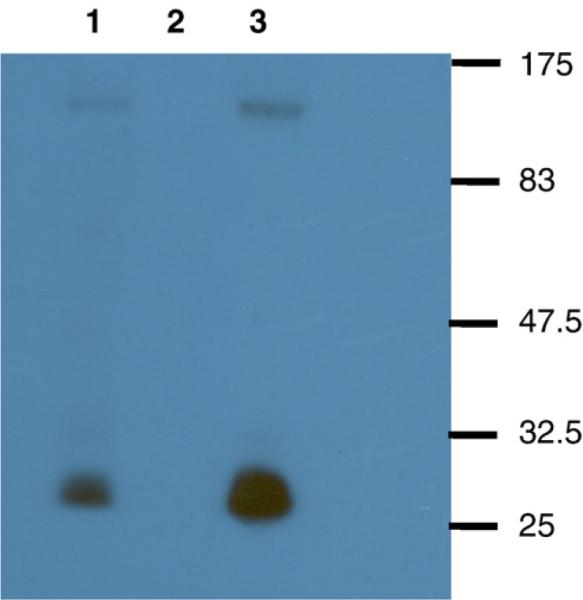
Envelope incorporation in SHIVKU-1bMC33 and SHIVSCVpu. C8166 cells were inoculated with either SHIVKU-1bMC33 or SHIVSCVpu for 6 days. At day 6, cultures were labeled as described in the Materials and methods section. The virus culture supernatants were centrifuged through a 20% sucrose cushion and lysed for immunoprecipitation. SHIV proteins were immunoprecipitated from cell lysates using plasma pooled from several pig-tailed monkeys infected previously with SHIV as described in the Materials and methods section. Uninfected C8166 cells served as a negative control. All immunoprecipitates were collected on protein A Sepharose, the beads were washed three times with RIPA buffer, and the samples were resuspended in sample reducing buffer. Samples were boiled and the SHIV specific proteins analyzed by SDS-PAGE (10% gel). (Lane 1) Immunoprecipitated SHIV proteins from SHIVSCVpu.(Lane 2) Immunoprecipitated SHIV proteins from uninfected C8166 cells. (Lane 3) Immunoprecipitated SHIV proteins from SHIVKU-1bMC33. Molecular weight markers are to the right of the gel.
The rate of CD4+ T-cell loss was decreased and viral loads lower in SHIVSCVpu inoculated macaques
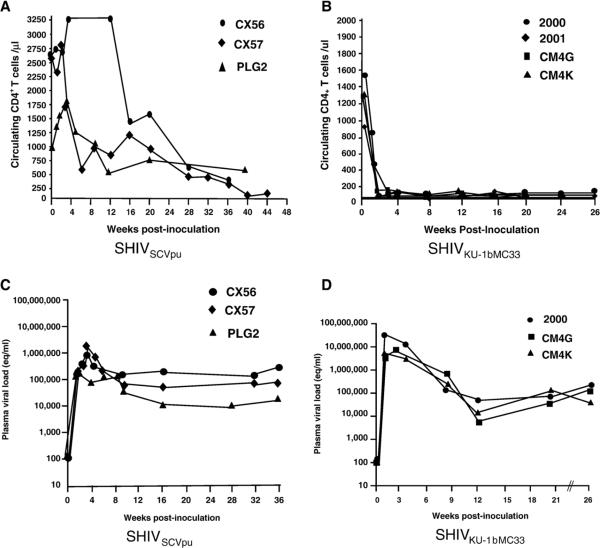
To determine whether the SHIVSCVpu was capable of causing disease in pig-tailed macaques, three macaques (CX56, CX57, and PLG2) were inoculated with SHIVSCVpu and CD4+ T cells, and viral loads followed for up to 44 weeks. Macaque CX56 and CX57 were euthanized in a moribund condition at 36 and 44 weeks post-inoculation. Macaque PLG2 was euthanized at 40 weeks post-inoculation following development of severe enteritis. The circulating CD4+ T-cell levels are shown in Fig. 8A and indicate that all three macaques developed a gradual decline in the levels of circulating CD4+ T cells during the course of their infection. The rate of circulating CD4+ T-cell loss in macaques inoculated with SHIVSCVpu was clearly decreased compared to parental SHIVKU-1bMC33 (Fig. 8B). The rate of CD4+ T-cell loss was found to be statistically significant (p<0.01). We also found that the early peak viral loads (between 1 and 3 weeks) in macaques inoculated with SHIVSCVpu were approximately 10-fold less than in macaques inoculated with the parental SHIVKU-1bMC33 (Figs. 8C and D).
Fig. 8.
Circulating CD4+ T-cell levels and plasma viral loads following inoculation of macaques with SHIVSCVpu and SHIVKU-1bMC33. (A) The levels of circulating CD4+ Tcells in macaques (CX56, •;CX57, ◆andPLG2, Δ) following inoculation with SHIVSCVpu. (B) The levels of circulating CD4+ Tcells in four macaques (2000,•; 2001,◆ CM4G,; and CM4K, ■) following inoculation with SHIVKU-1bMC33. (C) Plasma viral RNA levels in macaques (CX56, •; CX57, ◆and PLG2, Δ) inoculated with SHIVSCVpu. (D) Plasma viral RNA levels in three macaques (2000,•;2001,; andCM4K, Δ) inoculated with SHIVKU-1bMC33. Samples were subjected to real time RT-PCR and Taqman probe homologous to the SIV gag gene. Standard curves were generated using five dilutions of viral RNA of known concentration.
Sequence analysis of the vpu gene isolated from SHIVSCVpu inoculated macaques
We analyzed the sequence of the vpu genes amplified from PBMC at different times post-inoculation and three lymphoid tissues (spleen, thymus and mesenteric lymph node) at necropsy. In all three macaques, we found scattered amino acid substitutions at different times post-inoculation but no consensus amino acid substitutions (data not shown).
The pathology in lymphoid organs at necropsy was similar to the parental SHIVKU-1bMC33
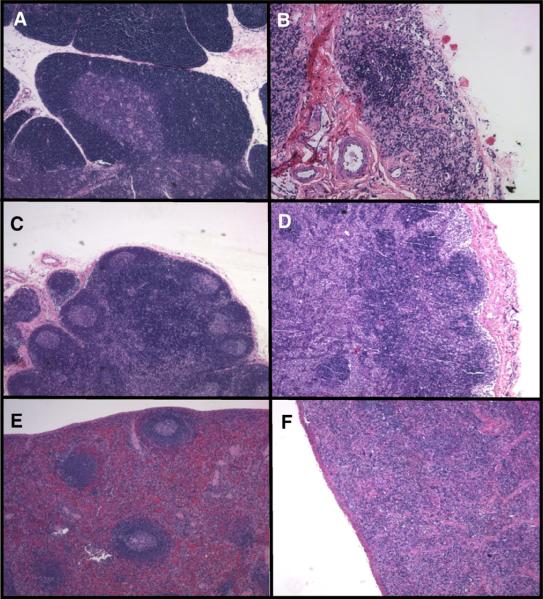
We examined the histological sections from tissues of macaques inoculated with SHIVSCVpu at necropsy and compared the lesions to those observed following inoculation with parental SHIVKU-1bMC33. No significant histological lesions were observed in the 15 regions of the brain and spinal cord (central nervous system; CNS), the heart, liver, lungs, kidney, or pancreas. The lack of histological lesions in the CNS is not surprising as the parental SHIVKU-1bMC33 is not a neuropathogenic virus. As shown in Fig. 9, the SHIVSCVpu-inoculated macaque CX56 developed severe atrophy of the thymus, marked lymphoid depletion with little follicular activity in the lymph nodes, and moderate lymphoid depletion in the spleen. These lesions were similar to that seen in macaques inoculated with parental SHIVKU-1bMC33. Macaque CX57 developed similar lymphoid depletion in the thymus and lymph nodes but no significant lesions were found in the spleen. Macaque PLG2 developed severe enteritis and protracted diarrhea. This macaque developed thymus atrophy and moderate depletion in the lymph nodes. The distribution of virus in the various visceral organs was determined by PCR for viral gag and 2-LTR sequences. All three macaques were positive for gag sequences in all 13 visceral organs despite exsanguination with 1 l of saline (data not shown). We also examined tissue RNA for the presence of viral RNA. As shown in Fig. 10, viral RNA sequences were detected in RNA sequences isolated from 7 of 13, 7 of 13, and 6 of 13 visceral organs from CX56, CX57, and PLG2, respectively. The majority of the tissue RNAs that were positive for the presence of viral RNA sequences were the lymphoid organs. We also analyzed the RNA samples of tissues from 15 regions of the CNS for viral RNA sequences. Macaques CX56, CX57, and PLG2 had no regions of the brain and spinal cord that were positive for viral RNA sequences (data not shown).
Fig. 9.
Histopathology associated with SHIVSCVpu infection of macaques. Hematoxylin and eosin stains of sections from the thymus (A and B), mesenteric lymph node (C and D) and spleen (E and F) from a non-infected age-matched macaque (A, C, and E) and macaque CX56 (B, D, and F).
Fig. 10.
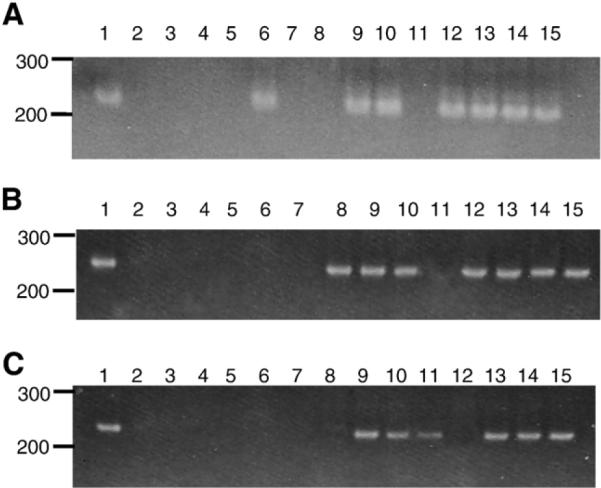
Distribution of virus in macaques CX56, CX57, and PLG2. Viral sequences were amplified using RT-PCR from RNA samples from visceral organs from macaques CX56 (A), CX57 (B), and PLG2 (C). The order of the gels are: (A) Positive control (RNA from SHIV infected C8166 cells). (Lane 2) Negative control-lymph node from an uninoculated macaque. (Lane 3) Heart. (Lane 4) Liver. (Lane 5) Lung. (Lane 6) Kidney. (Lane 7) Pancreas. (Lane 8) Salivary gland. (Lane 9) Mesenteric lymph node. (Lane 10) Axillary lymph node. (Lane 11) Inguinal lymph node. (Lane 12) Small intestine. (Lane 13) Spleen. (Lane 14) Thymus. (Lane 15) Tonsil.
Discussion
The vast majority of HIV-1 infections are in Sub-Saharan Africa, where an estimated 60-70% of infected people reside (WHO website). Within the HIV-1 group M pandemic, subtype C has increased in prevalence over the past 10 years, accounting for approximately 50% of the infections worldwide (Essex, 1999; Hemelaar et al., 2006; Takebe et al., 2004), and is most prominent in east Africa, south Africa, India and parts of China. In one study, both NSI/R5 and SI/X4 subtype C HIV-1 isolates were found to be significantly less fit in PBMC competition assays compared to all other group M isolates of the same phenotype (Ball et al., 2003). More recently, in a study that evaluated the replicative fitness of representative strains from subtypes A, B, C, D and CRF01_AE, the subtype C viruses had less replicative fitness in PBMC compared to the other subtypes. However, the subtype C isolates still replicated 100-fold higher than HIV-2 or group O isolates (Arien et al., 2005, 2007). Although less fit for replication in PBMC, the subtype C viruses were found to be similar to subtype B viruses for replicative fitness in skin-derived Langerhans cells, suggesting that these viruses might be more efficiently transmitted (Ball et al., 2003). In a recent study, subtype C HIV-1 was associated with the increased vaginal shedding of virus (John-Stewart et al., 2005). These data suggest that the relatively poor replication efficiency of subtype C in PBMCs (and possibly lymphoid organs) may be related to slower disease progression, hence, longer survival of the human host, which in turn could lead to increased time for transmission (Ball et al., 2003; Quinones-Mateu et al., 2001).
The subtype C Vpu protein has structural motifs that differ from the well-studied subtype B Vpu protein, and these may be associated with different biological properties of these two proteins (Hout et al., 2004). The subtype C Vpu proteins have an additional potential CK-II site (TMVD) downstream from the two consensus sites. A second, potentially interesting sequence motif unique to the subtype C Vpu proteins is a canonical dileucine motif (E/D-X-X-X-L-L/I) towards the carboxyl terminus of the Vpu protein. An acidic residue located at position 4, 5, or 6 upstream of the dileucine motif has been shown by investigators to be required for the efficient internalization of several proteins (Bonifacino and Traub, 2003). Dileucine motifs are known to interact with adaptor protein complexes AP-1, AP-2, and AP-3, resulting in the recruitment of cargo into clathrin-coated pits and clathrin-coated vesicles (Bonifacino and Traub, 2003). Previous studies have implicated dileucine-based motifs in the sorting of membrane proteins to cellular sites such as the trans-Golgi network (TGN), endosomes, lysosomes, and plasma membrane (Bonifacino and Traub, 2003). The HIV-1 Nef protein, associates with the inner side of the cell plasma membrane via its myristylated amino terminus. Importantly, the HIV-1 Nef, which also interacts with and down-regulates CD4 from the surface of infected cells, has a dileucine motif at the carboxyl terminus. This dileucine motif is required for CD4 down-regulation (Aiken et al., 1994; Bresnahan et al., 1998; Craig et al., 1998; Goldsmith et al., 1995). Our analysis indicates that the subtype C Vpu proteins from Indian and African subtype C isolates have “DM-GRLRLL” or “DMDGLRLL” motifs close to the carboxyl terminus, which closely fit the consensus motif described above. Thus, if these sequences are found to be responsible for altered biological properties, it has potential significance to HIV-1 pathogenesis, since subtype C isolates represent approximately 50% of the HIV-1 infections worldwide (Takebe et al., 2004).
In addition to differences regarding structural motifs, the subtype B and C have different biological properties. Using a reporter system that fused the Vpu protein to enhanced green fluorescent protein (EGFP), we previously showed that the subtype B Vpu protein is predominantly localized in the Golgi complex of the cell while the subtype C Vpu protein was transported to the cell plasma membrane (Pacyniak et al., 2005). In our data presented here, we used flow cytometric analysis to compare the efficiency of subtype C Vpu proteins at down-regulating CD4 from the cell surface. These studies revealed that the efficiency of subtype B Vpu CD4 down-regulation was statistically different than four subtype C Vpu proteins. These results indicate an additional difference in the biological properties of the subtype C and B Vpu proteins.
In previous studies using a pathogenic molecular clone of simian-human immunodeficiency virus known as SHIVKU-1bMC33, we showed that the subtype B Vpu protein contributes to the profound CD4+ T-cell loss following inoculation into pig-tailed macaques (Hout et al., 2005; Singh et al., 2001, 2003; Stephens et al., 2002). These studies showed that the two casein kinase II sites and the transmembrane domain contribute to the rapid CD4+ T-cell loss following inoculation into macaques. Furthermore, we showed that the Vpu transmembrane domain could be replaced with the TM domain of the M2 protein of influenza A virus and retain pathogenicity in pig-tailed macaques (Hout et al., 2006). Thus, this model has been useful in the molecular analysis of those domains of Vpu that are necessary for disease progression. Attempts have been made to construct pathogenic SHIVs expressing the env gene from subtype C HIV-1 (Chen et al., 2000; Ndung'u et al., 2001. In one of these studies, investigators constructed a SHIV expressing an R5 envelope glycoprotein (including the C-terminal end of the Vpu protein which contains the most amino acid divergence between the subtype B and C Vpu proteins) from a subtype C HIV-1 isolate (strain CHN19). These investigators found that this virus was capable of replicating in pig-tailed macaque PBMC but not rhesus macaque PBMC. They observed that with serial passage in pig-tailed macaques, the virus replicated more efficiently. However, these macaques never developed significant CD4+ T-cell loss and functionality of this chimeric Vpu was not determined (Chen et al., 2000). In another study, a SHIV was constructed with the majority of the subtype C env gene (the gp120 region starting just past the vpu ORF and part of gp41) into a SHIV89.6P based virus (Ndung'u et al., 2001). This virus was found to replicate in both rhesus and pig-tailed macaques. This virus was capable of replicating to high peak viral loads in rhesus macaques but did not result in significant CD4+ T-cell loss. In our studies presented here, we have concentrated on the contribution of the subtype C Vpu in the pathogenic SHIV/macaque model system. We hypothesized that if a SHIV expressing a Vpu protein from another subtype of HIV-1 (in this case, subtype C) still resulted in severe CD4+ T-cell loss, it would suggest that the divergent sequence of this Vpu (particularly the carboxyl terminus) was not a factor in disease progression. Our results presented here indicate that a SHIV constructed with the Vpu from a subtype C Vpu (SHIVSCVpu) significantly differed in the rate of CD4+ T-cell loss compared to parental pathogenic SHIVKU-1bMC33.As SHIVSCVpu and parental SHIVKU-1bMC33 are identical with the exception of the vpu gene and the overlapping env sequences (which do not appear to significantly affect envelope glycoprotein synthesis and processing), these results suggest that the efficiency of the CD4 down-regulation by the Vpu protein correlates with the rate of CD4+ T-cell loss in vivo. A potential caveat of this study is that the expressed Vpu may not be reflective of all subtype C Vpu proteins. Although a valid concern, the subtype C Vpu protein we used to construct SHIVSCVpu is similar to other subtype C Vpu sequences from Botswana (data not shown). Further, our experiments examining the down-regulation of CD4 from the cell surface revealed similarities among the four subtype C Vpu proteins (both from Botswana and India) when compared to the HXB2. Whether the decreased rate of CD4+ T-cell loss in SHIVSCVpu-inoculated macaques was due to the rate of turnover of the Vpu protein in cells in vivo, the magnitude of CD4 down-regulation or the ability to influence virus release within infected cells remains to be determined.
Materials and methods
Cells, viruses, and vectors
C8166 cell line was used as the indicator cells to measure infectivity and cytopathicity of the viruses used in this study. C8166 cells were maintained in RPMI-1640, supplemented with 10mM Hepes buffer pH 7.3, 2 mM glutamine, 5 μg per ml gentamicin and 10% fetal bovine serum (R10FBS). The 293 cell line was maintained in Dulbecco's minimal essential medium supplemented with 10% fetal bovine serum and 5 μg per ml gentamicin. The derivation and pathogenicity of SHIVKU-1bMC33 has been described (McCormick-Davis et al., 2000b; Singh et al., 2003; Stephens et al., 2002). Vectors expressing the subtype B (pcvpuegfp) and C (pcvpuscegfp) Vpu proteins fused to enhanced green fluorescent protein (EGFP) have been previously described (Gomez et al., 2005; Pacyniak et al., 2005; Singh et al., 2003). In addition to these two vectors, vectors were constructed as described above that expressed the vpu from three additional subtype C isolates from HIV-1 patients (IN21301; C.96BW04.07; and C.96BW06.H51).
Analysis of the efficiency of CD4 down-regulation
For analysis of the efficiency of CD4 down-regulation, HeLa CD4+ cells were seeded into six-well plates such that at 24 h the cells would be 70-80% confluent. Cells were transfected with plasmids expressing EGFP or various Vpu proteins fused to EGFP. Cultures were monitored for 48 h, then removed from the six-well plate and stained with PE-Cy5 conjugated anti-CD4 (BD Bioscience). Cells were analyzed using an LSR II flow cytometer, determining mean fluorescence intensity (MFI) of PE-Cy5 for transfected (EGFP positive) and untransfected (EGFP negative) cells within the same well. An MFI ratio was calculated for each; EGFP was normalized to 1. Normalized ratios from at least three separate experiments were averaged and the standard deviation calculated. All groups were compared using one-way ANOVA (performed using SAS software, alpha=0.5) to determine variance. Post hoc analysis was performed using Tukey's HSD test.
Construction of SHIVSCVpu
A plasmid containing the 96BW16B01 env and vpu was used in a PCR with oligonucleotides that introduced a NsiI siteatthe5′ end of the vpu (5′-GTAATGCATAGTTTAATAGAAAAAGTAGAT-3′ and a KpnI site in the env gene (5′-CACAGGTACCTATGCTTTAGCATC-3′). The 258-bp amplicon was gel purified, according to a Millipore isolation protocol, and ligated into pGEM-T Easy Vector (Promega, Madison, WI). The resulting transformants were screened for an insert and the plasmids sequenced to ensure that restriction sites as well as the subtype C vpu was intact. This plasmid was designated as pvpuSC.
Insertion of the subtype C vpu gene into SHIV was accomplished in a three-step process. In the first step, the pUC19ΔSN#12 plasmid, which has the SphI/KpnIfragment of SHIVKU1bMC33 in a pUC19 background and a NsiIsiteintroduced at the beginning of the vpu was digested with NsiI/KpnI and gel purified to remove the NsiI/KpnI fragment (containing the vpu and beginning of env). This plasmid was ligated with the NsiI/KpnI fragment isolated from plasmid pvpuSC using T4 DNA ligase. The resulting plasmid, pUCvpusc, was sequenced with M13 reverse primer (M13R) to ensure that tat, rev,subtypeC vpu, and env were intact and no mutations were introduced during the subcloning process. For introduction of the subtype C vpu into p3′ SHIVKU1bMC33, both pUCvpusc and p3′-SHIVKU1bMC33 were digested to completion with SphIand KpnI, gel purified, and ligated as previously described (McCormick-Davis et al., 2000a,b; Stephens et al., 2002; Singh et al., 2003; Hout et al., 2005, 2006). The resulting plasmid, known as p3′-SHIVSCvpu,was sequenced with appropriate oligonucleotide primers to insure that the tat, rev, vpusc and env genes were intact. For the production of SHIVSCvpu,′-SHIVSCvpu, and p5′-SHIV-4 were digested with SphI overnight, ligated, and used to transfect C8166 cells as previously described (Hout et al., 2005, 2006; McCormick-Davis et al., 2000a,b; Singh et al., 2003; Stephens et al., 2002). Stocks were prepared, titrated in C8166 cells and frozen at-86 °C until used.
Immunoprecipitation and pulse-chase analysis
To determine if the Vpu protein was expressed in the SHIVSCVpu-inoculated cultures, C8166 cells were inoculated with 103 TCID50 of either SHIVSCVpu or SHIVKU-1bMC33.At 5 days post-infection, the medium was removed and infected cells were incubated in methionine/cysteine-free Dulbecco's modified Eagle's medium (DMEM) for 2 h. The cells were then radiolabeled for 12 h with 500 μCi per ml of 35S-Translabel (methionine and cysteine, MP Biomedical, Costa Mesa, CA). Cell lysates were prepared by lysing radiolabeled cells in radio-immunoprecipitation buffer (RIPA: 50 mM Tris-HCl, pH 7.5; 50 mM NaCl; 0.5% deoxycholate; 0.2% SDS; 10 mM EDTA) on ice for 30 min, and the nuclei removed by centrifugation in a microcentrifuge. The Vpu proteins were immunoprecipitated with either a rabbit antiserum directed the subtype B Vpu cytoplasmic domain (from the NIH AIDS Reagents Branch, NIH) or a rabbit antiserum directed against the subtype C Vpu cytoplasmic domain. All immunoprecipitates were collected on protein A Sepharose, the beads washed three times with RIPA buffer, and the samples resuspended in sample reducing buffer. Samples were boiled and the SHIV specific proteins analyzed by SDS-PAGE. Proteins were visualized by standard autoradiographic techniques.
For pulse-chase analyses, cultures were inoculated with either SHIVSCVpu or SHIVKU-1bMC33 as described above. At 7 days post-infection, the medium was removed and infected cultures were incubated in methionine/cysteine-free Dulbecco's modified Eagle's medium (DMEM) for 2 h and then radiolabeled with 1 mCi per ml of 35S-methionine for 30 min. The radiolabel was chased for various periods in DMEM containing 100× unlabeled methionine/cysteine. At each time point, the cell culture medium was clarified (16,000×g) for 30 s. The supernatant was transferred to a new microfuge tube and made 1× with respect to RIPA buffer. SHIV proteins were immunoprecipitated with 10 μl of a pooled serum from several monkeys that were inoculated with non-pathogenic SHIV-4. For immunoprecipitation of cell-associated SHIV proteins, cell lysates were prepared as described previously (McCormick-Davis et al., 2000a,b; Hout et al., 2004; Stephens et al., 1995, 1997, 2002) prior to incubation with antiserum. Lysates were centrifuged in a microcentrifuge to remove nuclei prior to the addition of antibody. Cell lysates and culture medium were incubated with antibody for 16 h at 4 °C. Immunoprecipitates were collected and analyzed as described above.
Analysis of Env incorporation into viral particles
To determine if the exchanging the subtype B and C vpu genes would affect the incorporation of envelope glycoprotein into viral particles, virus was partially purified and used in immunoprecipitations. Cultures of C8166 cells were inoculated with either SHIVSCVpu or SHIVKU-1bMC33 for 6 days. Cultures were centrifuged at 300×g to pellet the cells and starved for methionine/cysteine for 2 h. The cells were radiolabeled with 35S-methionine/cysteine for 12 h. The cultures were centrifuged at 300×g for 5 min to remove the cells and the culture supernatant centrifuged at 10,000×g for 1 min. The supernatant from this centrifugation step was layered onto 20% sucrose (prepared in PBS, pH 7.4) and subjected to centrifugation at 100,000×g for 2 h (SW55Ti rotor, 35,000 rpm) to pellet the virus. The pellet was resuspended in 1× RIPA buffer and SHIV proteins immunoprecipitated as described above. The immunoprecipitated proteins were separated by SDSPAGE (10% gel) and visualized by standard autoradiographic techniques. The ratio of Gag p27 to Env gp120 was determined by densitometry.
p27 growth curve assays
Standard p27 assays (Beckman Coulter, SIV core antigen kit) were used to assess release of viral particles from cells infected with SHIVSCVpu or parental SHIVKU-1bMC33.Culturesof106 C8166 cells were inoculated with equivalent amounts infectious cell free virus (1×103) for 4 h. At the end of 4 h, the cells were centrifuged at 400×g for 10 min and the pellet washed with 10 ml of medium. This was repeated two additional times. The cells were resuspended in RPMI-1640 supplemented with 10% FBS and antibiotics and this was considered the 0 time point of the assay. Cultures were incubated at 37 °C and aliquots of the culture were removed at 0, 1, 3, 5, 7, and 10 days with fresh media added to cultures at days 3 and 6. The culture medium was separated from the cells by centrifugation and assayed for p27 according to the manufacturer's instructions.
Electron microscopy
To determine the site of intracellular maturation, infected cells were examined by transmission electron microscopy. Cultures of C8166 cells were inoculated with SHIVKU-1bMC33 or SHIVM2 at a multiplicity of infection of 0.01. Cells were incubated for 7 days at which time cells were pelleted at 400×g for 10 min. Cells were washed three times with 10 ml of phosphate buffered saline (pH 7.4) and fixed in 2% glutaraldehyde overnight at 4 °C. Cells were postfixed in 2% osmium tetroxide (OsO4) for 1 h. The cells were washed twice with water and dehydrated through a series of alcohols (30-100%) followed by embedding in Embed 812 resin. Thin sections were cut at 80 A, stained with uranyl acetate and lead citrate and examined under a JEOL 100CXII transmission electron microscope.
Macaques and inoculations
Three pig-tailed macaques (CX56, CX57, and PLG2) were inoculated intravenously with 104 TCID50 of SHIVSCVpu. These macaques were euthanized at 36 to 44 weeks post-inoculation. We previously reported on the circulating CD4+ T-cell levels and virus loads in macaques inoculated with pathogenic SHIVKU-1bMC33 (Stephens et al., 2002; Singh et al., 2003).Animals were housed in the AAALAC-approved animal facility at the University of Kansas Medical Center. Heparinized blood was collected weekly for 6 weeks, then at 3-week intervals for the next 6 weeks, and thereafter at monthly intervals.
Processing of blood samples
The PBMCs were prepared by centrifugation on Ficoll-Hypaque gradients as described previously (Joag et al., 1994, 1996). Ten-fold dilutions of PBMCs (106 cells/ml) were inoculated into replicate cultures and were examined for development of cytopathic effects as previously described (McCormick-Davis et al., 2000b; Stephens et al., 2002). Alterations in CD4+ lymphocytes after experimental inoculations were monitored sequentially by flow cytometric analysis (Becton Dickinson). T-lymphocyte subsets were labeled with OKT4 (CD4; Ortho Diagnostics Systems, Inc), SP34 (CD3; Pharmingen) or FN18 (CD3; Biosource International) monoclonal antibodies.
Processing of tissue samples at necropsy
At the time of euthanasia, animals were anesthetized by administration of 10 mg/kg ketamine (IM) followed by an intravenous administration of sodium pentobarbital at 20- 30 mg/kg. A laparotomy was performed on the animal exsanguinated by aortic cannulation. The chest was opened, the left ventricle cannulated, the right atrium nicked, and the animal perfused with 1 l of cold pyrogen-free Ringer's saline. All aspects of the animal studies were performed according to the institutional guidelines for animal care and use at University of Kansas Medical Center. At necropsy, tissues from the heart, kidney, liver, lungs, axillary lymph nodes (LN), mesenteric LN, inguinal LN, pancreas, salivary gland, small intestine, spleen, thymus, and tonsil were fixed in 10% neutral buffered formalin and embedded in paraffin. Sections (5 μm) were stained with hematoxylin and eosin for routine histological examination by a veterinary pathologist. In addition, the right half of the brain and spinal cord were dissected into frontal, motor, parietal, temporal and occipital cortices, corpus callosum, thalamus, basal ganglia, midbrain, pons, medulla, cerebellum, cervical, thoracic and lumbar spinal cord and were also processed as above. In addition to each of the visceral organs, regions of the CNS were also frozen in liquid nitrogen for DNA isolation.
PCR amplification and sequence analysis of gag
DNA was extracted from the visceral organs and different regions of the CNS as previously described (McCormick-Davis et al., 2000b) and used to amplify viral gag sequences. The oligo-nucleotides used for the first round of amplification were 5′-GATGGGCGTGAGAAACTCCGTCTT-3′(sense) and 5′-CCTCCTCTGCCGCTAGATGGTGCTGTTG-3′ (antisense) corresponding to the region 1052-1075 and 1423-1450 of the SIVmac239 gag gene, respectively (Regier and Desrosiers, 1990). The nested SIVmac239 primers used were 5′-GTTGAAGCATGTAGTATGGGCAGC-3′ (sense) and 5′-CACCACTAGGTGTCTCTGCACTATCTG-3′ (antisense), which are complementary to bases 1142-1165 and 1356-1382 of SIVmac239, respectively.
We also analyzed tissues for the presence of viral RNA, indicative of actively replicating virus. RNA was extracted from approximately 30 mg of each tissue from each of the visceral organs using the RNAEasy kit (Qiagen) and the manufacturer's instructions. RNA samples were digested with DNase I for 30 min. Samples were run on agarose formaldehyde gels before and after DNase treatment to check for the presence of contaminating DNA. RNA samples were amplified by using primers for the SHIV gag gene (using the primers described above) and the Easy-A One-Tube RT-PCR System (Stratagene, La Jolla, CA). One microgram of total RNA was used in the one-step reaction containing the manufacturer's buffer and enzyme mix, and oligonucleotide primers. The reactions were performed with an Eppendorf Gradient Thermal Cycler with the following thermal profile: 42 °C for 30 min, 1 cycle; 94 °C for 5 min, 1 cycle; 94 °C for 30 s, 55 °C for 30 s, and 68 °C for 45 s, 25 cycles. One microliter of the initial reaction mixture was added to a nested PCR mixture containing gag primers and a further 35 cycles were performed with the following thermal profile: 95 °C for 1 min, 55 °C for 2 min, and 72 °C for 5 min, 1 cycle; 95 °C for 30 s, 55 °C for 30 s, 72 °C for 45 s, 33 cycles. The amplified gag fragment is 240 base pairs.
Plasma virus loads
Plasma viral RNA loads were determined on RNA extracted from EDTA-treated plasma. Virus was pelleted and RNA extracted using the Qiagen viral RNA kit (Qiagen, Valencia, CA). RNA samples were analyzed by real-time RT-PCR using gag primers and a 5'FAM and 3'TAMRA labeled Taqman probe that was homologous to the SIV gag gene as previously described (Hofmann-Lehmann et al., 2000). Standard curves were prepared using a series of six 10-fold dilutions of viral RNA of known concentration. The sensitivity of the assay was 100 RNA equivalents per milliliter. Samples were analyzed in triplicate and the number of RNA equivalents were calculated per milliliter of plasma.
Sequence analysis of the vpu gene
The vpu was amplified from DNA samples isolated from several tissues taken at necropsy to examine the sequence of vpu. For amplification of the vpu, we used oligonucleotide primers 5′-CCTAGACTAGAGCCCTGGAAGCATCC-3′ (sense) and 5′-GTACCTCTGTATCATATGCTTTAGCAT-3′ (antisense), which are complementary to nucleotides 5845-5870 and 6393-6420 of the HIV-1 (HXB2) genome (Ratner et al., 1985), respectively. One microgram of genomic DNA was used in the PCR with Taq DNA polymerase and the conditions described above. For the second round of amplification, we used oligonucleotide primers 5′-TTAGGCATCTCCTATGGCAGGAAGAAG-3′ (sense) and 5′ CACAAAATAGAGTGGTGGTTGCTTCCT-3′ (antisense), which are complementary to nucleotides 5956-5984 and 6386-6413 of the HIV-1 (HXB2) genome (Ratner et al., 1985), respectively. The conditions for amplification were identical to those described above. For sequence analysis, the PCR products from three separate PCRs were separated by electrophoresis in a 1% agarose gel, isolated, and each PCR reaction directly sequenced. Cycle sequencing reactions were done using the BigDye Terminator Cycle Sequencing Ready Reaction Kit with AmpliTaq DNA polymerase, FS (PE Applied Biosystems, Foster City, CA) and sequence detection was conducted with an Applied Biosystems 377 Prism XL automated DNA sequencer and visualized using the ABI Editview program. Sequences were compared to the intact sequences from SHIVKU-1bMC33. Sequences showing differences were confirmed by molecularly cloning the PCR fragments into pGEM-T EZ vector followed by sequencing as described above.
Acknowledgments
The work reported here is supported by grants NIH grants AI51981 to E.B.S. The anti-Vpu serum was provided by the NIH AIDS Research and Reference Reagent Program. We thank Barbara Fegley at the KUMC Electron Research Microscope Laboratory for her excellent technical assistance with the electron microscopy and Dr. Joyce Slusser for assistance with the flow cytometry. We also thank the KUMC Biotechnology Support Facility for the oligonucleotide and sequence analyses.
References
- Aiken C, Konner J, Landau NR, Lenburg ME, Trono D. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell. 1994;76:853–864. doi: 10.1016/0092-8674(94)90360-3. [DOI] [PubMed] [Google Scholar]
- Arien KK, Abraha A, Quinones-Mateu ME, Kestens L, Vanham G, Arts EJ. The replicative fitness of primary human immunodeficiency virus type 1 (HIV-1) group M, HIV-1 group O, and HIV-2 isolates. J. Virol. 2005;79:8979–8990. doi: 10.1128/JVI.79.14.8979-8990.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arien KK, Vanham G, Arts EJ. Is HIV-1 evolving to a less virulent form in humans? Nat. Rev., Microbiol. 2007;5:141–151. doi: 10.1038/nrmicro1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball SC, Abraha A, Collins KR, Marozsan AJ, Baird H, Quinones-Mateu ME, Penn-Nicholson A, Murray M, Richard N, Lobritz M, Zimmerman PA, Kawamura T, Blauvelt A, Arts EJ. Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C. J. Virol. 2003;77:1021–1038. doi: 10.1128/JVI.77.2.1021-1038.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow KL, Ajao AO, Clewley JP. Characterization of a novel simian immunodeficiency virus (SIVmonNG1) genome sequence from a mona monkey (Cercopithecus mona) J. Virol. 2003;77:6879–6888. doi: 10.1128/JVI.77.12.6879-6888.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- Bresnahan PA, Yonemoto W, Ferrell S, Williams-Herman D, Geleziunas R, Greene WC. A dileucine motif in HIV-1 Nef acts as an internalization signal for CD4 downregulation and binds the AP-1 clathrin adaptor. Curr. Biol. 1998;8:1235–1238. doi: 10.1016/s0960-9822(07)00517-9. [DOI] [PubMed] [Google Scholar]
- Chen Z, Huang Y, Zhao X, Skulsky E, Lin D, Ip J, Gettie A, Ho DD. Enhanced infectivity of an R5-tropic simian/human immunodeficiency virus carrying human immunodeficiency virus type 1 subtype C envelope after serial passages in pig-tailed macaques (Macaca nemestrina) J. Virol. 2000;74:6501–6510. doi: 10.1128/jvi.74.14.6501-6510.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes FS, Kukol A, Forrest LR, Arkin IT, Sansom MS, Fischer WB. The structure of the HIV-1 Vpu ion channel: modelling and simulation studies. Biochim. Biophys. Acta. 2001;1512:291–298. doi: 10.1016/s0005-2736(01)00332-7. [DOI] [PubMed] [Google Scholar]
- Courgnaud V, Salemi M, Pourrut X, Mpoudi-Ngole E, Abela B, Auzel P, Bibollet-Ruche F, Hahn B, Vandamme AM, Delaporte E, Peeters M. Characterization of a novel simian immunodeficiency virus with a vpu gene from greater spot-nosed monkeys (Cercopithecus nictitans) provides new insights into simian/human immunodeficiency virus phylogeny. J. Virol. 2002;76:8298–8309. doi: 10.1128/JVI.76.16.8298-8309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courgnaud V, Abela B, Pourrut X, Mpoudi-Ngole E, Loul S, Delaporte E, Peeters M. Identification of a new simian immunodeficiency virus lineage with a vpu gene present among different cercopithecus monkeys (C. mona, C. cephus, and C. nictitans) from Cameroon. J. Virol. 2003;77:12523–12534. doi: 10.1128/JVI.77.23.12523-12534.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig HM, Pandori MW, Guatelli JC. Interaction of HIV-1 Nef with the cellular dileucine-based sorting pathway is required for CD4 down-regulation and optimal viral infectivity. Proc. Natl. Acad. Sci. U. S. A. 1998;95:11229–11234. doi: 10.1073/pnas.95.19.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dazza MC, Ekwalanga M, Nende M, Shamamba KB, Bitshi P, Paraskevis D, Saragosti S. Characterization of a novel vpu-harboring simian immunodeficiency virus from a Dent's Mona monkey (Cercopithecus mona denti) J. Virol. 2005;79:8560–8571. doi: 10.1128/JVI.79.13.8560-8571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essex M. Human immunodeficiency viruses in the developing world. Adv. Virus Res. 1999;53:71–88. doi: 10.1016/s0065-3527(08)60343-7. [DOI] [PubMed] [Google Scholar]
- Ewart GD, Sutherland T, Gage PW, Cox GB. The Vpu protein of human immunodeficiency virus type 1 forms cation-selective ion channels. J. Virol. 1996;70:7108–7115. doi: 10.1128/jvi.70.10.7108-7115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewart GD, Mills K, Cox GB, Gage PW. Amiloride derivatives block ion channel activity and enhancement of virus-like particle budding caused by HIV-1 protein Vpu. Eur. Biophys. J. 2002;31:26–35. doi: 10.1007/s002490100177. [DOI] [PubMed] [Google Scholar]
- Ewart GD, Nasr N, Naif H, Cox GB, Cunningham AL, Gage PW. Potential new anti-human immunodeficiency virus type 1 compounds depress virus replication in cultured human macrophages. Antimicrob. Agents Chemother. 2004;48:2325–2330. doi: 10.1128/AAC.48.6.2325-2330.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K, Omura S, Silver J. Rapid degradation of CD4 in cells expressing human immunodeficiency virus type 1 Env and Vpu is blocked by proteasome inhibitors. J. Gen. Virol. 1997;78:619–625. doi: 10.1099/0022-1317-78-3-619. [DOI] [PubMed] [Google Scholar]
- Gomez LM, Pacyniak E, Mulcahy ER, Flick M, Gomez M, Nerrient E, Ayouda A, Santiago M, Hahn B, Stephens EB. Vpu mediated CD4 down-regulation and degradation is conserved among highly divergent SIVcpz strains. Virology. 2005;335:46–60. doi: 10.1016/j.virol.2005.01.049. [DOI] [PubMed] [Google Scholar]
- Grice AL, Kerr ID, Sansom MS. Ion channels formed by HIV-1 Vpu: a modelling and simulation study. FEBS Lett. 1997;405:299–304. doi: 10.1016/s0014-5793(97)00198-1. [DOI] [PubMed] [Google Scholar]
- Goldsmith MA, Warmerdam MT, Atchison RE, Miller MD, Greene WC. Dissociation of the CD4 down-regulation and viral infectivity enhancement functions of human immunodeficiency virus type 1 Nef. J. Virol. 1995;69:4112–4121. doi: 10.1128/jvi.69.7.4112-4121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet T, Cheynier R, Meyerhans A, Roelants G, Wain-Hobson S. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature. 1990;345:356–359. doi: 10.1038/345356a0. [DOI] [PubMed] [Google Scholar]
- Joag SV, Stephens EB, Adams RJ, Foresman L, Narayan O. Pathogenesis of SIVmac infection in Chinese and Indian rhesus macaques: effects of splenectomy on virus burden. Virology. 1994;200:436–446. doi: 10.1006/viro.1994.1207. [DOI] [PubMed] [Google Scholar]
- Joag SV, Li Z, Foresman L, Stephens EB, Zhao LJ, Adany I, Pinson DM, McClure HM, Narayan O. Chimeric simian/human immunodeficiency virus that causes progressive loss of CD4+ T cells and AIDS in pig-tailed macaques. J. Virol. 1996;70:3189–3197. doi: 10.1128/jvi.70.5.3189-3197.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John-Stewart GC, Nduati RW, Rousseau CM, Mbori-Ngacha DA, Richardson BA, Rainwater S, Panteleeff DD, Overbaugh J. Subtype C is associated with increased vaginal shedding of HIV-1. J. Infect. Dis. 2005;192:492–496. doi: 10.1086/431514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20:W13–W23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- Hofmann-Lehmann R, Swenerton RK, Liska V, Leutenegger CM, Lutz H, McClure HM, Ruprecht RM. Sensitive and robust one-tube real-time reverse transcriptase-polymerase chain reaction to quantify SIV RNA load: comparison of one- versus two-enzyme systems. AIDS Res. Hum. Retrovir. 2000;16:1247–1257. doi: 10.1089/08892220050117014. [DOI] [PubMed] [Google Scholar]
- Hout DR, Mulcahy ER, Pacyniak E, Gomez LM, Gomez M, Stephens EB. Vpu: a multifunctional protein that enhances the pathogenesis of human immunodeficiency virus type 1. Cur. HIV-1 Res. 2004;2:255–270. doi: 10.2174/1570162043351246. [DOI] [PubMed] [Google Scholar]
- Hout DR, Gomez ML, Pacyniak E, Gomez LM, Inbody SH, Mulcahy ER, Culley N, Pinson DM, Powers MF, Wong SW, Stephens EB. Scrambling of the amino acids within the transmembrane domain of Vpu results in a simian-human immunodeficiency virus (SHIV™) that is less pathogenic for pig-tailed macaques. Virology. 2005;339:56–69. doi: 10.1016/j.virol.2005.04.038. [DOI] [PubMed] [Google Scholar]
- Hout DR, Gomez ML, Pacyniak E, Gomez LM, Fegley B, Mulcahy ER, Hill MS, Culley N, Pinson DM, Nothnick W, Powers MF, Wong SW, Stephens EB. Substitution of the transmembrane domain of Vpu in simian-human immunodeficiency virus (SHIVKU1bMC33) with that of M2 of influenza A results in a virus that is sensitive to inhibitors of the M2 ion channel and is pathogenic for pig-tailed macaques. Virology. 2006;344:541–559. doi: 10.1016/j.virol.2005.08.022. [DOI] [PubMed] [Google Scholar]
- Klimkait T, Strebel K, Hoggan MD, Martin MA, Orenstein JM. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J. Virol. 1990;64:621–629. doi: 10.1128/jvi.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli F, Chen MY, Willey RL, Strebel K. Human immunodeficiency virus type 1 Vpu protein is an oligomeric type I integral membrane protein. J. Virol. 1993;67:5056–5061. doi: 10.1128/jvi.67.8.5056-5061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick-Davis C, Dalton SB, Singh DK, Stephens EB. Comparison of Vpu sequences from diverse geographical isolates of HIV type 1 identifies the presence of highly variable domains, additional invariant amino acids, and a signature sequence motif common to subtype C isolates. AIDS Res. Hum. Retrovir. 2000a;16:1089–1095. doi: 10.1089/08892220050075363. [DOI] [PubMed] [Google Scholar]
- McCormick-Davis C, Dalton SB, Hout DR, Singh DK, Berman NE, Yong C, Pinson DM, Foresman L, Stephens EB. A molecular clone of simian-human immunodeficiency virus (ΔvpuSHIVKU-1bMC33) with a truncated, non-membrane-bound vpu results in rapid CD4+ T cell loss and neuroAIDS in pig-tailed macaques. Virology. 2000b;272:112–126. doi: 10.1006/viro.2000.0333. [DOI] [PubMed] [Google Scholar]
- Ndung'u T, Lu Y, Renjifo B, Touzjian N, Kushner N, Pena-Cruz V, Novitsky VA, Lee TH, Essex M. Infectious simian/human immunodeficiency virus with human immunodeficiency virus type 1 subtype C from an African isolate: rhesus macaque model. J. Virol. 2001;75:11417–11425. doi: 10.1128/JVI.75.23.11417-11425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacyniak E, Gomez ML, Mulcahy ER, Jackson MJ, Hout DR, Wisdom BJ, Stephens EB. Identification of a region within the cytoplasmic domain of the subtype B Vpu protein of human immunodeficiency virus type 1 (HIV-1) that is responsible for retention in the Golgi complex and its absence in the Vpu protein from subtype C HIV-1. AIDS Res. Hum. Retrovir. 2005;21:379–394. doi: 10.1089/aid.2005.21.379. [DOI] [PubMed] [Google Scholar]
- Park SH, Mrse AA, Nevzorov AA, Mesleh MF, Oblatt-Montal M, Montal M, Opella SJ. Three-dimensional structure of the channel-forming trans-membrane domain of virus protein “u” (Vpu) from HIV-1. J. Mol. Biol. 2003;333:409–424. doi: 10.1016/j.jmb.2003.08.048. [DOI] [PubMed] [Google Scholar]
- Paul M, Jabbar MA. Phosphorylation of both phosphoacceptor sites in the HIV-1 Vpu cytoplasmic domain is essential for Vpu-mediated ER degradation of CD4. Virology. 1997;232:207–216. doi: 10.1006/viro.1997.8541. [DOI] [PubMed] [Google Scholar]
- Quinones-Mateu ME, Ball SC, Marozsan AJ, Torre VS, Albright JL, Vanham G, van Der Groen G, Colebunders RL, Arts EJ. A dual infection/competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J. Virol. 2001;74:9222–9233. doi: 10.1128/jvi.74.19.9222-9233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner L, Haseltine W, Patarca R, Livak K, Starcich J, Josephs B, Doran F, Rafalski R, Whitehorn A, Baumeister A, et al. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- Regier DA, Desrosiers RC. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res. Hum. Retrovir. 1990;6:1221–1231. doi: 10.1089/aid.1990.6.1221. [DOI] [PubMed] [Google Scholar]
- Schubert U, Ferrer-Montiel AV, Oblatt-Montal M, Henklein P, Strebel K, Montal M. Identification of an ion channel activity of the Vpu transmembrane domain and its involvement in the regulation of virus release from HIV-1-infected cells. FEBS Lett. 1996a;398:12–18. doi: 10.1016/s0014-5793(96)01146-5. [DOI] [PubMed] [Google Scholar]
- Schubert U, Bour S, Ferrer-Montiel AV, Montal M, Maldarell F, Strebel K. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J. Virol. 1996b;70:809–819. doi: 10.1128/jvi.70.2.809-819.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Anton LC, Bacik I, Cox JH, Bour S, Bennink JR, Orlowski M, Strebel K, Yewdell JW. CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway. J. Virol. 1998;72:2280–2288. doi: 10.1128/jvi.72.3.2280-2288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Felber BK, Fenyo EM, Pavlakis GN. Env and Vpu proteins of human immunodeficiency virus type 1 are produced from multiple bicistronic mRNAs. J. Virol. 1990;64:5448–5456. doi: 10.1128/jvi.64.11.5448-5456.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DK, McCormick C, Pacyniak E, Lawrence K, Dalton SB, Pinson DM, Sun F, Berman NE, Calvert M, Gunderson RS, Wong SW, Stephens EB. A simian human immunodeficiency virus with a nonfunctional Vpu (ΔvpuSHIVKU-1bMC33) isolated from a macaque with neuroAIDS has selected for mutations in env and nef that contributed to its pathogenic phenotype. Virology. 2001;282:123–140. doi: 10.1006/viro.2000.0821. [DOI] [PubMed] [Google Scholar]
- Singh DK, Griffin DM, Pacyniak E, Jackson M, Werle MJ, Wisdom B, Sun F, Hout DR, Pinson DM, Gunderson RS, Powers MF, Wong SW, Stephens EB. The presence of the casein kinase II phosphorylation sites of Vpu enhances the CD4+ T celllosscausedbythe simian-human immunodeficiency virus SHIVKU-lbMC33 in pig-tailed macaques. Virology. 2003;313:435–451. doi: 10.1016/s0042-6822(03)00339-8. [DOI] [PubMed] [Google Scholar]
- Stephens EB, McClure HM, Narayan O. The proteins of lymphocyte- and macrophage-tropic strains of simian immunodeficiency virus are processed differently in macrophages. Virology. 1995;206:535–544. doi: 10.1016/s0042-6822(95)80070-0. [DOI] [PubMed] [Google Scholar]
- Stephens EB, Mukherjee S, Sahni M, Zhuge W, Raghavan R, Singh DK, Leung K, Atkinson B, Li Z, Joag SV, Liu ZQ, Narayan O. A cell-free stock of simian-human immunodeficiency virus that causes AIDS in pig-tailed macaques has a limited number of amino acid substitutions in both SIVmac and HIV-1 regions of the genome and has offered cytotropism. Virology. 1997;231:313–321. doi: 10.1006/viro.1997.8534. [DOI] [PubMed] [Google Scholar]
- Stephens EB, McCormick C, Pacyniak E, Griffin D, Pinson DM, Sun F, Nothnick W, Wong SW, Gunderson R, Berman NE, Singh DK. Deletion of the vpu sequences prior to the env in a simian-human immunodeficiency virus results in enhanced Env precursor synthesis but is less pathogenic for pig-tailed macaques. Virology. 2002;293:252–261. doi: 10.1006/viro.2001.1244. [DOI] [PubMed] [Google Scholar]
- Takebe Y, Kusagawa S, Motomura K. Molecular epidemiology of HIV: Tracking AIDS pandemic. Ped. Int. 2004;46:236–244. doi: 10.1046/j.1442-200x.2004.01869.x. [DOI] [PubMed] [Google Scholar]
- Willey RL, Maldarelli F, Martin MA, Strebel K. Human immunodeficiency virus type 1 Vpu protein regulates the formation of intracellular gp160-CD4 complexes. J. Virol. 1992;66:226–234. doi: 10.1128/jvi.66.1.226-234.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]