Abstract
Post-translational modifications such as phosphorylation are known to play an important role in the gene regulation by the transcription factors including the nuclear hormone receptor superfamily of which the glucocorticoid receptor (GR) is a member. Protein phosphorylation often switches cellular activity from one state to another. Like many other transcription factors, the GR is a phosphoprotein, and phosphorylation plays an important role in the regulation of GR activity. Cell signaling pathways that regulate phosphorylation of the GR and its associated proteins are important determinants of GR function under various physiological conditions. While the role of many phosphorylation sites in the GR is still not fully understood, the role of others is clearer. Several aspects of transcription factor function, including DNA binding affinity, interaction of transactivation domains with the transcription initiation complex, and shuttling between the cytoplasmic compartments, have all been linked to site-specific phosphorylation. All major phosphorylation sites in the human GR are located in the N-terminal domain including the major transactivation domain, AF1. Available literature clearly indicates that many of these potential phosphorylation sites are substrates for multiple kinases, suggesting the potential for a very complex regulatory network. Phosphorylated GR interacts favorably with critical coregulatory proteins and subsequently enhances transcriptional activity. In addition, the activities and specificities of coregulators may be subject to similar regulation by phosphorylation. Regulation of the GR activity due to phosphorylation appears to be site-specific and dependent upon specific cell signaling cascade. Taken together, site-specific phosphorylation and related kinase pathways play an important role in the action of the GR, and more precise mechanistic information will lead to fuller understanding of the complex nature of gene regulation by the GR- and related transcription factors. This review provides currently available information regarding the role of GR phosphorylation in its action, and highlights the possible underlying mechanisms of action.
Keywords: glucocorticoid receptor, phosphorylation, transactivation activity, gene regulation, coactivators
Introduction
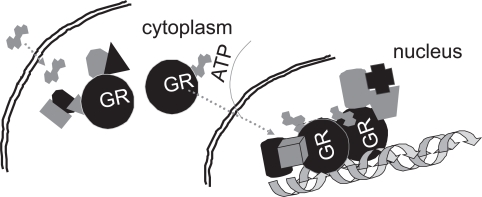
The glucocorticoid receptor (GR) belongs to the superfamily of the hormone-activated intracellular transcription factors, the nuclear hormone receptors (NHRs). Most of the biological effects of glucocorticoids are mediated through the GR and occur at the level of regulation of gene transcription (Evans 1988; Beato 1989; Yamamoto 1985). The role of the GR, which acts in a ligand-, cell type-, and promoter-specific manner is important in the transcriptional activation of genes involving the regulated assembly of multiprotein complexes on enhancers and promoters (Simons 1994; Kumar et al 1999a; Rogatsky et al 2003). The classical mechanism of steroid/hormone action (Figure 1) (Dean et al 1996; Kumar et al 1999b; Nissen et al 2000; Mendelson 2004) states that the GR in its inactive form is located in the cytoplasm, and after entry into the cell the steroid/hormone finds the receptor as part of a large heteromeric complex consisting of several proteins including HSP90, HSP70, Immunophilins, FKBPs, CyP-40, P23, and possibly others (Housley et al 1985; Sanchez et al 1987; Pratt et al 1997, 2003; Silverstein et al 1999; Shikama et al 2000; Davies et al 2002; Morishima et al 2003). Steroid binding leads to the disassembling of the complex through conformational changes in the receptor molecule and the activated receptor enters the nucleus, where it interacts with critical regulatory sites on the relevant genes (Hollenberg et al 1987; Orti et al 1992; Henriksson et al 1997; Horwitz et al 1996; Starr et al 1996; Lefstin et al 1998; Yamamoto et al 1998; McKenna et al 1999; Kumar et al 2003; Rogatsky et al 2003; Weigel et al 2007a, 2007b). The classic model for steroid action states that the DNA bound receptor collects a variety of ancillary factors which modify chromatin structure and/or contact the primary transcription complex machinery proteins such that transcription from the relevant promoter can be either enhanced or repressed. However, this has led to the problem of understanding how the receptor molecule makes such large protein surface areas available to accommodate specific sites for all these factors. The molecular mechanisms to explain this phenomenon have not been fully defined.
Figure 1.
Classical action of the glucocorticoid signaling mediated by the GR. Unliganded receptor is located in the cytosol associated with several heat shock and other chaperone proteins including HSP90, HSP70, CyP-40, P23, and FKBPs (shown by different shapes and shades in the cytosol). Ligand binding leads to conformational alterations in the GR, and by doing so GR dissociates from these associated proteins, and ligand bound GR is free to translocate to the nucleus. This process appears to be phosphorylation-dependent. Once in the nucleus, GR dimerizes and binds to site-specific DNA binding sequences and interacts with several other coregulatory proteins including coactivators and proteins from the basal transcription machinery including SRCs, CBP/p300, DRIP/TRAP, TBP, GRIP1, and several others (shown by different shapes and shades) in the nucleus, and subsequently leading to transcriptional regulation.
There are possibilities that some of these actions can be explained on the basis of post-translational modifications of specific NHRs. In spite of the fact that GR is a hormone-activated transcription factor, its expression and many physiological activities are highly regulated by post-translational modifications (Webster et al 1997; King et al 1998; Wallace et al 2001; Le Drean et al 2002; Tian et al 2002). One such important post-translational modification associated with GR function is site-specific phosphorylation (Wang et al 2002; Ismaili et al 2004; Duma et al 2006). Involvement of several known phosphorylation sites in the GR and variety of kinases facilitates integration between cell-signaling pathways and the GR action (Krstic et al 1997; Rogatsky et al 1998a). Most, if not all known phosphorylation sites in the GR are localized within the N-terminal domain (NTD) that contains a major transactivation domain, AF1 (Bodwell et al 1991, 1998; Blind et al 2004; Kumar et al 2004a).
Post-translational modifications including phosphorylation are generally an important phenomenon in regulation of protein function in eukaryotic cells, and are often concerned with switching of a cellular activity from one state to another (Auricchio 1989; Orti et al 1989; Kuiper et al 1994). It is now well accepted that for several transcription factors including GR, site-specific phosphorylation can modulate their DNA binding affinity, the interaction of transactivation domains of these transcription factors with components of the transcription initiation complex, and the shuttling of transcription factors between the cytoplasmic compartments (Bai et al 1995; Rogatsky et al 1998b; Wang et al 1999; Gioeli et al 2002). There are several reports showing that kinases can phosphorylate GR at multiple sites leading to altered GR activity (Blind et al 2004; Ismaili et al 2004). Depending upon the kinases involved, the GR activity can be both up- or down-regulated (Ismaili et al 2004). For example, phosphorylation of GR by cyclin-dependent kinases (Cdk1, Cdk2, and cdk5) leads to up regulate GR activity (Ismaili et al 2004; Weigel et al 2007a), whereas JNK and GSK3 inhibits GR activity through direct phosphorylation of GR (Rogatsky et al 1998a, 1998b). In contrast, certain kinases such as p38 MAPK are reported to both up- and down-regulate GR activity, these effects of p38 MAPK appears to be cell-specific (Szatmary et al 2004; Miller et al 2005).
Some of the GR phosphorylation sites are conserved across species, whereas others are unique to specific species (Ismaili et al 2004). Generally, GR phosphorylation is ligand-dependent, which suggests that this may be a determinant of promoter specificity, interaction with coregulatory proteins, and even GR stability (Webster et al 1997; Zhou et al 2005; Faus et al 2006). There are several kinases that phosphorylate the GR with the resultant phosphorylation enhancing GR transcriptional activity (Ismaili et al 2004). However, GR can also be phosphorylated in a ligand independent manner as well (Ismaili et al 2004, 2005). In fact, we have earlier shown that p38 MAPK can phosphorylate human GR fragment lacking the LBD, which enhances the transactivation activity of the GR and mediates GR-dependent apoptosis in cells transfected with this fragment (Miller et al 2005). We also found that p38 was activated probably through immediate upstream activators, MKK3 and MKK6, both specific to p38 (Miller et al 2005). In addition, MKK4, whose best-known substrate is JNK, can also phosphorylate p38. In recent years, it is becoming clearer that many receptor phosphorylation sites are substrates for multiple kinases, suggesting towards potential for very complex regulatory patterns. In this review article, we have summarized knowledge regarding the role of GR phosphorylation in the regulation of receptor function, and the possible underlying mechanisms of action.
The structural arrangements of the glucocorticoid receptor (GR)
The GR protein consists of a domain structure arrangement, typical of the superfamily of the NHRs (Yamamoto 1985; Evans 1988; Beato 1989; Simons 1994; Kumar et al 1999). This modular conceptual framework has been useful, but it has become clear that there are certain limits to the domain. This fundamental domain model divides the primary sequence into NTD, DNA-binding domain (DBD) (Hard et al 1990; Luisi et al 1991), and LBD with “activation function” (AF2) sub-domain that regulates transcription (Giguere et al 1986; Hollenberg et 1987; Bocquel et al 1989; Ikonen et al 1997; Metivier et al 2001; Bledsoe et al 2002). Ligand binding to the LBD results in conformational rearrangement of its AF2 sub-domain (usually helix 12) such that its surfaces are available for interactions with specific coregulatory proteins through LXXLL motifs. The GR and several other receptors in the family have long NTD (Kumar et al 2003). In these is found another, potent transactivation domain, AF1 (Godowski et al 1987; Miesfeld et al 1987). Cooperative binding and site specificity combine for GR:GRE tethering allows receptor’s interaction with the transcription initiation complex, directly by interactions between AF1/AF2 and the complex, or indirectly through specific coregulators (Thompson et al 2003). To a first approximation, this conceptual model is useful, but fails to explain several essential facts such as the role of post-transcriptional modifications, including sumoylation, acetylation, and site-specific phosphorylation. Our recent data and available literature from other laboratories clearly indicate an important role of GR phosphorylation and involvement of specific kinases (Rogatsky et al 1998a; Ismaili et al 2004; Miller et al 2005). Other data suggests that intramolecular signaling occurs as well between the amino-terminal and LBD regions (Ikonen et al 1997; Hittelman et al 1999; Tetel et al 1999; Kumar et al 1999; Bommer et al 2002). To fully understand the mechanisms of action of the GR, we must understand the answer to the above mentioned fundamental questions.
Phosphorylation regulates structure and functions of the GR
Like many other transcription factors, the GR is a phosphoprotein, which becomes hyperphosphorylated upon steroid binding, and it has been suggested that phosphorylation plays an important role in the regulation of GR activity (Ismaili et al 2004). Protein phosphorylation is generally an important regulatory mechanism for protein function in eukaryotic cells, and is often concerned with switching of a cellular activity from one state to another. Phosphorylation of the Pol II protein complex is correlated with the transition between transcriptional initiation and elongation (Nissen et al 2000; Pinhero et al 2004); thus phosphorylation may be involved in transcriptional regulation. In recent years, more and more evidence point towards the role of phosphorylation and other post-translational modifications in the gene expression and regulation of many transcription factors (Boyle et al 1991; Nichols et al 1992; Wisniewski et al 1999).
Cellular kinases play a prominent regulator role for the NHR (Ismaili et al 2004). Ligand binding, nuclear translocations, modulation of binding to REs, receptor dimerization, and interaction with general transcription factors, have all been linked to phosphorylation (Ismaili et al 2004). It has been reported that kinases enhance the transactivation activity of several steroid receptors both in a ligand-dependent and ligand-independent manner (Rogatsky et al 1998b; Ismaili et al 2004). For transcription factors, three main mechanisms of regulation by phosphorylation can be identified; 1) the DNA binding affinity of transcription factors can be modulated negatively or positively; 2) the interaction of transactivation domains of transcription factors with components of the transcription initiation complex can be affected; and 3) the shuttling of transcription factors between the cytoplasmic compartments can be influenced.
In the human GR five serine residues (S113, S141, S203, S211, and S226) have been identified (Ismaili et al 2004), whereas in the mouse GR there are eight phosphorylation residues identified. In mice, seven of the 8 phosphorylation sites are at serine residues (S122, S150, S212, S220, S234, S315, and S412) and one at threonine (T159) (Bodwell et al 1991; Bodwell 1998; Rogatsky et al 1998a). All the known phosphorylation sites identified in mouse GR are found in the NTD, in or near the AF1 domain (Mason et al 1993). Several kinases are reported to be involved in the phosphorylation of these sites, such as mitogen protein kinases (MAPK), cyclin-dependent kinase (CDK) (Krstic et al 1997), glycogen synthase kinase-3 (GSK-3), and c-Jun N-terminal kinases (JNK) (Rogatsky et al 1998a). Though, each of these kinases is reported to show distinct specificities for potential phosphorylation residues, full receptor activity may require synergistic effects of these kinases and their signaling cascades.
In the human GR AF1, major functionally important known phosphorylated residues are S203, S211, and S226 (ure 2). At least two of these (S211 and S226) are thought to be important for transcriptional activity of the GR (Rogatsky et al 1998b; Ismaili et al 2004; Miller et al 2005). The use of site-specific antibodies has shown that S203 phosphorylation is involved in the GR translocation (Ismaili et al 2004). It has also shown that site-specific (S211) phosphorylation of the GR AF1 enhances its interaction with a protein from the DRIP/TRIP complex, and subsequently GR activity (Blind et al 2004; Ismaili et al 2004). These observations clearly suggest that site-specific phosphorylation in GR AF1 may regulate GR function, in an AF1-dependent manner. However, it is not yet known exactly how phosphorylation influences the structure and functions of the GR AF1.
The AF1 can act constitutively in the absence of LBD, and is quite active in stimulating transcription from simple promoters containing cognate binding sites(s). Because GR AF1 exists in intrinsically disordered conformation(s), advances in understanding its structure and functions have been slowed (Dahlman-Wright et al 1995; Baskakov et al 1999; Kumar et al 2004b). It is known that for function AF1 interacts with specific coregulatory proteins such as CBP, TBP, TIF2, DRIP/TRAP, Ada among others, and the available data strongly suggest that AF1 must adopt an ordered conformation to optimize interactions and subsequent transcriptional activity (Kumar et al 2003). In recent years, it has become quite evident that eukaryotic genomes are highly enriched in intrinsically disorder proteins relative to prokaryotes, reflecting the greater need for signaling and regulation in nucleated cells (Crivici et al 1995; Namba 2001; Dyson et al 2002; Iakoucheva et al 2002; Tompa 2002; Romero et al 2004; Ward et al 2004; Liu et al 2006). These disordered protein regions/domains promote molecular recognition primarily through unique combination of high specificity and low binding affinity with their functional binding partners, recognize and bind a number of biological targets, and create propensity to form large interaction surfaces suitable for interactions with their specific binding partners (Crivici et al 1995; Namba 2001; Iakoucheva et al 2002; Dyson et al 2002; Tompa 2002; Romero et al 2004; Ward et al 2004; Liu et al 2006). We hypothesize that site-specific phosphorylation of the GR AF1 leads to changes in its conformations that are important for AF1’s interaction with other critical coregulatory proteins, and subsequent transcriptional activity.
There are also reports suggesting that phosphorylation may affect GR stability and thus alter transcriptional activity of the receptor (Zhou et al 2005). Co-transfection data from Cidlowski and colleagues (Webster et al 1997; Wallace et al 2001) have shown that phosphorylated mouse GR had a half-life of 8–9 h in the presence of agonist, whereas mutations of multiple GR phosphorylation sites increased its half-life to 32 h. All the GR phosphorylation sites that are conserved among human, mouse, and rat are located within the AF1 domain. Single mutations of these do not alter receptor activity in an MMTV-driven promoter-reporter construct, but a GR lacking all five sites that are well conserved in mouse, rat and human GR shows significantly decreased activity (Almlof et al 1995). Our recent published data however show that p38 and ERK2 are potent kinases for in vitro phosphorylation of S211 (located in AF1 domain) on the human GR (Miller et al 2005). The mutant lacking a phosphorylatable amino acid at position 211 (S221A) was considerably less potent in inducing the AF1-medited, GRE-driven reporter gene, and GR-mediated apoptosis induced by dexamethasone (Miller et al 2005). Garabedian and colleagues (Rogatsky et al 1998b; Blind et al 2004; Ismaili et al 2004; Kino et al 2007; Wang et al 2007) have also demonstrated that site-specific phosphorylation in GR, particularly S211 and S226 play an important role in gene regulation by the GR, and AF1 appears to be a main player in this process. Thus, in contrast to earlier study that suggested lack of specificity of phosphorylation (Almlof et al 1995), these more recent findings clearly indicate the role of site-specific phosphorylation in AF1-mediated GR functions (Ismaili et al 2004; Miller et al 2005).
Role of phosphorylation in protein: protein interactions involving the GR and specific coregulators
In the conceptual model of a GR:coactivator complexes, the ligand-bound receptor recruits one or more coactivators, which subsequently results in the recruitment of additional coactivators to the assembly. The histone acetylation and methylation activities of various constituents of the coactivator complex facilitate the relaxation of the chromatin architecture at the target gene promoter, thereby enhancing transcriptional activation. Analogous arguments can be made for GR:corepressors complexes (Horwitz et al 1996; Yamamoto et al 1998; McKenna et al 1999). The GR AF1 domain is known to play an important role in many of these interactions, via interaction with regions of the coactivator remote from the LXXLL motif (Kumar et al 2003). The ability of GR AF1 to interact with components of the general transcriptional machinery or with coregulator complexes provides a broad insight into the process of transcriptional initiation. CBP, TBP, AdA2, DRIP150, TSG101, and several other co-regulators have been shown to bind to the GR AF1 (Kumar et al 2003; Ismaili et al 2004). Direct binding of TBP with AF1, raises the possibility that this GR domain somehow directly influences the transcriptional machinery. In vitro transcription studies indicated that the holo-GR acts to stabilize the pre-initiation complex (Horwitz et al 1996; Yamamoto et al 1998; McKenna et al 1999), though other mechanisms to control transcription have recently been reported (Glass et al 1997; Yamamoto et al 1998; Fujita et al 2003; Loven et al 2003).
It has been reported that the interaction between the GR and TSG101 may be modulated through GR phosphorylation (Ismaili et al 2005). This study shows that TSG101 is preferentially recruited to the nonphosphorylated form of the GR (Ismaili et al 2005). Another GR AF1 coregulator DRIP150 is also been reported to be modulated through GR phosphorylation (Ismaili et al 2004). Our unpublished data (Garza and Kumar, in preparation) show that interaction of several GR AF1 coregulators including TBP, CBP, and SRC-1 are facilitated by GR phosphorylation. Moreover, phosphorylation of several transcription factors, including NHRs enhances recruitment of coregulatory proteins (Wu et al 2005). Thus, it can be concluded that site-specific phosphorylation of the AF1 domain of GR can either enhance or diminish recruitment of coregulators.
These contrasting effects of AF1/GR phosphorylation on recruitment of specific coregulators may reflect the biologic need for the GR to up- or down-regulate gene(s) in a cell- and promoter-specific manner. Recent studies have shown that GR site-specific phosphorylation can differentially regulate the expression of several target genes (Chen et al 2008; Davies et al 2008; Blind et al 2008). However, this phenomenon is not clearly understood, and further studies are needed to provide a more comprehensive explanation for this behavior. One clue to the complexity of this system is that SG101 stabilizes ligand-unbound GR in its unphosphorylated form to protect it from degradation. Our recent findings (Garza and Kumar, in preparation) suggest that phosphorylation of the GR stabilizes the conformation of otherwise unstructured AF1 such that AF1’s surfaces are available for its interaction with co-regulatory proteins. Thus, TSG101 interaction with GR may be important to keep unliganded GR protected from autodegradation until the GR becomes phosphorylated.
Recent studies suggest that in addition to the receptor phosphorylation, the activities and specificities of coregulators are also subject to similar regulation by phosphorylation. For example, the actions of coregulators including SRCs and CBP can be modified by phosphorylation (Tzukerman et al 1994; Chen et al 1999; Font de Mora et al 2000; Rowan et al 2000a; Lopez et al 2001; See et al 2001; Puigserver et al 2001; Yang et al 2001; Chevillard-Briet et al 2002; Kotaja et al 2002; Chauchereau et al 2003; Wu et al 2005). Several phosphorylation sites for SRC-1 and SRC-3 have recently been identified (Rowan et al 2000b; Wu et al 2004). Phosphorylation of these coregulators is induced by steroid hormones (Lopez et al 2001; Wu et al 2004, 2005), and phosphorylation is critical for optimal activities. These data suggest that phosphorylation of coregulators may influence their protein-protein interactions with GR. This concept raised the possibility that the phosphorylation patterns of specific coregulator(s) might serve as a signal integrator, permitting this complex network of GR and cofactors to accurately and specifically activate a broad range of promoters for transcriptional activation.
Role of phosphorylation on the transactivation activity of the GR
It has been demonstrated that the status of GR phosphorylation can alter its transcriptional activity (Mason et al 1993). There are reports showing that in human GR S203A and/or S211A mutations significantly repress GR activity, whereas S226A mutation has increased GR activity in a yeast system (Almlof et al 1995), suggesting that effects of phosphorylation on the GR transcriptional activity could be both up- or down-regulated depending upon the site of phosphorylation, and (due to site-specificity of the kinase) probably the specific kinase pathways involved (Ismaili et al 2004). Reduced GR activity and decreased level of phosphorylation status have been observed in cells deficient in the CDK inhibitor p27 (Rogatski et al 1997). However, other studies show that mutations of multiple GR phosphorylation sites do not significantly alter GRE-mediated GR activity in cell co-transfection studies (Almlof et al 1991). A number of explanations for these apparently conflicting results exist. For example, some studies were conducted using different promoters, and therefore it is quite possible that the influence of the GR phosphorylation on its transcriptional activation activity may be dependent on the promoter context of the target gene. It is also likely that site-specific phosphorylation of the GR would contribute to endogenous gene regulation, acting in a cell-type specific manner in which the use of kinases might differ (Zhou et al 2005). Another possible explanation is that individual phosphorylation sites may influence translocation to the nucleus (Webster et al 1997). However, the presence of conflicting data (Wang et al 2007) highlights the complexity of this system, and the need for more studies to establish underlying mechanisms.
We have shown that a mutation of Ser211 to Ala residue reduced GR-mediated transcriptional activation and apoptosis in a human leukemia cell line, suggesting a role for p38 MAPK signaling in glucocorticoid-induced apoptosis of lymphoid cells. With respect to the consequences of p38 MAPK phosphorylation, it has further been shown that S211 residue of AF1 is a specific substrate site for p38 MAPK, and that mutation S211A, which prevents phosphorylation at this position, diminishes apoptosis driven by the constitutively active GR lacking the ligand binding domain, suggesting a possible role of AF1-mediated GR transcriptional activity (Miller et al 2005). However, it is not known whether other phosphorylation sites (eg, S203 and S226) are also involved in this process. Further studies in our laboratory are exploring a variety of possibilities to understand the role of phosphorylation in GR function. Similar effects have been shown in other laboratories, using different cells and kinase pathways (Ismaili et al 2004). Thus, in contrast to earlier study (Almlof et al 1995), these recent findings clearly indicate the critical role of site-specific phosphorylation of AF1 in regulating GR functions. Activity of steroid bound GR may also be reduced due to site-specific phosphorylation (Ismaili et al 2004). For example, activation of JNK induces rat GR phosphorylation at S246 which significantly reduces GR activity; mutation of S246A, eliminating the potential phosphorylation site, diminishes inhibitory effects of JNK (Ismaili et al 2004). These results suggest that phosphorylation at S246 either increases its affinity for a corepressor or decreases its interaction with a coactivator. Thus, site-specific phosphorylation and the particular kinase pathways involved in different cell types may dictate the pattern of GR regulation of specific target gene(s). Further, these effects may be synergistic in nature.
Additional studies are needed to clarify the underlying mechanisms to determine these effects. A possible role of phosphatases has also been reported to regulate GR phosphorylation and its subsequent effects of the transcriptional activity (Ismaili et al 2004). Perturbations in protein phosphatase activity have been shown to affect GR function. Treatment of cells with protein phosphatase inhibitor results in enhanced phosphorylation of GR, accumulation of GR in the cytoplasm, and subsequent reduction in GR-mediated transcriptional activation (DeFranco et al 1991; Sommers et al 1992). However, the effects of specific phosphatases (and by extension specific phosphatase inhibitor) on site-specific GR phosphorylation and function have not been well established. Collectively, the effect of phosphatases on GR signaling and transcriptional regulation is complex, and requires further examination. Thus, it becomes of great importance to determine the pattern, kinetics, extent, and effects of phosphorylation/dephosphorylation in modulating GR action. Subsequently, it will be critical to establish the mechanisms by which modulation of GR activity occurs, and determine whether AF1 phosphorylation, in a site-specific manner, is an integrating phenomenon for GR regulation. Although short of actual proof, it seems apparent that both kinases and phosphatases may be interacting in complex ways to determine phosphorylation of specific sites within the GR AF1 that may well be cell- and promoter-specific in nature.
Summary and perspectives
Steroids have been frontline therapy for decades in the treatment of malignancies and inflammatory disorders; however the mechanism by which steroid receptors pass signals from ligand to regulate specific genes is not fully understood. Recent progress combined with classical understanding of the steroid receptors action, and the availability of the structure of individual domains of several members of the NHR superfamily including the GR has provided new understanding of the function of the GR. Recent studies have suggested that under physiological conditions, there are many factors that influence the conformation of the GR such that malleable protein surfaces are available for interaction with appropriate coactivators/corepressors. However, it remains to be determined how precisely these GR:protein interactions may differ with different cell- and promoter-specific conditions, and how the receptor communicates with transcription initiation machinery. Another important problem that remains to be solved is the precise mechanism by which AF1 and AF2 synergize with each other in the holo-receptor. Post-translational modifications including phosphorylation, ubiquitination, and sumoylation have all been shown to affect functions of NHR family members.
In recent years it has become clear that the cross-talk exists between GR signaling and other receptor cascades including inflammatory kinases (MAPKs, ERK, p38 and JNK) as well as the cAMP-driven PKA pathways. There is evidence that that differential phosphorylation is a potential regulator of species-specific actions of the GR. However, current knowledge reveals that the role of GR phosphorylation is a remarkably complex phenomenon. It is evident that the GR is a target for multiple kinases, and that many GR functions are regulated by phosphorylation. Several outstanding questions remain to be answered. For example: 1) what are the relative levels of phosphorylation of individual sites under physiological conditions; 2) how do phosphatases influence the cellular machinery; and 3) what is the correlation between cell based studies and in vivo animal models. However, it is clear that GR-mediated glucocorticoid signaling is a multifaceted process involving crosstalk with various regulatory kinase pathways. Thus, signaling cascades that induce phosphorylation of the GR and its coactivator proteins are critical factors in determining the physiological actions of the GR. Further studies that clarify the regulation of endogenous target genes by specific phosphorylation site(s) should lead to target- and perhaps tissue-specific requirements for phosphorylation. There are several other avenues of post-translational modifications that might also affect the actions of GR. Ubiquitination-mediated degradation regulates glucocorticoid signaling by controlling the degradation rates of GR (Kinyamu et al 2005). Another post-translational modification, sumoylation, can also regulate the GR functions (Le Drean et al 2002). There are at least three known sumoylation sites in the human GR. Two of them are located in the NTD (K277 and K293) and one (K703) in the LBD (Faus et al 2006). Acetylation and methylation are other possible modifications that may influence GR actions; however, there is not enough evidence to suggest their direct role in modulating GR functions. At least one recent study has shown a ligand-dependent acetylation of the human GR at K494 and K495 residues, but the functional consequences of this modification are not clear (Ito et al 2006).
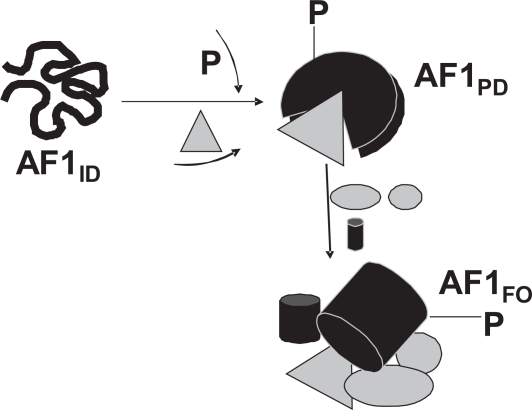
We propose that under physiological conditions, site-specific phosphorylation plays a crucial role in allowing the AF1 domain of the GR to adopt functionally active conformation(s) in vivo (Figure 3). The resulting structurally modified forms of AF1 suit it for its varied interactions with other critical coregulatory proteins, and possibly additional modulations in receptor structure essential for gene regulation by the GR. These interactions give a set of functionally active folded structure to AF1 and form the basis for the multiprotein assemblies involved in GR-mediated regulation of transcription. How site-specific phosphorylation leads to such AF1 conformation(s) and what kind of functional folded conformation it adopts are open questions, and we and others have been pursuing answers to these long-standing problems. Knowledge of the conformational changes in AF1 and/or other domains/regions of the GR due to site-specific phosphorylation and subsequently gene regulation will lead to an understanding of the role of this important phenomenon in the transcription process, information essential to understanding how glucocorticoids affect gene regulation.
Figure 3.
A hypothetical model of the role of site-specific phosphorylation on the structure and functions of the AF1 domain. The GR AF1 domain exists in an intrinsically disordered (ID) conformation ie, as an ensemble of conformers that collectively appear to lack any significant secondary/tertiary structure. Phosphorylation of one or more sites within the AF1 alters its conformation in such a way that AF1 adopts a partially folded (PD) conformation that facilitates AF1’s interaction with one or more of the coregulatory proteins. This protein:protein interaction allows structurally modified forms of AF1 to suit for its varied interactions with other critical coregulatory proteins, and possibly additional modulations in receptor structure essential for gene regulation by the GR. This AF1:coregulators assembly provides optimal ordered conformation in the AF1 for subsequent transactivation activity. Alternatively, one or more of the coregulator proteins directly interact with one or more unphosphorylated AF1 conformers that may trigger phosphorylation of AF1. The resultant assembly of proteins may depend upon the cellular environment, specific promoters used, and the kinase pathway involved. All the available phosphorylation sites in the GR could be involved simultaneously or there may be a coordinated synergy between each site. In the full length GR, AF1:coregulators assembly may also be influenced by other factors such as DNA binding to its DBD, ligand binding and the cross talk between AF1 and AF2. In addition to the level of AF1 phosphorylation, phosphorylation of different coregulators may also influence the outcome, and certain coregulators may be included or excluded from the assembly depending upon the interacting surfaces of the AF1 and/or coregulator proteins.
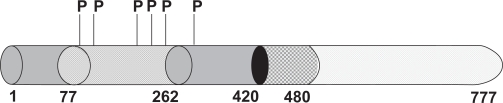
Figure 2.
A topological diagram of the human GR (amino acids 1–777) showing its modular structure and known phosphorylation sites in it (based on ref. 38). Numbers on the bottom indicate amino acid positions of different functional domains. 1–420, NTD; 77–262, AF1; 421–481; DBD; and remaining C-terminal part, the LBD are shown. P denotes known phosphorylation sites in the NTD of the human GR. Shown from left to right: S113, S141, S203, S211, S226, and S308. Corresponding amino acids in the rat GR are S134, S162, S224, S232, S246, and S329, whereas in the mouse GR these correspond to S122, S150, S212, S220, S234, and S315. In the rat and mouse GR, there is one Threonine residue (T171 in rat and T159 in mouse) that is known to be phosphorylable, and is not conserved in the human GR. In the human GR except for S308, all other residues are located within the AF1 domain, and S203, S211, and S226 are reported to have to have some functional roles in the action of the GR. Phosphorylation of these sites are kinase- and cell-dependent.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- Almlof T, Wright AP, Gustafsson JA. Role of acidic and phosphorylated residues in gene activation by the glucocorticoid receptor. J Biol Chem. 1995;270:17535–40. doi: 10.1074/jbc.270.29.17535. [DOI] [PubMed] [Google Scholar]
- Auricchio F. Phosphorylation of steroid receptors. J Steroid Biochem. 1989;32:613–22. doi: 10.1016/0022-4731(89)90397-x. [DOI] [PubMed] [Google Scholar]
- Bai W, Weigel NL. Phosphorylation and steroid hormone action. Vit Hormones. 1995;51:289–313. doi: 10.1016/s0083-6729(08)61042-0. [DOI] [PubMed] [Google Scholar]
- Baskakov IV, Kumar R, Srinivasan G, et al. Trimethylamine N-Oxide-induced cooperative folding of an intrinsically unfolded transcription-activating fragment of human glucocorticoid receptor. J Biol Chem. 1999;274:10693–6. doi: 10.1074/jbc.274.16.10693. [DOI] [PubMed] [Google Scholar]
- Beato M. Gene regulation by steroid hormones. Cell. 1989;56:335–4. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- Bledsoe RK, Montana VG, Stanley TB, et al. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell. 2002;110:93–105. doi: 10.1016/s0092-8674(02)00817-6. [DOI] [PubMed] [Google Scholar]
- Blind RD, Garabedian MJ.2004Regulation of glucocorticoid receptor-mediated transcriptional activation: The role of receptor phosphorylation and cofactor recruitmentAbstract Book, KeyStone Symposia on Nuclear Receptors.
- Blind RD, Garabedian MJ. Differential recruitment of glucocorticoid receptor phosphoisoforms to glucocorticoid-induced genes. J Steroid Biochem Mol Biol. 2008;109:150–7. doi: 10.1016/j.jsbmb.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocquel MT, Kumar V, Stricker C, et al. The contribution of the N- and C-terminal regions of steroid receptors to activation of transcription is both receptor and cell-specific. Nucleic Acids Res. 1989;17:2581–95. doi: 10.1093/nar/17.7.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodwell JE, Webster JC, Jewell CM, et al. Glucocorticoid receptor phosphorylation: overview, function and cell cycle-dependence. J Steroid Biochem Mol Biol. 1998;65:91–9. doi: 10.1016/s0960-0760(97)00185-4. [DOI] [PubMed] [Google Scholar]
- Bodwell JE, Orti E, Coull JM, et al. Identification of phosphorylated sites in the mouse glucocorticoid receptor. J Biol Chem. 1991;266:7549–55. [PubMed] [Google Scholar]
- Bommer M, Benecke A, Gronemeyer H. TIF2 mediates the synergy between RARα1 activation functions AF-1 and AF-2. J Biol Chem. 2002;277:37961–6. doi: 10.1074/jbc.M206001200. [DOI] [PubMed] [Google Scholar]
- Boyle WJ, Smeal T, Defize LHK, et al. Activation of protein kinas C decreases phosphorylation of c-jun at sites that negatively regulate its DNA-binding activity. Cell. 1991;64:573–84. doi: 10.1016/0092-8674(91)90241-p. [DOI] [PubMed] [Google Scholar]
- Chauchereau A, Amazit L, Quesne M. Sumoylation of the progesterone receptor and of the steroid receptor coactivator SRC-1. J Biol Chem. 2003;278:12335–43. doi: 10.1074/jbc.M207148200. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Xie W, et al. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–86. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- Chen W, Dang T, Blind RD, et al. 2008Glucocorticoid receptor phosphorylation differentially affects target gene expression Mol Endocrinol2008 May 15. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevillard-Briet M, Trouche D, Vandel L. Control of CBP co-activating activity by arginine methylation. EMBO J. 2002;21:5457–66. doi: 10.1093/emboj/cdf548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crivici A, Ikura M. Molecular and structural basis of target recognition by calmodulin. Annu Rev Biophys Biomol Struct. 1995;24:85–116. doi: 10.1146/annurev.bb.24.060195.000505. [DOI] [PubMed] [Google Scholar]
- Dahlman-Wright K, Baumann H, McEwan IJ, et al. Structural characterization of a minimal functional transactivation domain from the human glucocorticoid receptor. Proc Natl Acad Sci U S A. 1995;92:1699–703. doi: 10.1073/pnas.92.5.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies TH, Ning YM, Sánchez ER. A new first step in activation of steroid receptors: hormone-induced switching of FKBP51 and FKBP52 immunophilins. J Biol Chem. 2002;277:4597–600. doi: 10.1074/jbc.C100531200. [DOI] [PubMed] [Google Scholar]
- Davies L, Karthikeyan N, Lynch JT, et al. Crosstalk of signaling pathways in the regulation of the glucocorticoid receptor function. Mol Endocrinol. 2008;22:1331–44. doi: 10.1210/me.2007-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean DM, Sanders MM. Ten years after: reclassification of steroid-responsive genes. Mol Endocrinol. 1996;10:1489–95. doi: 10.1210/mend.10.12.8961259. [DOI] [PubMed] [Google Scholar]
- DeFranco DB, Qi M, Borror KC, et al. Protein phosphatase types 1 and/or 2A regulate nucleocytoplasmic shuttling of glucocorticoid receptors. Mol Endocrinol. 1991;5:1215–28. doi: 10.1210/mend-5-9-1215. [DOI] [PubMed] [Google Scholar]
- Duma D, Jewell CM, Cidlowski JA. Multiple glucocorticoid receptor isoforms and mechanisms of post-translational modification. J Steroid Biochem Mol Biol. 2006;102:11–21. doi: 10.1016/j.jsbmb.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–5. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faus H, Haendler B. Post-translational modifications of steroid receptors. Biomed Pharmacother. 2006;60:520–8. doi: 10.1016/j.biopha.2006.07.082. [DOI] [PubMed] [Google Scholar]
- Font de Mora J, Brown M. AIB1 is a conduit for kinase-mediated growth factor signaling to the estrogen receptor. Mol Cell Biol. 2000;20:5041–7. doi: 10.1128/mcb.20.14.5041-5047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Kobayashi Y, Wada O, et al. Full activation of estrogen receptor alpha activation function-1 induces proliferation of breast cancer cells. J Biol Chem. 2003;278:26704–14. doi: 10.1074/jbc.M301031200. [DOI] [PubMed] [Google Scholar]
- Giguere V, Hollenberg SM, Rosenfeld MG. Functional domains of the human glucocorticoid receptor. Cell. 1986;46:545–52. doi: 10.1016/0092-8674(86)90339-9. [DOI] [PubMed] [Google Scholar]
- Gioeli D, Ficarro SB, Kwiek JJ, et al. Androgen receptor phosphorylation. J Biol Chem. 2002;32:29304–14. doi: 10.1074/jbc.M204131200. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rose DW, Rosenfeld MG. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–32. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- Godowski PJ, Rusconi S, Miesfeld R, et al. Glucocorticoid receptor mutants that are constitutive activators of transcriptional enhancement. Nature. 1987;325:365–8. doi: 10.1038/325365a0. [DOI] [PubMed] [Google Scholar]
- Hard T, Kellenbach E, Boelens R, et al. Solution structure of the DNA binding domain of the glucocorticoid receptor. Science. 1990;249:157–60. doi: 10.1126/science.2115209. [DOI] [PubMed] [Google Scholar]
- Henriksson A, Almlof T, Ford J, et al. Role of Ada adapter complex in gene activation by the glucocorticoid receptor. Mol Cell Biol. 1997;17:3065–73. doi: 10.1128/mcb.17.6.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittelman AB, Burakov D, Iniguez-Lluhi JA. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 1999;18:5380–8. doi: 10.1093/emboj/18.19.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg SM, Giguere V, Segui P, et al. Colocalization of DNA-binding and transcriptional activation functions in the human glucocorticoid receptor. Cell. 1987;49:39–46. doi: 10.1016/0092-8674(87)90753-7. [DOI] [PubMed] [Google Scholar]
- Horwitz KB, Jackson TA, Bain DL, et al. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–77. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- Housley PR, Sanchez ER, Westphal HM, et al. The molybdate-stabilized L-cell glucocorticoid receptor isolated by affinity chromatography or with a monoclonal antibody is associated with a 90–92-kDa nonsteroid-binding phosphoprotein. J Biol Chem. 1985;260:13810–17. [PubMed] [Google Scholar]
- Iakoucheva LM, Brown CJ, Lawson JD, et al. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–84. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- Ikonen T, Palvimo JJ, Janne OA. Interaction between the amino- and carboxy-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J Biol Chem. 1997;272:29821–8. doi: 10.1074/jbc.272.47.29821. [DOI] [PubMed] [Google Scholar]
- Ismaili N, Garabedian MJ. Modulation of glucocorticoid receptor function via phosphorylation. Ann NY Acad Sci. 2004;1024:86–101. doi: 10.1196/annals.1321.007. [DOI] [PubMed] [Google Scholar]
- Ismaili N, Blind R, Garabedian MJ. Stabilization of the unliganded glucocorticoid receptor by TSG101. J Biol Chem. 2005;280:11120–6. doi: 10.1074/jbc.M500059200. [DOI] [PubMed] [Google Scholar]
- Ito K, Yamamura S, Essilfie-Quaye S, et al. Histone deacetylase 2-mediated deacetylation of the glucocorticoid receptor enables NF-kappaB suppression. J Exp Med. 2006;203:7–13. doi: 10.1084/jem.20050466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KL, Cidlowski JA. Cell cycle regulation and apoptosis. Annu Rev Physiol. 1998;60:601–17. doi: 10.1146/annurev.physiol.60.1.601. [DOI] [PubMed] [Google Scholar]
- Kino T, Ichijo T, Amin ND, et al. Cyclin-dependent kinase 5 differentially regulates the transcriptional activity of the glucocorticoid receptor through phosphorylation: Clinical implications for the nervous system response to glucocorticoids and stress. Mol Endocrinol. 2007;21:1552–68. doi: 10.1210/me.2006-0345. [DOI] [PubMed] [Google Scholar]
- Kinyamu HK, Chen J, Archer TK. Linking the ubiquitin–proteasome pathway to chromatin remodeling/modification by nuclear receptors. J Mol Endocrinol. 2005;34:281–97. doi: 10.1677/jme.1.01680. [DOI] [PubMed] [Google Scholar]
- Kotaja N, Karvonen U, Janne OA, et al. The nuclear receptor interaction domain of GRIP1 is modulated by covalent attachment of SUMO-1. J Biol Chem. 2002;277:30283–8. doi: 10.1074/jbc.M204768200. [DOI] [PubMed] [Google Scholar]
- Krstic MD, Rogatsky I, Yamamoto KR, et al. Mitogen-activated and cyclin-dependent protein kinases selectively and differentially modulate transcriptional enhancement by the glucocorticoid receptor. Mol Cell Biol. 1997;17:3947–54. doi: 10.1128/mcb.17.7.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper G, Brinkmann AO. Steroid hormone receptor phosphorylation: is there a physiological role? Mol Cell Endocrinol. 1994;100:103–7. doi: 10.1016/0303-7207(94)90287-9. [DOI] [PubMed] [Google Scholar]
- Kumar R, Thompson EB. Structure and functions of the nuclear hormone receptors. Steroids. 1999a;64:310–19. doi: 10.1016/s0039-128x(99)00014-8. [DOI] [PubMed] [Google Scholar]
- Kumar R, Baskakov IV, Srinivasan G, et al. Interdomain signaling in a two-domain fragment of human glucocorticoid receptor. J Biol Chem. 1999b;274:24737–41. doi: 10.1074/jbc.274.35.24737. [DOI] [PubMed] [Google Scholar]
- Kumar R, Thompson EB. Transactivation functions of the N-terminal domains of nuclear hormone receptors: Protein folding and coactivator interactions. Mol Endocrinol. 2003;17:1–10. doi: 10.1210/me.2002-0258. [DOI] [PubMed] [Google Scholar]
- Kumar R, Johnson BH, Thompson EB. Overview of the structural basis for transcription regulation by nuclear hormone receptors. Essay Biochem. 2004a;40:27–39. doi: 10.1042/bse0400027. [DOI] [PubMed] [Google Scholar]
- Kumar R, Volk DE, Li J, et al. TBP binding induces structure in the recombinant glucocorticoid receptor AF1 domain. Proc Natl Acad Sci U S A. 2004b;101:16425–30. doi: 10.1073/pnas.0407160101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Drean Y, Mincheneau N, Le Goff P, et al. Potentiation of glucocorticoid receptor transcriptional activity by sumoylation. Endocrinology. 2002;143:3482–9. doi: 10.1210/en.2002-220135. [DOI] [PubMed] [Google Scholar]
- Lefstin JA, Yamamoto KR. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–8. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- Liu J, Perumal NB, Oldfield CJ, et al. Intrinsic disorder in transcription factors. Biochem. 2006;45:6873–88. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisi BF, Xu WX, Otwinowski Z, et al. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- Lopez GN, Turck CW, Schaufele F, et al. Growth factors signal to steroid receptors through mitogen-activated protein kinase regulation of p160 coactivator activity. J Biol Chem. 2001;276:22177–82. doi: 10.1074/jbc.M010718200. [DOI] [PubMed] [Google Scholar]
- Loven AM, Muster N, Yates JR, et al. A novel estrogen receptor alpha-associated protein, template-activating factor Ibeta, inhibits acetylation and transactivation. Mol Endocrinol. 2003;17:67–78. doi: 10.1210/me.2002-0280. [DOI] [PubMed] [Google Scholar]
- Mason SA, Housley PR. Site-directed mutagenesis of the phosphorylation sites in the mouse glucocorticoid receptor. J Biol Chem. 1993;268:21501–4. [PubMed] [Google Scholar]
- McKenna NJ, Lanz RB, O’Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocrine Rev. 1999;20:321–44. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- Mendelson CR. Mechanism of hormone action. In: Griffin JE, Ojeda SR, editors. Text Book of Endocrine Physiology. 5th ed. New York: Oxford University Press; 2004. pp. 49–88. [Google Scholar]
- Metivier R, Penot G, Flouriot G. Synergism between ERα transactivation function 1 (AF-1) and AF-2 mediated by steroid receptor coactivator protein-1: requirement for the AF-1 α-helical core and for a direct interaction between the N- and C-terminal domains. Mol Endocrinol. 2001;15:1953–70. doi: 10.1210/mend.15.11.0727. [DOI] [PubMed] [Google Scholar]
- Miesfeld R, Godowski PJ, Maler BA, et al. Glucocorticoid receptor mutants that define a small region sufficient for enhancer activation. Science. 1987;236:423–7. doi: 10.1126/science.3563519. [DOI] [PubMed] [Google Scholar]
- Milller AL, Webb MS, Copik AJ, et al. p38 MAP kinase is a key mediator in glucocorticoid-induced apoptosis of lymphoid cells: Correlation between p38 MAPK activation and site-specific phosphorylation of the human glucocorticoid receptor at serine 211. Mol Endocrinol. 2005;19:1569–83. doi: 10.1210/me.2004-0528. [DOI] [PubMed] [Google Scholar]
- Morishima Y, Kanelakis KC, Murphy PJM, et al. The Hsp90 cochaperone p23 is the limiting component of the multiprotein Hsp90/Hsp70-based chaperone system in vivo where it acts to stabilize the client protein Hsp90 complex. J Biol Chem. 2003;278:48754–63. doi: 10.1074/jbc.M309814200. [DOI] [PubMed] [Google Scholar]
- Namba K. Roles of partly unfolded conformations in macromolecular self-assembly. Genes Cells. 2001;6:1–12. doi: 10.1046/j.1365-2443.2001.00384.x. [DOI] [PubMed] [Google Scholar]
- Nichols M, Weih F, Schmid W, et al. Phosphorylation of CREB affects its binding to high and low affinity sites: implications for cAMP induced gene transcription. EMBO J. 1992;11:3337–46. doi: 10.1002/j.1460-2075.1992.tb05412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen RM, Yamamoto KR. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 2000;14:2314–29. doi: 10.1101/gad.827900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orti E, Bodwell JE, Munck A. Phosphorylation of steroid hormone receptors. Endocri Rev. 1992;13:105–28. doi: 10.1210/edrv-13-1-105. [DOI] [PubMed] [Google Scholar]
- Orti E, Mendel DB, Smith LI, et al. A dynamic model for glucocorticoid receptor phosphorylation and cycling in insect cells. J Steroid Biochem. 1989;34:85–96. doi: 10.1016/0022-4731(89)90069-1. [DOI] [PubMed] [Google Scholar]
- Pinhero R, Liaw P, Bertens K. Three cyclin-dependent kinases preferentially phosphorylate different parts of C-terminal domain of the large subunit of RNA polymerase II. Euro J Biochem. 2004;271:1004–14. doi: 10.1111/j.1432-1033.2004.04002.x. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endo Rev. 1997;18:306–60. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med. 2003;228:111–33. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Lin J, et al. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARγ coactivator-1. Mol Cell. 2001;8:971–82. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- Rogatsky I, Wang JC, Derynck MK, et al. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc Natl Acad Sci U S A. 2003;100:13845–50. doi: 10.1073/pnas.2336092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogatsky I, Waase CL, Garabedian MJ. Phosphorylation and inhibition of rat glucocorticoid receptor transcriptional activation by glycogen synthase kinase-3 (GSK-3). Species-specific differences between human and rat glucocorticoid receptor signaling as revealed through GSK-3 phosphorylation. J Biol Chem. 1998a;273:14315–21. doi: 10.1074/jbc.273.23.14315. [DOI] [PubMed] [Google Scholar]
- Rogatsky I, Logan SK, Garabedian MJ. Antagonism of glucocorticoid receptor transcriptional activation by the c-Jun N-terminal kinase. Proc Natl Acad Sci U S A. 1998b;95:2050–5. doi: 10.1073/pnas.95.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogatsky I, Trowbridge JM, Garabedian MJ. Glucocorticoid receptor-mediated cell cycle arrest is achieved through distinct cell-specific transcriptional regulatory mechanisms. Mol Cell Biol. 1997;17:3181–93. doi: 10.1128/mcb.17.6.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P, Obradovic Z, Dunker AK. Natively disordered proteins: functions and predictions. Appl Bioinformatics. 2004;3:105–13. doi: 10.2165/00822942-200403020-00005. [DOI] [PubMed] [Google Scholar]
- Rowan BG, Weigel NL, O’Malley BW. Phosphorylation of steroid receptor coactivator-1. Identification of phosphorylation sites and phosphorylation through the mitogen-activated protein kinase pathway. J Biol Chem. 2000a;275:4475–83. doi: 10.1074/jbc.275.6.4475. [DOI] [PubMed] [Google Scholar]
- Rowan BG, Garrison N, Weigel NL, et al. 8-Bromo-cyclic AMP induces phosphorylation of two sites in SRC-1 that facilitate ligand-independent activation of the chicken progesterone receptor and are critical for functional cooperation between SRC-1 and CREB binding protein. Mol Cell Biol. 2000b;20:8720–30. doi: 10.1128/mcb.20.23.8720-8730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez ER, Meshinchi S, Tienrungroz W, et al. Relationship of the 90 kDa murine heat shock protein with the untransformed and transformed states of the cell glucocorticoid receptor. J Biol Chem. 1987;262:6986–91. [PubMed] [Google Scholar]
- See RH, Calvo D, Shi Y, et al. Stimulation of p300-mediated transcription by the kinase MEKK1. J Biol Chem. 2001;276:16310–7. doi: 10.1074/jbc.M008113200. [DOI] [PubMed] [Google Scholar]
- Shikama N, Chan HM, Krstic-Demonacos M, et al. Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators. Mol Cell Biol. 2000;20:8933–43. doi: 10.1128/mcb.20.23.8933-8943.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein AM, Galigniana MD, Kanelakis KC, et al. Different regions of the immunophilin FKBP52 determine its association with the glucocorticoid receptor, hsp90, and cytoplasmic dynein. J Biol Chem. 1999;274:36980–6. doi: 10.1074/jbc.274.52.36980. [DOI] [PubMed] [Google Scholar]
- Simons SS., Jr . Function/activity of specific amino acids in glucocorticoid receptors. In: Litwack G, editor. Vitamins and Hormones Volume 49: Steroids. New York: Academic Press; 1994. pp. 49–130. [DOI] [PubMed] [Google Scholar]
- Somers JP, DeFranco DB. Effects of okadaic acid, a protein phosphatase inhibitor, on glucocorticoid receptor-mediated enhancement. Mol Endocrinol. 1992;6:26–34. doi: 10.1210/mend.6.1.1310797. [DOI] [PubMed] [Google Scholar]
- Starr DB, Matsui W, Thomas JR, et al. Intracellular receptors use a common mechanism to interpret signaling information at response elements. Genes Devel. 1996;10:1271–83. doi: 10.1101/gad.10.10.1271. [DOI] [PubMed] [Google Scholar]
- Szatmary Z, Garabedian MJ, Vilcek J. Inhibition of glucocorticoid receptor-mediated transcriptional activation by p38 mitogen-activated protein (MAP) kinase. J Biol Chem. 2004;279:43708–15. doi: 10.1074/jbc.M406568200. [DOI] [PubMed] [Google Scholar]
- Tetel MJ, Giangrande PH, Leonhardt SA, et al. Hormone-dependent interaction between the amino- and carboxyl- terminal domains of progesterone receptor in vitro and in vivo. Mol Endocrinol. 1999;13:910–24. doi: 10.1210/mend.13.6.0300. [DOI] [PubMed] [Google Scholar]
- Thompson EB, Kumar R. DNA binding of nuclear hormone receptors influences their structure and function. Biochem Biophys Res Commun. 2003;306:1–4. doi: 10.1016/s0006-291x(03)00877-5. [DOI] [PubMed] [Google Scholar]
- Tian S, Poukka H, Palvimo JJ, et al. Small ubiquitin-related modifier-1 (SUMO-1) modification of the glucocorticoid receptor. Biochem J. 2002;367:907–11. doi: 10.1042/BJ20021085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–33. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- Tzukerman MT, Esty A, Santiso-Mere D. Human estrogen receptor transactivational capacity is determined by both cellular and promoter context and mediated by two functionally distinct intramolecular regions. Mol Endocrinol. 1994;8:21–30. doi: 10.1210/mend.8.1.8152428. [DOI] [PubMed] [Google Scholar]
- Wallace AD, Cidlowski JA. Proteasome-mediated glucocorticoid receptor degradation restricts transcriptional signaling by glucocorticoids. J Biol Chem. 2001;276:42714–21. doi: 10.1074/jbc.M106033200. [DOI] [PubMed] [Google Scholar]
- Wang LG, Liu MX, Kreis W, et al. Phosphorylation/dephosphorylation of androgen as a determinant of androgen agonistic or antagonistic activity. Biochem Biophys Res Commun. 1999;259:21–8. doi: 10.1006/bbrc.1999.0655. [DOI] [PubMed] [Google Scholar]
- Wang Z, Chen W, Kono E, et al. Modulation of glucocorticoid receptor phosphorylation and transcriptional activity by a C-terminal-associated protein phosphatase. Mol Endocrinol. 2007;21:625–34. doi: 10.1210/me.2005-0338. [DOI] [PubMed] [Google Scholar]
- Wang Z, Frederick J, Garabedian MJ. Deciphering the phosphorylation “code” of the glucocorticoid receptor in vivo. J Biol Chem. 2002;277:26573–80. doi: 10.1074/jbc.M110530200. [DOI] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, et al. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–45. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Webster JC, Jewell CM, Bodwell JE, et al. Mouse glucocorticoid receptor phosphorylation status influences multiple functions of the receptor protein. J Biol Chem. 1997;272:9287–93. doi: 10.1074/jbc.272.14.9287. [DOI] [PubMed] [Google Scholar]
- Weigel NL, Moore NL. Kinases and protein phosphorylation as regulators of steroid hormone action. Nucl Recept Signal. 2007a;5:e005. doi: 10.1621/nrs.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel NL, Moore NL. Cyclins, cyclin dependent kinases, and regulation of steroid receptor action. Mol Cell Endocrinol. 2007b;265–66:157–61. doi: 10.1016/j.mce.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Szewczuk Z, Petry I, et al. Constitutive phosphorylation of the acidic tails of the high mobility group 1 proteins by casein kinase II alters their conformation, stability, and DNA binding specificity. J Biol Chem. 1999;274:20116–22. [PubMed] [Google Scholar]
- Wu RC, Smith CL, O’Malley BW. Transcriptional regulation by steroid receptor coactivator phosphorylation. Endocr Rev. 2005;26:393–9. doi: 10.1210/er.2004-0018. [DOI] [PubMed] [Google Scholar]
- Wu RC, Qin J, Yi P, et al. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic responses to multiple cellular signaling pathways. Mol Cell. 2004;15:937–49. doi: 10.1016/j.molcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Yamamoto KR. Steroid receptor regulated transcription of specific genes and gene network. Annu Rev Genet. 1985;19:209–52. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]
- Yamamoto KR, Darimont BD, Wagner RL, et al. Building transcriptional regulatory complexes: signals and surfaces. Cold Spring Harb Symp Quant Biol. 1998;63:5875–98. doi: 10.1101/sqb.1998.63.587. [DOI] [PubMed] [Google Scholar]
- Yang W, Hong YH, Shen XQ, et al. Regulation of transcription by AMP-activated protein kinase: phosphorylation of p300 blocks its interaction with nuclear receptors. J Biol Chem. 2001;276:38341–4. doi: 10.1074/jbc.C100316200. [DOI] [PubMed] [Google Scholar]
- Zhou J, Cidlowski JA. The human glucocorticoid receptor: one gene, multiple proteins and diverse responses. Steroids. 2005;70:407–17. doi: 10.1016/j.steroids.2005.02.006. [DOI] [PubMed] [Google Scholar]