SUMMARY
Ameloblastin (AMBN) is the second most abundant extracellular matrix protein produced by the epithelial cells called ameloblasts and is found mainly in forming dental enamel. Inactivation of its expression by gene knockout results in absence of the enamel layer and its replacement by a thin layer of dysplastic mineralized matrix. The objective of this study was to further characterize the enamel organ and mineralized matrix produced in the AMBN knockout mouse. However, in the course of our study, we unexpectedly found that this mouse is in fact a mutant that does not express the full-length protein but that produces a truncated form of AMBN. Mandibles from wild type and mutant mice were processed for morphological analyses and immunolabeling. Microdissected enamel organs and associated matrix were also prepared for molecular and biochemical analyses. In incisors from mutants, ameloblasts lost their polarized organization and the enamel organ detached from the tooth surface and became disorganized. A thin layer of dysplastic mineralized material was deposited onto dentin, and mineralized masses were present within the enamel organ. These mineralized materials generated lower backscattered electron contrast than normal enamel, and immunocytochemistry with colloidal gold revealed the presence of amelogenin, bone sialoprotein and osteopontin. In addition, the height of the alveolar bone was reduced, and the junctional epithelium lost its integrity. Immunochemical and RT–PCR results revealed that the altered enamel organ in the mutant mice produced a shorter AMBN protein that is translated from truncated RNA missing exons 5 and 6. These results indicate that absence of full-length protein and/or expression of an incomplete protein have direct/indirect effects beyond structuring of mineral during enamel formation, and highlight potential functional regions on the AMBN molecule.
Keywords: Ameloblastin, Enamel organ, Junctional epithelium, Animal model, Mineralization
1. INTRODUCTION
Ameloblastin (AMBN) is a member of the secretory calcium-binding phosphoprotein (SCPP) gene cluster of evolutionally-related molecules that regulate skeletal mineralization (Kawasaki and Weiss, 2003). It is the second most abundant matrix protein produced by ameloblasts and is generally believed to be located exclusively in forming enamel (reviewed in Hu et al., 2005). However, transient expression of AMBN was also found in differentiating odontoblasts (Bègue-Kirn et al., 1998, Hao et al., 2005, Simmons et al., 1998), during tooth root formation (Fong et al. 1996) and craniofacial bone development (Spahr et al., 2006). AMBN is short-lived and rapidly undergoes major C-terminal processing (Murakami et al., 1997; Uchida et al., 1997). The cleaved fragments leave the enamel layer while the N-terminal portions persist for longer periods (Nanci et al., 1998, Uchida et al., 1997). This unique protein contains potential sites for cell adhesion and for posttranslational modifications such as O-linked glycosylation, sulfation and phosphorylation (Cerny et al., 1996; Krebsbach et al., 1996). The phosphorylation modification includes a site for casein kinase II that is shared by other proteins involved in mineralization such as bone sialoprotein (BSP) and osteopontin (OPN) (Krebsbach et al., 1996). Newly secreted AMBN temporarily accumulates at enamel growth sites where crystals actively elongate (Nanci et al., 1998; Uchida et al., 1997). This has led to the suggestion that it may regulate crystal elongation (Hu et al., 2005, Nanci et al., 1998) perhaps as a result of its calcium-binding properties (Vymetal et al., 2008). For reasons that are still unknown, AMBN continues to be expressed throughout the maturation stage, long after the entire thickness of the enamel layer has been deposited (Lee et al., 2003, Nanci et al., 1998).
There are presently two animal models involving genetic manipulation of AMBN, a knockout (KO) mouse (Fukumoto et al., 2004) and a transgenic mouse overexpressing AMBN (Paine et al., 2003). In the AMBN KO mouse model, ameloblasts (and associated cells layers of the enamel organ) detach from the tooth surface as they enter the secretory stage. This directly or indirectly causes them to stop their typical differentiation sequence and abort enamel formation (Fukumoto et al., 2004). Instead, only a very thin layer of dysplastic mineralized material covers the dentin surface; the origin and nature of this material are presently unknown. Interestingly, overproduction of AMBN in transgenic mice leads to formation of thinner and more porous enamel, with disrupted rod patterns and abnormal crystallites (Paine et al., 2003). Together, these findings suggest that AMBN acts, in part, as a promoter of the process of enamel formation, but that it may also act as an inhibitor of enamel formationwhenpresent in quantities greater than its normal basal levels.
To broaden our understanding of the role of AMBN, we have therefore further examined the structure of the enamel organ and characterized the nature of the dysplastic mineralized matrix produced in the KO mouse (Fukumoto et al., 2004). Since the enamel organ is implicated in formation of the junctional epithelium (reviewed by Bosshardt and Lang, 2005, Shimono et al., 2003), we have also investigated whether detachment and disorganization of the enamel organ in this mouse model has consequences for development of a normal junctional epithelium when the tooth erupts. While the strategy used to create the AMBN KO mouse model was initially believed to fully abrogate gene and protein expression, we have now found that a truncated mRNA is still produced from the targeted allele. We therefore also carried out immunolabeling, biochemical and molecular analyses to characterize the expressed protein. Our results show that AMBN influences both cells and matrix events and that absence of the full-length protein and/or expression of an incomplete protein has effects beyond the structuring of mineral during enamel formation. Identification of the portion of AMBN protein still produced in this mouse model provides new insights into the functional domains that may be implicated in these activities.
2. RESULTS
2.1. Genotype and physical characteristics of mice in which exons 5 and 6 were targeted to disrupt expression of full-length AMBN (AMBNΔ5–6)
The genotype of mice used in this study was confirmed by PCR analysis. Both AMBN heterozygote (AMBNHET) and AMBNΔ5–6 mice had no obvious health or fertility problems. In addition, their weight matched that of wild type mice (AMBNWT) from the same age group. By qualitative macroscopic examination, incisor and molar teeth from AMBNHET and AMBNWT mice appeared indistinguishable (data not shown). However, obvious enamel organ and enamel defects were apparent on both the mandibular and maxillary incisors, as well as molars, of AMBNΔ5–6 mice.
2.2. Characterization of the enamel in AMBNΔ5–6 mice by scanning electron microscopy (SEM) and X-ray microtomography
Absence of expression of full-length AMBN resulted in enamel defects on maxillary incisors. The labial surface of upper and lower incisors in AMBNWT and AMBNHET mice was covered with a smooth layer of enamel, while in AMBNΔ5–6 mice a dysplastic layer covering dentin exhibited a rough surface texture (compare Figs. 1a and b). This dysplastic material did not show any of the prismatic organization (no rods) that was readily apparent in enamel from normal or AMBNHET animals (compare Figs. 1c and d).
Fig. 1.
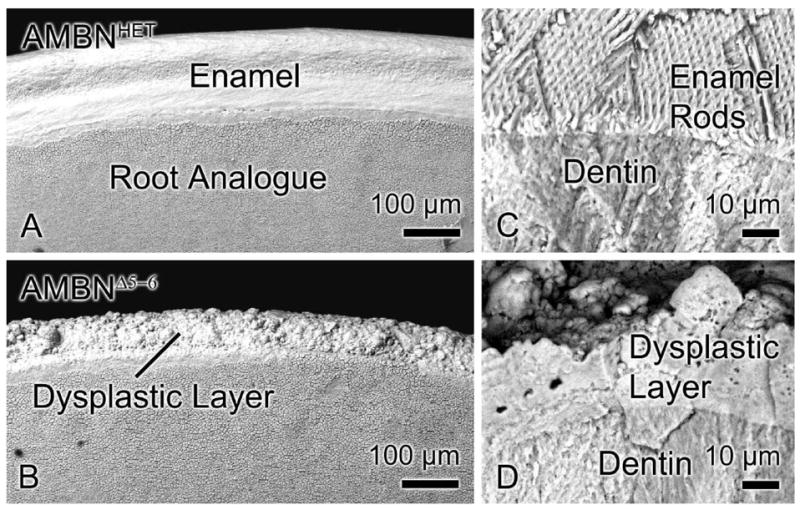
SEM images of maxillary incisors from AMBNHET (A, C) and AMBNΔ5–6 (B, D) mice. (A, B) Surface and (C, D) fractured views. The enamel layer in AMBNHET mice exhibits (A) a smooth surface and (C) a complex rod and interrod pattern. (B, D) Incisors from AMBNΔ5–6 animals instead show a dysplastic layer composed of irregular mineralized masses with no defined structural pattern.
Comparative backscatter electron imaging (BEI) on calcified hemimandible sections, in the region corresponding to the secretory stage of amelogenesis in normal mice, revealed the absence of an enamel layer in AMBNΔ5–6 mice and deposition of a thin layer of mineralized material with a globular appearance along the dentin surface (compare Figs. 2a and c). This dysplastic material yielded a backscatter signal that was visibly higher (brighter) than that of normal forming enamel and lower than mature enamel (compare Figs. 2c with a and b). The signal of the enamel layer from the erupted portion of the incisor increased significantly but that of the dysplastic layer in AMBNΔ5–6 mice did not change (compare Figs. 2b and d). Globular masses of mineralized material, with a similar backscatter signal as the dysplastic layer, were also found throughout the region of the enamel organ (Fig. 2c). In AMBNΔ5–6 mice, the tip of the incisor appeared rounded, suggesting excessive occlusal wear, and the gingival margin was displaced with respect to the tip of the tooth and the crest of the alveolar bone (compare Figs. 2b and d).
Fig. 2.

Backscatter SEM images of sagittally cut incisors from AMBNHET (A, B) and AMBNΔ5–6 (C, D) mice from corresponding late secretory (A, C) and erupted (B, D) portions of the tooth. The absence of full-length AMBN (B) results in lack of formation of an enamel layer and its replacement by a thin, dysplastic mineralized layer along the dentin surface (arrow), and the presence of mineralized masses throughout the disorganized enamel organ (EO). (D) A shift of the gingival margin with respect to the tip of the incisor and the alveolar bone is also observed. CT, connective tissue.
Transverse X-ray microtomographic scans clearly revealed the enamel layer on molars and incisor of AMBNHET mice (Figs. 3b–d). In the region examined, the enamel layer over the labial aspect of the tooth had not yet fully mineralized and appeared less radio-opaque than the completely mineralized enamel on the crowns of the erupted molars. In AMBNΔ5–6 mice, comparable scans could barely discriminate the thin, dysplastic mineralized layer which formed over the dentin (Figs. 3f–h). Microtomography also showed an attrition of the tooth crown in these mice (Figs. 3f–h), as well as possible differences in alveolar bone thickness in certain areas (Figs. 3g and h). These alterations may have resulted from the changes in occlusal forces caused by absence of a hard enamel layer.
Fig. 3.

Sagittal (A, E) and transverse (B–D, F–H) microtomography (micro-CT) views of the mandibles from AMBNHET (A–D) and AMBNΔ5–6 (E–H) mice. (A, E) The dotted lines indicate the approximate positions of the transverse micro-CT X-ray image slices. (B–D) Enamel covers the coronal dentin surface of molars and the labial aspect of the incisor where it gradually increases in thickness along the length of the tooth (from apical to incisal end). (E–H) In AMBNΔ5–6 mice, the material covering the dentin on both molars and incisors is barely visible (arrows). The molars also exhibit attenuated occlusal surfaces.
2.3. Histological characterization of amelogenesis in the absence of full-length AMBN
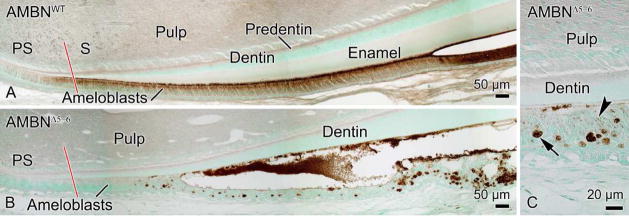
AMBNWT, AMBNHET and AMBNΔ5–6 mice presented a similar histological organization in the apical portion of the incisors, where differentiation of ameloblasts and odontoblasts takes place and dentin formation begins (Figs. 4a and b). However, from the position where active deposition of the enamel layer starts in normal animals, incisors from AMBNΔ5–6 mice exhibited evident problems with amelogenesis (compare Figs. 4a and b). A typical initial enamel layer failed to build up, and Tomes’ processes did not develop (compare Figs. 4c and d with e). Instead, there was vacuolation at the interface between the ameloblasts and dentin that contained a material that stained faintly with toluidine blue (Fig. 4b, inset). Ultrastructurally, ameloblasts in AMBNΔ5–6 mice showed rudimentary apical extensions that interdigitated with a layer of a fine granular material (compare Figs. 4c and d with e). Thereafter, the enamel organ detached from the tooth surface, and ameloblasts gradually lost their columnar appearance (Fig. 4b). The enamel organ became disorganized and ameloblasts were no longer readily distinguishable (Figs. 4b and 5a).
Fig. 4.
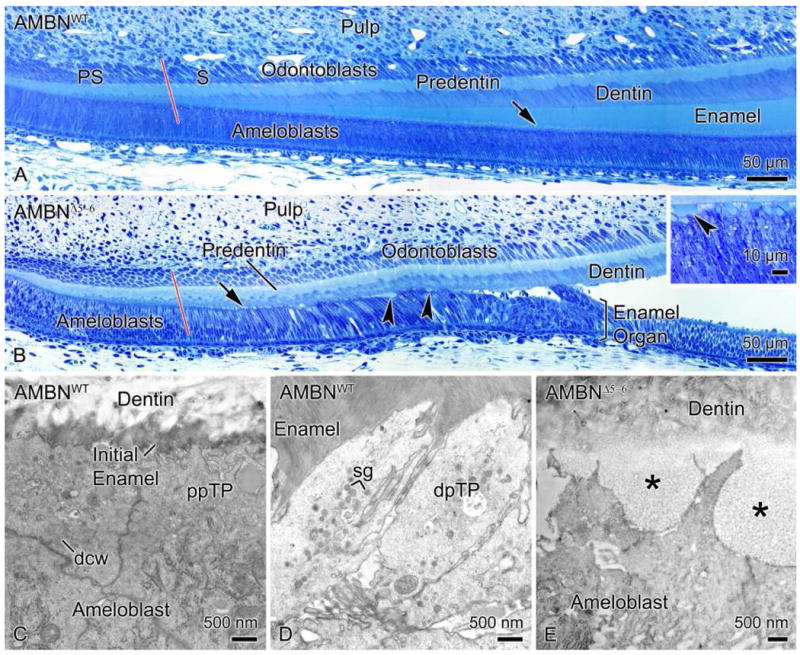
Light micrographs illustrating the histological appearance of the presecretory (PS) and secretory (S) stages of amelogenesis in AMBNWT (A) and corresponding regions in AMBNΔ5–6 (B) mouse incisors. (A, B) The red line delineates the approximate boundary between the two regions. (A) In AMBNWT mice, enamel normally starts building up as the cells enter the secretory stage. Tomes’ processes (arrow) located at the apical extremity of ameloblasts develop and interdigitate with enamel. (B) The PS stage appears normal in AMBNΔ5–6 mice. However, ameloblasts thereafter fail to build up an enamel layer and to create a Tomes’ process (arrow). (B, Inset) Instead, a vacuolation (arrowheads) appears between dentin and ameloblasts and the enamel organ detaches from the tooth surface. Electron micrographs showing (C) the initial enamel deposited by ameloblasts in the early secretory stage and (D) the distal portions of Tomes’ processes (dpTP) that interdigitate with enamel in AMBNWT mice. (E) In AMBNΔ5–6 mice, Tomes’ processes remain rudimentary and a finely granular material is deposited instead of enamel (*). dcw, Distal cell web; ppTP, proximal portion of Tomes’ process; sg, secretory granules.
Fig. 5.

Light micrographs of the position corresponding to the maturation stage of amelogenesis in AMBNΔ5–6 mice incisors. (A) In the absence of full-length AMBN, the enamel organ is disorganized, and mineralized masses (arrowhead) of variable appearance gradually appear within it. In some cases, connective tissue (CT) forms between the enamel organ and dentin, and there is a metachromatic interfacial line (black arrow) between it and the material overlying the dentin (inset). (B) In AMBNΔ5–6 mice, the enamel organ is generally directly in contact with the dysplastic mineralized matrix (white arrow) that forms on the dentin surface.
The focus of this study was on enamel formation and we therefore did not examine in detail the impact of absence of full-length AMBN on other tissues where it has been reported, such as bone and dentin. However, survey histological analysis did not reveal any obvious alteration in these tissues as was observed for enamel (data not shown).
In some cases, connective tissue formed between the disorganized enamel organ and dentin (Fig. 5a, inset) and on rare occasions, portions of the enamel organ were missing and replaced by connective tissue. It should be noted that the surface layer of dysplastic matrix observed by SEM in calcified teeth and microdissected for biochemical analyses, was not always seen in decalcified preparations. Also, tooth samples that were incompletely decalcified frequently exhibited such a layer (Fig. 5b). Taken together, these observations suggest that the dysplastic matrix is, at certain sites, lost during decalcification. Matrix accumulation with variable appearance, size and texture also gradually formed throughout the disorganized enamel organ (Figs. 2c and 5a) and their number and size increased incisally. These correspond to the mineralized masses seen by BEI (Fig. 2c).
In AMBNWT mice, at the start of the maturation stage, a basal lamina reappeared at the interface between ameloblasts and enamel (data not shown). At the corresponding position in AMBNΔ5–6 mice, there was no well-defined basal lamina between cells and the dysplastic material formed along the dentin surface (Fig. 6a). Instead, a deeply metachromatic layer, reminiscent of laminae limitantes in bone (Kawaguchi et al., 1993), was frequently observed at this interface (Fig. 5a, inset). However, there was a basal lamina-like structure at the interface between cells and some of the matrix masses found throughout the enamel organ (Fig. 6b).
Fig. 6.
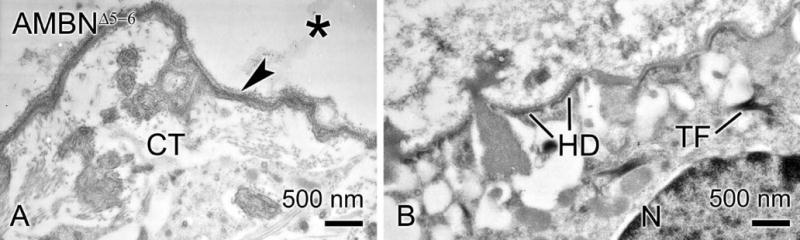
Electron micrographs from AMBNΔ5–6 mice. (A) Connective tissue (CT) elements are separated from material overlying the dentin (*) by an electron dense, interfacial line (arrowhead). (B) There is a basal lamina-like structure between cells of the enamel organ and the masses of mineralized matrix within it. HD, hemidesmosome; N, nucleus; TF, tonofilament.
2.4. Immunocytochemical characterization of the dysplastic material
Immunogold labeling revealed that albumin (Fig. 7a), amelogenin (AMEL) (Figs. 7b–d) and the mesenchymal noncollagenous proteins, BSP and OPN (Figs. 7e and f) were present in the dysplastic mineralized material located along the dentin surface (Figs. 7a and b) and throughout the disorganized enamel organ (Fig. 7c).
Fig. 7.
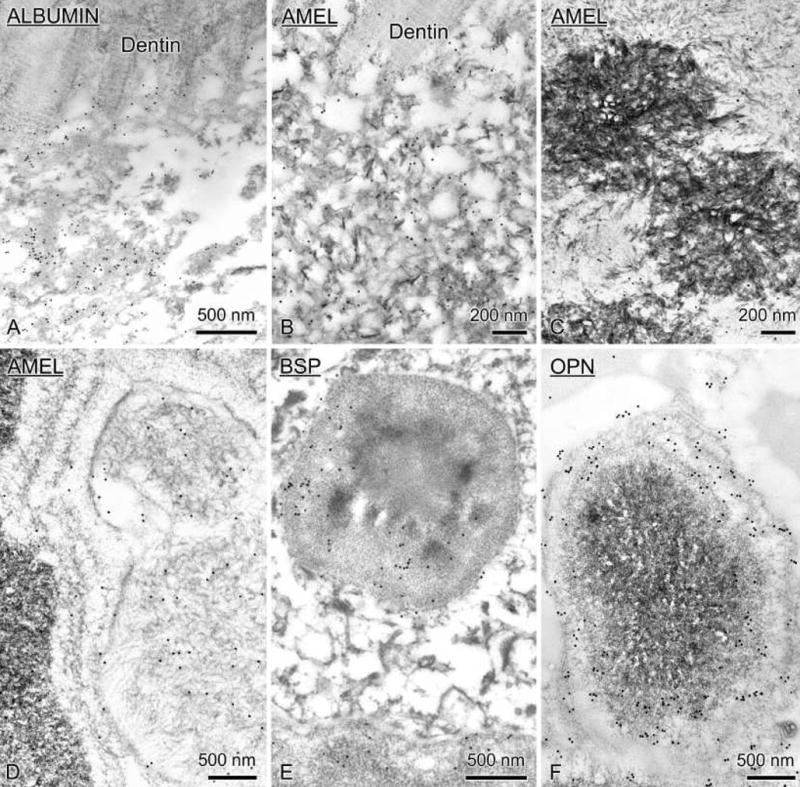
Electron micrographs of immunocytochemical preparations from AMBNΔ5–6 mice. Sections were incubated with antibodies against albumin (A), amelogenin (AMEL, B–D), bone sialoprotein (BSP, E) and osteopontin (OPN, F). These various proteins were immunodetected in the dysplatic mineralized matrix layer overlying the dentin and in the mineralized masses found throughout the enamel organ.
2.5. Histochemical detection of AMBN antigenicity in AMBNΔ5–6 mice
In AMBNWT mice, immunohistochemical detection of AMBN showed that the protein was produced by ameloblasts early during the secretory stage (Fig. 8a). The labeling intensity in ameloblasts increased gradually as the enamel layer increased in thickness. Labeling was also present along the surface of the enamel layer. Antigenicity for AMBN was unexpectedly also detected in AMBNΔ5–6 mice (Fig. 8b). Staining was initiated more incisally than in AMBNWT mice and was found along the dentin surface and over most, but not all, of the matrix masses present throughout the disorganized enamel organ (Figs. 8b and c). Some staining was also seen over ameloblasts before they lost their columnar appearance (Fig. 8c).
Fig. 8.
Immunohistochemical preparations for AMBN in AMBNWT (A) and AMBNΔ5–6 (B, C) mandibular incisors. The red line delineates the presecretory (PS) and secretory (S) stages of amelogenesis. (A) In AMBNWT teeth, labeling starts towards the end of the PS stage ameloblasts and its intensity increases in the S stage. (B) In AMBNΔ5–6, the matrix along the dentin surface and, most but not all, mineralized masses throughout the disorganized enamel organ show reactivity. (C) Labeling (arrowhead) is also seen within the cells associated with these masses (arrow).
2.6. Detection of a truncated AMBN protein and mRNA in AMBNΔ5–6 mice
Figure 9 illustrates comparative immunoblots of cell (enamel organ) and corresponding mineralized matrix extracts from AMBNWT and AMBNΔ5–6 mice. In wild type mice, antibodies against the C-terminal portion of AMBN (Fig. 9a, left panel) and the full-length protein (Fig. 9b, left panel) both revealed protein bands of apparent molecular weights near 65 kDa and 58 kDa and between ~14–50 kDa. The intensity of the bands weakened from secretion to maturation (incisal direction). The cell samples from AMBNΔ5–6 incisors showed two major stained bands that migrated at apparent molecular weights of ~32 and 38 kDa and whose intensity also decreased in an incisal direction and was less than that of the major bands observed in AMBNWT mice (Figs. 9a and b, right panels). No significant staining was seen in the dysplastic matrix samples under the loading conditions used. Since in immunohistochemical preparations both cells and mineralized masses throughout the disorganized enamel organ were reactive (see Fig. 8), these masses likely contributed to the cell staining profile seen on immunoblots of AMBNΔ5–6 mice.
Fig. 9.
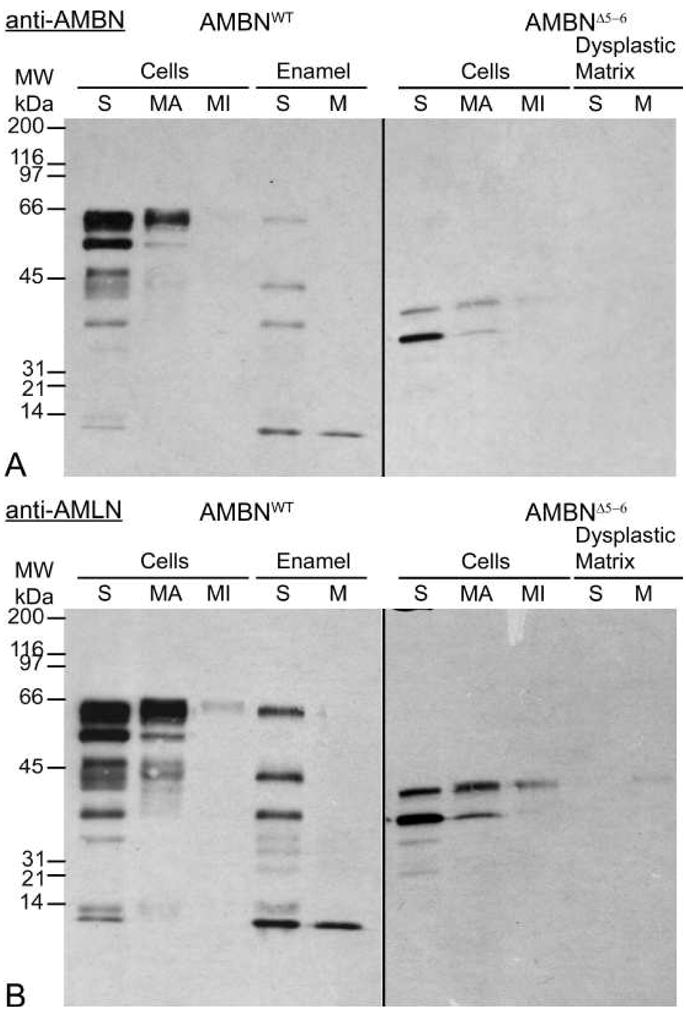
Immunoblotting with anti-AMBN antibody for the C-terminal half of the AMBN protein (A) and anti-AMLN (amelin, other name for AMBN) for the full-length AMBN protein (B). Enamel organs and corresponding mineralized matrix from incisors were dissected into a series of sequential strips relative to the position of secretory (S) and maturation (M; apical portion, MA; incisal portion, MI) stages of amelogenesis in AMBNWT mice. Truncated forms of AMBN are produced by AMBNΔ5–6 enamel organ cells from regions corresponding to secretion and maturation. These peptides have estimated molecular weights between 32–38 kDa. Standard broad-range molecular weight marker proteins (MW in kDa) are given on the left.
In order to confirm that the peptides produced were a fragment of full-length AMBN, and to determine the portion of the molecule expressed, RT-PCR analyses were done on total RNA extracted from secretion stage enamel organs using different primer sets covering different portion of the entire AMBN cDNA (Table 1, Fig. 10b and c). Similarly to AMBNWT mice, a product of 651 bp was observed in AMBNΔ5–6 mice with oligonucleotides (F2+R2) corresponding to the 3′ half of the coding portion of the cDNA (Fig. 10d). Likewise, primers covering the most 5′ part of the cDNA (F5+R5 and F6+R5) gave identical product sizes (211 bp and 151 bp, respectively) in the AMBNWT and AMBNΔ5–6. Other primer pairs (F5+R4 and F6+R4) designed to include exon 6 gave rise to two bands of the expected sizes due to alternative splicing of a 45 bp portion within exon 6 in AMBNWT mice (Fig. 10b). However, in AMBNΔ5–6 mice, the use of primers F5+R4 and F6+R4 produced smaller products of 265 bp and 205 bp, respectively (Fig. 10d). Sequencing of the F5+R4 265 bp PCR product revealed that it only lacked exons 5 and 6 (Fig. 10a). These results suggest that the mRNA transcribed from the targeted allele presumably arose by skipping of exons 5 and 6. This prompted us to re-examine the map of the targeting vector used to create AMBN KO mice (Fukumoto et al., 2004). Indeed, we confirmed that the targeting vector replaced the XbaI-EcoRI genomic segment containing exons 5 and 6, where the XbaI site is within intron 4, instead of within exon 5, which had been described in the previous paper (Fukumoto et al., 2004) (Ambntm1Nid in Mouse Genome Informatics, http://www.informatics.jax.org/). The exon 5 and 6 skipping results in an in-frame translation. Assuming translation initiation from the ATG present in exon 2, which is in a better Kozak consensus than the one present in exon 1, the AMBN protein precursor produced in the AMBNΔ5–6 mice would be missing 117 residues and give rise to a 295 amino acid protein (Fig. 10e). Upon cleavage of the signal peptide, the secreted AMBNΔ5–6 form would have 279 amino acids compared to 396 for the wild type, with computed molecular masses of 29.7 and 42.5 kDa, respectively. The AMBN antibody recognition site was preserved on the truncated protein. Sequence analysis (data not shown) indicated that the truncated AMBN fragment was missing a phosphorylation site for protein tyrosine kinase, a SH3 binding region, and an O-linked glycosylation present on the full-length protein (Kobayashi et al., 2007, Krebsbach et al., 1996, Yamakoshi et al., 2001).
Table 1.
List of PCR primer pairs
| Product size (bp) | ||||
|---|---|---|---|---|
| Target | Primer Pairsa | Primer sequence (5′ → 3′) | AMBNWT | AMBNΔ5–6 |
| Primer pairs used for AMBN mRNA characterization | ||||
| Exons 2–5 | F5 | CTGGGAGCACAGTGAATGTC | 211 | 211 |
| R5 | GAGAAAGTGCGTTGAGTCC | |||
| Exons 2–8 | F5 | CTGGGAGCACAGTGAATGTC | 571 and 616 | 265 |
| R4 | CGATCTGGAACACGGTTG | |||
| Exons 3–5 | F6 | CTGTTCCTGTCCCTAGTG | 151 | 151 |
| R5 | GAGAAAGTGCGTTGAGTCC | |||
| Exons 3–8 | F6 | CTGTTCCTGTCCCTAGTG | 511 and 556 | 205 |
| R4 | CGATCTGGAACACGGTTG | |||
| Exons 9–11 | F2 | GGACCAATGGCACACAAC | 651 | 651 |
| R2 | TCAGGGCTCTTGGAAACGC | |||
| Primer pairs used for mice genotyping | ||||
| Exon 5 | F7 | ACCCATTCCACACAAACCTG | 187 | |
| R6 | TGTTCCCTTGGTCCTATCC | |||
| Neomycin | neo-F1 | GGTGGAGAGGCTATTCGGC | 540 | |
| neo-R2 | TTCGGCAAGCAGGCATCGC | |||
F stands for forward (5′-side region of DNA) and R for reverse (3′-side).
Fig. 10.
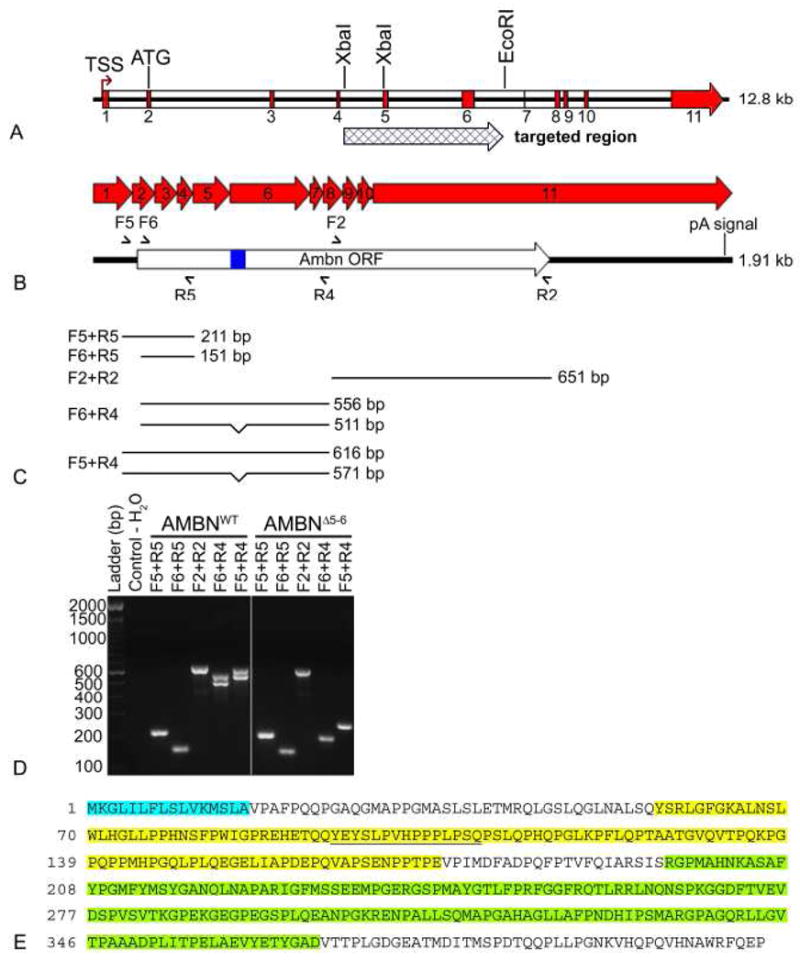
Details of the AMBN gene and RT-PCR analyses in AMBNWT and AMBNΔ5–6 mice. (A) Schematic representation of the AMBN gene and the replacement region with the targeting cassette. The pGK-Neo selection cassette replaced an AMBN segment from the XbaI site present in intron 4 to the EcoRI within intron 6. The targeting event eliminates exons 5 and 6 and results in an in-frame translation product. (B) Five different primer pairs were used for analyzing mRNA expression in the secretory stage of amelogenesis in AMBNWT and AMBNΔ5–6 mice. Semi-arrowhead above and below indicate the location of the primers used for RT-PCR. The alternative splicing region is represented by a blue rectangle. (C) The expected products for each primer pairs in AMBNWT mice are illustrated and reported in Table 1. Alternative splicing in exon 6 give rises to two bands for primer pairs F6+R4 or F5+R4. (D) PCR products were separated on a 2% agarose gel and stained with ethidium bromide. In AMBNΔ5–6 mice, only F6+R4 and F5+R4 primer pairs gave fragments different than the expected ones. A single band, around 205 bp and 265 bp respectively, are detected while two bands are found in AMBNWT mice (571 bp and 616 bp; 511 bp and 556 bp respectively). The first lane represents the 100 bp DNA ladder and the second lane is a negative control. (E) Sequencing the F5+R4 PCR product allowed to deduce the amino acid sequence of the AMBN produced in the KO which is missing the portions encoded by exons 5 and 6 (highlighted in yellow). Signal peptide sequence is highlighted cyan blue. The alternative splicing region is underlined. The anti-AMBN antibody reacts against the internal portion of AMBN extending from residues 196 to 368 (highlighted in green). F, forward primer (5′-side region of DNA); ORF, open reading frame; pA signal, polyadenylation signal; R, reverse primer (3′-side region of DNA); TSS, Transcription start signal.
2.7. Absence of full-length AMBN results in structural changes in the junctional epithelium and the alveolar bone
Scanning electron microscope observation of hemimandibles revealed an increase in the distance between the cervical extremity of the molar crown and the crest of the alveolar bone in AMBNΔ5–6 mice (compare Figs. 11a and b with c and d). Changes in the interdental bony septum were also noted in these mice, and the periodontal ligament overall appeared shifted to a lower position on the roots of the teeth (compare Figs. 11b and d).
Fig. 11.
SEM images of the molars and alveolar bone in AMBNWT (A, B) and AMBNΔ5–6 mice (C, D). The absence of full-length AMBN expression affects crown morphology, which shows excessive wear and changes in the occlusal plane of the molars. The periodontal ligament (PDL) and mandibular bone are positioned lower on the molar roots of these animals (compare the length of the red double headed arrow in B and D). The interdental bone (*) also appears more porous.
Histological analysis revealed differences in structural organization and positioning of the junctional epithelium in AMBNWT and AMBNΔ5–6 mice (Compare Figs. 12a and b with c and d). In the absence of full-length AMBN, the interdental junctional epithelium exhibited an irregular outline, and cells at the surface appeared to desquamate and migrated onto cementum (Figs. 12d and e). The junctional epithelium along the labial and lingual aspects of molars also extended deeper along the root surface, however its structural integrity seemed less affected (Fig. 12f). As seen by SEM, the periodontal ligament also shifted lower on the root surface (Figs. 12c and 12d). No exacerbated inflammatory reaction was seen in the periodontal tissues, and, based on survey qualitative observations, there was also no obvious increase in osteoclasts.
Fig. 12.
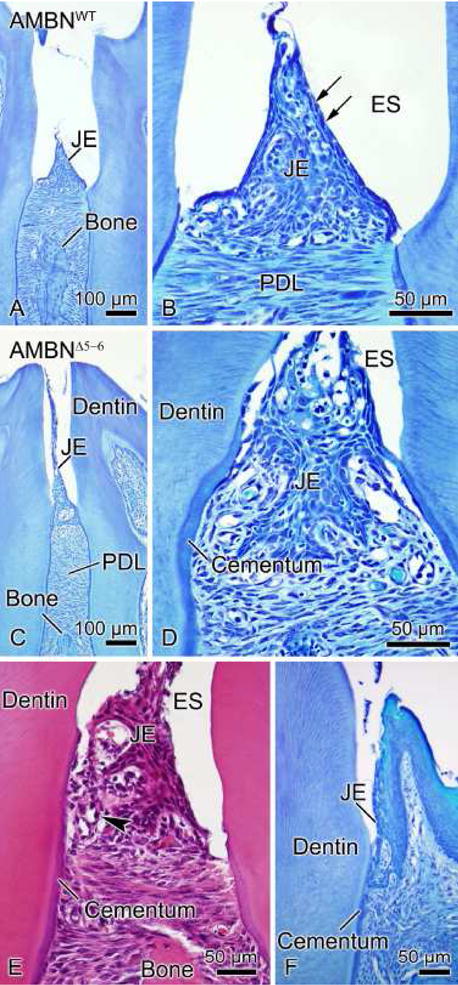
Light micrographs of the junctional epithelium (JE) in AMBNWT (A, B) and AMBNΔ5–6 mice (C–F). (A, B) The JE (arrows) is a stratified squamous non-keratinizing epithelium whose surface cells appear flattened and oriented parallel to the tooth surface. (C, D, E) Absence of expression of full-length AMBN gives rise to several structural defects in the interdental JE including detachment from the tooth surface and its repositioning onto cementum. The periodontal ligament (PDL) is also found lower on the root of the tooth. Inflammatory cells (arrowhead) infiltrated the JE of AMBNΔ5–6 mice. (F) Alterations in organization are less evident at the level of JE surrounding the labial and lingual aspects of molars. ES, enamel space.
3. DISCUSSION
The present data show that in the mouse model previously reported as an AMBN KO (Fukumoto et al., 2004), protein expression was not completely inactivated. While expression of full-length AMBN is abrogated, a truncated RNA missing a portion of exons 5 and 6 is still translated, resulting in protein bands migrating between ~32–38 kDa on SDS-PAGE gels. This mouse genotype therefore represents a form of mutant and the generated phenotype cannot be unequivocally linked to absence of the full-length protein, as would occur in a “complete” KO, and may also perhaps result from the production of a defective, mutant protein. Indeed, the resulting phenotype may be due to dominant negative effects, partial loss of function and/or neomorphic changes in gene function. However, since the amount of the truncate protein produced in AMBNΔ5–6 teeth is apparently less than in AMBNWT teeth, it is likely that dominant negative effects of the truncated protein are minimal. Because of the deletion of exons 5 and 6 in the AMBN allele, we therefore suggest referring to this model as an AMBNΔ5–6 mutant mouse. Irrespective of the exact cause, this model is particularly interesting because it results in epithelial defects that abolish structured enamel formation and that lead directly or indirectly to establishment of a defective attachment of the gingiva to the tooth surface. It also uniquely highlights potentially active region on the AMBN molecule whose absence may be responsible for the observed phenotype.
Immunochemical analyses reveal that enamel organ cells in the AMBNΔ5–6 mouse produce both AMEL and fragmented forms of AMBN. Both proteins accumulate in the dysplastic mineralized material that forms instead of enamel on the surface of the dentin, and among cells of the enamel organ. Unexpectedly, this dysplastic material also contains BSP, OPN and serum albumin. One possible explanation for the presence of these exogenous proteins is that, when the enamel organ detaches and loses its integrity, unintentional mineral precipitation occurs in these areas. Then, serum proteins such as albumin (Nanci et al., 1996) and various bone matrix proteins normally found in tissue fluids (Nanci et al., 2004) can now freely enter the area and associate with mineral. However, there could also be active local secretion of BSP and OPN by enamel organ cells, since these cells, under certain circumstances, are capable of producing mesenchymal proteins (Bosshardt and Nanci, 1997).
Based on the phenotypic changes observed in the AMBNΔ5–6 mouse, one might expect that transgenic mice overexpressing AMBN should have a thicker layer of enamel. Instead, a generally thinner enamel with abnormal crystallites has been reported in these animals (Paine et al., 2003). This can only occur if the capabilities of AMBN depend upon its “effective” concentration and/or if its expression level has an impact on production of other matrix proteins and/or on cellular function. For example, it was reported that targeting of the AMBN gene causes a reduction in expression levels of AMEL, with no apparent change in other proteins normally expressed by ameloblasts (enamelin and enamelysin expression levels all appear normal) (Fukumoto et al., 2004) which may affect enamel formation. Indeed, hammerhead ribozyme, designed to cleave AMEL mRNA, caused a partial reduction of AMEL gene expression, which was sufficient to regionally abolish enamel formation (Lyngstadaas et al., 1995). Therefore, it cannot be concluded with certainty that the loss of full-length AMBN production is the sole and direct reason why an enamel layer does not develop in AMBNΔ5–6 mice. One possibility is that AMEL expression occurs at basal levels in differentiating ameloblasts (presecretion) and that normally it is upregulated when intense enamel matrix protein production is required for rapid growth of an enamel layer. If AMEL production is affected, as happens in the AMBNΔ5–6 mice and in the ribozyme-treated mice, then AMEL expression remains at low levels and there is no buildup of an enamel layer. One interesting observation consistent with this hypothesis is that during the presecretory stage, when enamel matrix proteins start accumulating extracellularly but no signs of mineralization are yet detectable, ameloblasts have secretory granules containing predominantly just AMEL (Zalzal et al., 2008). This changes when active buildup of the enamel layer begins, at a point near where the enamel organ detaches from dentin in AMBNΔ5–6 mice, at which time the majority of secretory granules being released contain both AMEL and AMBN. Taken together, these observations lead to the possibility that there may be a functional secretory dependency between AMEL and AMBN (Zalzal et al., 2008).
In normal animals, expression of AMBN unexpectedly continues after the enamel layer is fully formed and structured (maturation stage) (Lee et al., 2003, Nanci et al., 1998). The reason for this protracted expression, while expression of AMEL ceases, remains to be addressed but clearly it is not related to building the thickness of the enamel layer. In addition, the alterations of the junctional epithelium observed in AMBNΔ5–6 raise the question of whether AMBN may also be implicated in events beyond enamel formation. When the tooth erupts into the oral cavity, the reduced enamel organ fuses with the oral mucosa at sites of future gingival attachments to the tooth, thereby contributing to formation of the primordial junctional epithelium (reviewed in Bosshardt and Lang, 2005). While the junctional epithelium alterations in AMBNΔ5–6 mice may be a direct consequence of lack of expression of full-length AMBN and its replacement by a truncated form, they may also result because the enamel organ is defective in these animals and its fusion to gingiva creates an improper relationship. Similarly, despite reports that AMBN is expressed during embryogenesis in certain bones (Spahr et al., 2006), the changes in alveolar bone in AMBNΔ5–6 mice cannot be directly related to the protein, as they could arise from other factors such as changes in occlusal forces in teeth without enamel. Similarly, the junctional epithelium alterations may reflect a secondary periodontal adaptation.
In conclusion, this study has demonstrated that the mouse model originally described as an AMBN KO is actually a mutant in which the full-length protein is replaced by a truncated form. It should therefore correctly be referred to as an AMBNΔ5–6 mutant mouse and, while the in vivo effects reported by Fukumuto et al.(2004) have been confirmed, these cannot be ascribed to the complete absence of AMBN as was previously assumed. The present study, however, has no bearing on the in vitro cell binding activity with recombinant AMBN reported by these authors. Indeed, our results suggest that the capacity of AMBN to interact with cells may reside in the deleted portion of the molecule. The change in AMBN expression in mutant mice affects the relationship that ameloblasts maintain with the tooth surface and also has an impact on likely various cellular/extracellular events that lead to the formation of a highly-mineralized and structured enamel layer. In addition, it also directly or indirectly alters the relationship that the gingiva and alveolar bone have with the tooth. The complex nature of AMBN and its multifunctional roles are consistent with its bipolar nature, which exhibits two domains with different structures and functions (Vymetal et al., 2008). It is also reinforced by recent reports showing that AMBN is present in developing bone, can enhance pulp repair and induces cementum regeneration (Kanazashi et al., 2006, Nakamura et al., 2006, Spahr et al., 2006). Creation of a complete KO mouse will be needed to better understand the role of this unique protein.
4. EXPERIMENTAL PROCEDURES
4.1. Generation and genotyping of mice lacking full-length ameloblastin
The original AMBN KO mouse model was created and obtained from Dr. Y. Yamada (Fukumoto et al., 2004). Since, in these mice exons 5 and 6 were targeted to disrupt expression of full-length AMBN, we will hereafter refer to this mouse model as AMBNΔ5–6. The colony was maintained using the 1 male/2 females mating approach for heterozygote-heterozygote crosses. Genotyping was done by PCR analysis of genomic DNA extracted from mouse tail clips, using neomycin primers to detect the targeted allele, and F7 and R6 primers to detect the wild type allele (Table 1). All experimental protocols and animal handling were approved by the Comité de déontologie de l′expérimentation sur les animaux of Université de Montréal.
4.2. Tissue processing for light and electron microscopy
AMBNWT (Charles Rivers Canada; St-Constant, QC, Canada), AMBNHET and AMBNΔ5–6 mice were anesthetized and sacrificed. Hemimandibles were fixed by immersion in a fixative solution consisting of 4% paraformaldehyde (BDH; Toronto, ON, Canada) and 0.1% glutaraldehyde (Electron Microscopy Sciences, Washington, PA) in 0.08M sodium cacodylate (Electron Microscopy Sciences) buffer containing 0.05% calcium chloride (Sigma-Aldrich Canada Ltd, Oakville, ON, Canada), pH 7.2, for 3 hours at 4°C. They were decalcified for 7 days, at 4°C, in Planck-Rychlo solution consisting of 0.13M aluminium chloride hexahydrate (Sigma-Aldrich Canada Ltd), 0.2N hydrochloric acid (Fisher Scientific, Whitby, ON, Canada), 1.35% formic acid (Fisher Scientific) (Schroeder, 1991). The decalcifying solution was changed daily. Decalcified tissues were washed for 24 hours in 0.1M cacodylate buffer containing 0.05% calcium chloride, pH 7.2. They were then processed for embedding in paraffin, LR White resin (London Resin Company; Berkshire, UK) and Epoxy resin (Electron Microscopy Sciences).
Seven to ten μm thick paraffin sections were prepared with a Leica RM2155 microtome (Leica Microsystems Canada Inc, Richmond Hill, ON, Canada), mounted on Superfrost®/Plus slides (Fisher Scientific), deparaffinized and stained with toluidine blue or haematoxylin and eosin. One μm thick semi-thin sections were cut with glass knives on a Reichert Jung Ultracut E ultramicrotome, stained with toluidine blue and observed on an Axiophot light microscope (Carl Zeiss, Oberkochen, Germany).
4.3. Scanning electron microscope observations (SEM) and X-ray microtomography analysis
The following preparations from the various experimental groups were examined using backscattered electron imaging (BEI) from a JEOL JSM-6460LV variable pressure SEM operated at 25 kV and 40–70 Pa (JEOL, Tokyo, Japan): (1) calcified hemimandibles fixed for histological analysis; (2) calcified incisors fractured at several sites along their length; and (3) sagittal sections obtained from undecalcified incisors embedded in Epoxy resin prepared using an Isomet® low-speed saw and a diamond disk (Buehler Canada, Markham, ON, Canada). Grey scale values achieved by backscatter signals provide information on mineral distribution and density within tissues (Boskey, 2006). Other fixed calcified hemimandibles were analyzed in the wet state with a SkyScan-1072 X-ray microtomography system operated at 80 kV (SkyScan, Antwerpen, Belgium).
4.4. Immunocytochemistry
Ultrathin 80–100 nm sections were cut with a diamond knife, transferred onto Formvar®-coated (polyvinyl formate) 200-mesh nickel grids, and processed at room temperature for postembedding colloidal gold immunolabeling as previously described (Nanci et al., 1998). The following polyclonal rabbit primary antibodies were used: anti-recombinant M179 mouse AMEL, anti-rat albumin, anti-human OPN and anti-human BSP (Table 2). All grids were stained with 4% aqueous uranyl acetate for 8 min and with lead citrate for 2 min, and examined at 80 kV with a JEM-2010 transmission electron microscope (JEOL).
Table 2.
Primary antibodies and dilutions used
| Primary Antibody | Dilution/Time | Source |
|---|---|---|
| Rabbit polyclonal anti-rat ameloblastin(against C-terminal portion of ameloblastin extending from residues 196 to 368) | 1:500/3h | P.H. Krebsbach (Krebsbach et al. 1996) |
| Rabbit polyclonal anti-rat amelin (against full-length ameloblastin) | 1:500/3h | L. Hammarström (Fong et al. 1996) |
| Rabbit polyclonal anti-recombinant mouse amelogenin (M179) | 1:150/3h | J. Simmer (Simmer and Fincham 1995) |
| Rabbit polyclonal anti-rat albumin | 1:60/3h | ICN Pharmaceutical (Aurora, OH) |
| Rabbit polyclonal anti-human osteopontin (LF123) | 1:100/3h | L.W. Fisher (Fisher et al. 1995) |
| Rabbit polyclonal anti-human bone sialoprotein (LF6) | 1:50/3h | L.W. Fisher (Fisher et al. 1995) |
4.5. Immunohistochemistry
Immunohistochemistry was performed on 7–10 μm thick sections mounted on Superfrost®/Plus (Fisher Scientific) slides. Briefly, deparaffinized sections were first blocked for 20 min, with a solution consisting of 0.01M PBS, pH 7.2, containing 0.05% Tween 20 (Fisher Scientific) (0.01M PBS - Tween 20) and 5 % skim milk (Carnation; Nestle, Don Mills, ON, Canada). They were then incubated with anti-AMBN or anti-Amelin (anti-AMLN) primary antibodies (Table 2), washed with 0.01M PBS - Tween 20 for 30 min, followed by treatment with the Dako EnVision™+ System, peroxidase kit (Dako Corporation, Carpinteria, CA) as recommended by the manufacturer. Visualization was performed with 3,3′-diaminobenzidine and sections were counterstained with methyl green (Dako Corporation). Negative controls included omission of primary antibody and incubation with pre-immune serum.
4.6. Sample preparation for biochemical and molecular analyses
AMBNWT and AMBNΔ5–6 mice were anesthetized and sacrificed; hemimandibles were dissected and directly immersed in liquid nitrogen for at least five hours, and then freeze-dried at −80°C for two days on a 12-liter cascade lyophilizer system (Labconco; Kansas City, MO). Enamel organs from incisors were dissected into a series of sequential strips relative to the position of secretory (S) and maturation (apical, MA; incisal, MI) stages of amelogenesis in normal mice (Smith and Nanci, 1989). Enamel and dysplastic matrix samples were also dissected from positions corresponding to the secretory (S) and maturation (M) stages in normal mice.
4.7. SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting
Freeze-dried microdissected enamel organ strips and corresponding enamel or dysplastic matrix, from a pool of five incisors from AMBNWT and AMBNΔ5–6 mice, were solubilized into 45 μl of sample preparation buffer containing 125 mM Tris (pH 6.8), 4% SDS, 20% glycerol, 200 mM beta-mercapthoethanol, and 0.2% bromophenol blue (final concentrations). Proteins were applied to individual lanes of Ready gel® precast 15% Tris-HCl polyacrylamide gels (format 8.6 cm × 6.8 cm × 1 mm; Bio-Rad, Mississauga, ON, Canada), and electrophoresis in the presence of SDS was carried out under discontinuous conditions (Nanci et al., 1998). Proteins were transferred onto 0.45μm nitrocellulose membranes (Schleicher & Schuell, distributed by Mandel Scientific Company Inc., Guelph, ON, Canada) and probed with polyclonal rabbit primary antibodies raised against recombinant rat AMBN and recombinant rat AMLN (Table 2), all in PBS-0.05% Tween 20 (Fisher Scientific) with 5% skim milk (Carnation; Nestle). Detection was performed with a secondary goat anti-rabbit antibody IgG-peroxidase conjugate (1:30,000, 1 hour; Sigma-Aldrich Canada Ltd), using the ECL plus™ Western blotting detection system (GE-Amersham Biosciences, QC, Canada) as per recommendations by the manufacturer. Broad range molecular weight proteins markers (Bio-Rad) were also loaded.
4.8. RNA extraction and RT-PCR
Freeze-dried microdissected enamel organ strips (S stage), from a pool of five incisors from AMBNWT and AMBNΔ5–6 mice, were placed in the Trizol® reagent (Invitrogen, Burlington, Ontario, Canada) and homogenized for total RNA extraction. RT-PCRs were carried out with 125 ng of total RNA using the Superscript™ III One-step RT-PCR System with Platinum Taq High Fidelity kit (Invitrogen). Oligonucleotide primers, listed in Table 1, were designed to cover various regions from exons 2 to 11 (Fig. 10b). Following the RT step at 55°C/30 min, a denaturation step was done at 94°C/2 min, PCR conditions were: 94°C/30 s, 57°C/30 s, 68°C/45 s for 35 cycles, and a final step at 68°C/5 min.. PCR products were separated on 2% agarose gel. A 100 bp DNA ladder (Invitrogen) was used as a marker. The 265 bp amplification product from primer set F5+R4 was purified on Minelute PCR purification kit column (QIAGEN, Mississauga, ON, Canada) and sequenced.
Acknowledgments
The authors are grateful to Ms Karine Sellin and Ms Micheline Fortin for technical assistance. We are thankful to Dr. Charles E. Smith for discussion and for assistance with freeze drying and for kindly preparing the microdissected samples, and Dr. Hiroichi Koba for his independent corroboration of the sequence of the targeting vector used to create the AMBNΔ5–6 mutant mice and RT-PCR analysis. We also thank SkyScan for performing the X-ray microtomographic scans. This work was supported by the Canadian Institutes of Health Research (CIHR), the Network for Oral and Bone Health Research (RSBO), and the Intramural Research Programof the National Institute of Dental and Craniofacial Research (NIDCR #DE000720-02), NIH, U.S.A. Dr. Pierre Moffatt is supported by the Shriners of North America.
ABBREVIATIONS
- AMBN
ameloblastin
- AMBNΔ5–6
AMBN mouse lacking exons 5 and 6
- AMBNHET
AMBN heterozygote mouse
- AMBNWT
AMBN wild type mouse
- AMEL
amelogenin
- AMLN
amelin
- BEI
backscattered electron imaging
- BSP
bone sialoprotein
- CT
connective tissue
- dcw
distal cell web
- dpTP
distal portions of Tomes’ processes
- EO
enamel organ
- ES
enamel space
- F
forward primer (5′-side region of DNA)
- HD
hemidesmosome
- JE
junctional epithelium
- M
maturation stage of amelogenesis
- MA
apical maturation
- MI
incisal maturation
- N
nucleus
- OPN
osteopontin
- ORF
open reading frame
- pA signal
polyadenylation signal
- PDL
periodontal ligament
- ppTP
proximal portion of Tomes’ process
- PS
presecretory stage of amelogenesis
- R
reverse primer (3′-side of DNA)
- RT-PCR
reverse transcriptase polymerase chain reaction
- S
secretory stage of amelogenesis
- SCPP
secretory calcium-binding phosphoprotein
- SDS-PAGE
SDS-polyacrylamide gel electrophoresis
- SEM
scanning electron microscope
- sg
secretory granules
- TF
tonofilament
- TSS
transcription start signal
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kawasaki K, Weiss KM. Mineralized tissue and vertebrate evolution: the secretory calcium-binding phosphoprotein gene cluster. Proc Natl Acad Sci U S A. 2003;100(7):4060–4065. doi: 10.1073/pnas.0638023100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu JC, Yamakoshi Y, Yamakoshi F, Krebsbach PH, Simmer JP. Proteomics and genetics of dental enamel. Cells Tissues Organs. 2005;181(3–4):219–231. doi: 10.1159/000091383. [DOI] [PubMed] [Google Scholar]
- 3.Fong CD, Slaby I, Hammarström L. Amelin: An enamel-related protein, transcribed in the cells of epithelial root sheath. J Bone Miner Res. 1996;11(7):892–898. doi: 10.1002/jbmr.5650110704. [DOI] [PubMed] [Google Scholar]
- 4.Simmons D, Gu TT, Krebsbach PH, Yamada Y, MacDougall M. Identification and characterization of a cDNA for mouse ameloblastin. Connect Tissue Res. 1998;39(1–3):307–316. doi: 10.3109/03008209809023907. [DOI] [PubMed] [Google Scholar]
- 5.Bègue-Kirn C, Krebsbach PH, Bartlett JD, Butler WT. Dentin sialoprotein, dentin phosphoprotein, enamelysin and ameloblastin: tooth-specific molecules that are distinctively expressed during murine dental differentiation. Eur J Oral Sci. 1998;106(5):963–970. doi: 10.1046/j.0909-8836.1998.eos106510.x. [DOI] [PubMed] [Google Scholar]
- 6.Spahr A, Lyngstadaas SP, Slaby I, Pezeshki G. Ameloblastin expression during craniofacial bone formation in rats. Eur J Oral Sci. 2006;114(6):504–511. doi: 10.1111/j.1600-0722.2006.00403.x. [DOI] [PubMed] [Google Scholar]
- 7.Hao J, He G, Narayanan K, Zou B, Lin L, Muni T, Ramachandran A, George A. Identification of differentially expressed cDNA transcripts from a rat odontoblast cell line. Bone. 2005;37(4):578–588. doi: 10.1016/j.bone.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Murakami C, Dohi N, Fukae M, Tanabe T, Yamakoshi Y, Wakida K, Satoda T, Takahashi O, Shimizu M, Ryu OH, Simmer JP, Uchida T. Immunochemical and immunohistochemical study of 27 and 29 kDa calcium binding proteins and related proteins in the porcine tooth germ. Histochem Cell Biol. 1997;107:485–494. doi: 10.1007/s004180050136. [DOI] [PubMed] [Google Scholar]
- 9.Uchida T, Murakami C, Dohi N, Wakida K, Satoda T, Takahashi O. Synthesis, secretion, degradation and fate of ameloblastin during the matrix formation stage of the rat incisor as shown by immunocytochemistry and immunochemistry using region-specific antibodies. J Histochem Cytochem. 1997;45(10):1329–1340. doi: 10.1177/002215549704501002. [DOI] [PubMed] [Google Scholar]
- 10.Nanci A, Zalzal S, Lavoie P, Kunikata M, Chen WY, Krebsbach PH, Yamada Y, Hammarström L, Simmer JP, Fincham AG, Snead ML, Smith CE. Comparative immunochemical analyses of the developmental expression and distribution of ameloblastin and amelogenin in rat incisors. J Histochem Cytochem. 1998;46(8):911–934. doi: 10.1177/002215549804600806. [DOI] [PubMed] [Google Scholar]
- 11.Krebsbach PH, Lee SK, Matsuki Y, Kozak CA, Yamada KM. Full-length sequence, localization, and chromosomal mapping of ameloblastin: a novel tooth-specific gene. J Biol Chem. 1996;271:4431–4435. doi: 10.1074/jbc.271.8.4431. [DOI] [PubMed] [Google Scholar]
- 12.Vymetal J, Slaby I, Spahr A, Vondrasek J, Lyngstadaas SP. Bioinformatic analysis and molecular modelling of human ameloblastin suggest a two-domain intrinsically unstructured calcium-binding protein. Eur J Oral Sci. 2008;116(2):124–134. doi: 10.1111/j.1600-0722.2008.00526.x. [DOI] [PubMed] [Google Scholar]
- 13.Lee SK, Kim SM, Lee YJ, Yamada KM, Yamada Y, Chi JG. The structure of the rat ameloblastin gene and its expression in amelogenesis. Mol Cells. 2003;15(2):216–225. [PubMed] [Google Scholar]
- 14.Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, Krebsbach PH, Nanci A, Kulkarni AB, Yamada Y. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 2004;167(5):973–983. doi: 10.1083/jcb.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paine ML, Wang HJ, Luo W, Krebsbach PH, Snead ML. A transgenic animal model resembling amelogenesis imperfecta related to ameloblastin overexpression. J Biol Chem. 2003;278(May 23):19447–19452. doi: 10.1074/jbc.M300445200. [DOI] [PubMed] [Google Scholar]
- 16.Bosshardt DD, Lang NP. The junctional epithelium: from health to disease. J Dent Res. 2005;84(1):9–20. doi: 10.1177/154405910508400102. [DOI] [PubMed] [Google Scholar]
- 17.Shimono M, Ishikawa T, Enokiya Y, Muramatsu T, Matsuzaka K, Inoue T, Abiko Y, Yamaza T, Kido MA, Tanaka T, Hashimoto S. Biological characteristics of the junctional epithelium. J Electron Microsc. 2003;52(6):627–639. doi: 10.1093/jmicro/52.6.627. [DOI] [PubMed] [Google Scholar]
- 18.Kawaguchi H, McKee MD, Okamoto H, Nanci A. Immunocytochemical and lectin-gold characterization of the interface between alveolar bone and implanted hydroxyapatite in the rat. Cells Mater. 1993;3:337–350. [Google Scholar]
- 19.Yamakoshi Y, Tanabe T, Oida S, Hu CC, Simmer JP, Fukae M. Calcium binding of enamel proteins and their derivatives with emphasis on the calcium-binding domain of porcine sheathlin. Arch Oral Biol. 2001;46(11):1005–1014. doi: 10.1016/s0003-9969(01)00070-x. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi K, Yamakoshi Y, Hu JC, Gomi K, Arai T, Fukae M, Krebsbach PH, Simmer JP. Splicing determines the glycosylation state of ameloblastin. J Dent Res. 2007;86(10):962–967. doi: 10.1177/154405910708601009. [DOI] [PubMed] [Google Scholar]
- 21.Nanci A, Fortin M, Ghitescu DL. Endocytotic functions of ameloblasts and odontoblasts: Immunocytochemical and tracer studies on the uptake of plasma proteins. Anat Rec. 1996;245:219–234. doi: 10.1002/(SICI)1097-0185(199606)245:2<219::AID-AR9>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 22.Nanci A, Wazen RM, Zalzal S, Fortin M, Goldberg HA, Hunter GK, Ghitescu DL. A tracer study with systemically and locally administered dinitrophenylated osteopontin. J Histochem Cytochem. 2004;52(12):1591–1600. doi: 10.1369/jhc.4A6452.2004. [DOI] [PubMed] [Google Scholar]
- 23.Bosshardt DD, Nanci A. Immunodetection of enamel- and cementum-related (bone) proteins at the enamel-free area and cervical portion of the tooth in rat molars. J Bone Miner Res. 1997;12(3):367–379. doi: 10.1359/jbmr.1997.12.3.367. [DOI] [PubMed] [Google Scholar]
- 24.Lyngstadaas SP, Risnes S, Sproat BS, Thrane PS, Prydz HP. A synthetic, chemically modified ribozyme eliminates amelogenin, the major translation product in developing mouse enamel in vivo. EMBO J. 1995;14(21):5224–5229. doi: 10.1002/j.1460-2075.1995.tb00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zalzal SF, Smith CE, Nanci A. Ameloblastin and amelogenin share a common secretory pathway and are co-secreted during enamel formation. Matrix Biol. 2008;27(4):352–359. doi: 10.1016/j.matbio.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura Y, Slaby I, Spahr A, Pezeshki G, Matsumoto K, Lyngstadaas SP. Ameloblastin fusion protein enhances pulpal healing and dentin formation in porcine teeth. Calcif Tissue Int. 2006;78(5):278–284. doi: 10.1007/s00223-005-0144-2. [DOI] [PubMed] [Google Scholar]
- 27.Kanazashi M, Gomi K, Nagano T, Tanabe T, Arai T, Fukae M. The 17-kDa sheath protein in enamel proteins induces cementum regeneration in experimental cavities created in a buccal dehiscence model of dogs. J Periodontal Res. 2006;41(3):193–199. doi: 10.1111/j.1600-0765.2005.00859.x. [DOI] [PubMed] [Google Scholar]
- 28.Schroeder HE. The rate of the eruption of human teeth. A review Schweiz Monatsschr Zahnmed. 1991;101(3):279–284. [PubMed] [Google Scholar]
- 29.Boskey AL. Assessment of bone mineral and matrix using backscatter electron imaging and FTIR imaging. Curr Osteoporos Rep. 2006;4(2):71–75. doi: 10.1007/s11914-006-0005-6. [DOI] [PubMed] [Google Scholar]
- 30.Smith CE, Nanci A. A method for sampling the stages of amelogenesis on mandibular rat incisors using the molars as a reference for dissection. Anat Rec. 1989;225:257–266. doi: 10.1002/ar.1092250312. [DOI] [PubMed] [Google Scholar]
- 31.Fisher LW, Stubbs JT, 3rd, Young MF. Antisera and cDNA probes to human and certain animal model bone matrix noncollagenous proteins. Acta Orthop Scand Suppl. 1995;266:61–65. [PubMed] [Google Scholar]
- 32.Simmer JP, Fincham AG. Molecular mechanisms of dental enamel formation. Crit Rev Oral Biol Med. 1995;6(2):84–108. doi: 10.1177/10454411950060020701. [DOI] [PubMed] [Google Scholar]