Abstract
Introduction
IFN-β is widely used as first-line immunomodulatory treatment for multiple sclerosis. Response to treatment is variable (30–50% of patients are nonresponders) and requires a long treatment duration for accurate assessment to be possible. Information about genetic variations that predict responsiveness would allow appropriate treatment selection early after diagnosis, improve patient care, with time saving consequences and more efficient use of resources.
Materials & methods
We analyzed 61 SNPs in 34 candidate genes as possible determinants of IFN-β response in Irish multiple sclerosis patients. Particular emphasis was placed on the exploration of combinations of allelic variants associated with response to therapy by means of a Markov chain Monte Carlo-based approach (APSampler).
Results
The most significant allelic combinations, which differed in frequency between responders and nonresponders, included JAK2–IL10RB–GBP1–PIAS1 (permutation p-value was pperm = 0.0008), followed by JAK2–IL10–CASP3 (pperm = 0.001).
Discussion
The genetic mechanism of response to IFN-β is complex and as yet poorly understood. Data mining algorithms may help in uncovering hidden allele combinations involved in drug response versus nonresponse.
Keywords: APSampler, Bayesian statistics, IFN-β, multiple sclerosis, pharmacogenomics, polymorphism
Multiple sclerosis (MS) is a demyelinating disease of the CNS mediated by autoreactive T cells. Long-term treatment of the condition by IFN-β aims to reduce relapses and slow disease progression [1]. Even while a series of novel MS drugs are being introduced or are currently in trials [2], IFN-β remains, for now, the first-line disease-modifying therapy of choice, and has been widely used in this quality over the last decade. Although its precise mechanism of action is not fully understood, it exerts beneficial effects resulting from a combination of immunomodulatory, anti-inf lammatory and antiproliferative activities. Notwithstanding the conceived efficacy of IFN-β as demonstrated by clinical trials, some 30–50% of patients do not respond to the treatment [1,3,4]. Determination of response status requires clinical evaluation over a relatively long treatment period. Unfortunately this means that accumulation of disability may have occurred before a patient is realized to be a nonresponder (NR). It is particularly important that NRs are identified and receive an alternative treatment as soon as possible. Early indicators of responsiveness would enable rational provision of this medicine and improve patient care. As a consequence, this would additionally lead to more efficient use of resources. Genetic variants that alter drug response are biomarkers that can be quickly and easily determined before treatment is commenced.
The pharmacogenomic analysis of IFN-β treatment in MS is still somewhat in its infancy. Compared with the wealth of publications examining genetic susceptibility to MS, only a relatively small number have examined response to IFN-β. In the first study to examine IFN-β pharmacogenomics in MS patients, it was hypothesized that the confirmed susceptibility locus in the MHC class II region may also alter treatment responsiveness [5]. However, neither this study nor others have identified any association with the HLA class II alleles [5,6]. Genes encoding the subunits of the IFN type I receptor, IFNAR1 and IFNAR2, have also been investigated as candidate response genes. Sriram et al. analyzed a total of eight SNPs and two micro-satellites in 57 responders (Rs) and 48 NRs. A SNP in IFNAR1 (rs1012334) showed a trend towards relapse-free status [7]. In another study, three SNPs in the IFN type I receptor genes were examined in 104 Rs and 43 NRs but were not associated with response [8]. In our own laboratory we screened the interferon (IFN)-stimulated response elements (ISREs) of 100 putative response-modifying genes for polymorphisms by sequencing pooled DNA. A total of 54 polymorphisms in 32 genes were detected in these regions with 15 genes showing allelic peak distortions between the R and NR pools. To confirm these putative associations the polymorphisms were genotyped in individual patient samples (94 Rs, 68 NRs). Significant (uncorrected) associations were found for four candidate genes; MX1, IFNAR1, LMP7 and CTSS [9]. Weinstock-Guttman et al. examined SNPs in MX1 in 179 relapsing MS patients receiving IFN-β treatment, but none of the markers were found to affect response in terms of clinical or MRI measures [10]. The first genome-wide screen examining response to IFN-β in 206 MS patients has recently been published [11]. Whereas previous studies have used the candidate gene approach, this is the first thorough and objective analysis of IFN-β pharmacogenomics and the first study to clearly substantiate a polygenic underlying mechanism [12]. Moreover, this study highlighted a previously overlooked group of genes related to ion channels and signal transduction pathways as significantly associated with response phenotype [11]. Evidence for putative biomarkers of IFN-β response in MS may be obtained from various methodologies including pharmacogenomics, transcriptomics and proteomics. Moreover, given the shared pathways for receptor binding, signaling and biological effector function of IFN-α and IFN-β (type I IFNs), polymorphisms that modify response to IFN-α treatment in hepatitis may provide clues as to markers of IFN-β response in MS (for review of type I IFN pharmacogenomics, see [13]).
In this study we sought to determine genetic markers of response in a group of Irish MS patients receiving IFN-β treatment. We screened 61 polymorphisms in 34 candidate response genes (Table 1), based on notions of their known function and the literature, with particular emphasis on a seminal transcriptomics paper by Baranzini et al. [14]. We selected SNPs in the promoter/5´-UTR regions, hypothesizing that polymorphisms may modify IFN inducibility, and hence, the expression profile differences observed in Baranzini et al. [14]. In addition, nonsynonymous SNPs and those potentially affecting splicing were also selected. Some HapTag SNPs were selected for genes that were of particular interest but that had no known functional polymorphisms, in order to capture a large proportion of the genetic variability (Table 1). Polymorphisms were typed in DNA samples from 255 Irish patients classified as R or NR. As it is proposed that IFN responsiveness in MS patients may constitute a polygenic trait [9,11–13], the main focus of this study was to explore allelic combinations associated with response to treatment.
Table 1.
SNPs selected for analysis
| Chr. | Gene symbol | SNP ID | Allele | Position relative from ATG | SNP location | Ref. for gene selection |
|---|---|---|---|---|---|---|
| 1 | VCAM1 | rs1409419 | C/T | −2021 | Promoter | [25,26] |
| 1 | JAK1 | rs310249 | C/G | −8189 | Promoter | |
| 1 | JAK1 | rs310199 | C/T | −994 | Intron 1 | |
| 1 | JAK1 | rs310200 | C/G | −689 | Intron 1 | |
| 1 | JAK1 | rs310202 | C/T | −416 | Intron 1 | |
| 1 | IL10 | rs1800896 | A/G | −1117 | Promoter | [27–31] |
| 1 | IL10 | rs1800871 | C/T | −854 | Promoter | |
| 1 | IL10 | rs1800872 | A/C | −627 | Promoter | |
| 1 | IL12RB2 | rs11209046 | C/T | −12403 | Exon 1 | [14] |
| 1 | GBP1 | rs1536670 | C/T | −2475 | Promoter | [17,18] |
| 1 | GBP1 | rs12089335 | C/T | −1508 | Intron 1 | |
| 1 | GBP1 | rs6682273 | C/T | −1060 | Intron 1 | |
| 1 | GBP1 | rs10493822 | A/G | −763 | Intron 1 | |
| 2 | ITGA4 | rs155141 | A/T | −5180 | Promoter | [25,32] |
| 2 | ITGA4 | rs4667319 | A/G | 72963 | Exon 24 | |
| 2 | STAT1 | rs1914408 | A/G | 34753 | Intron 23/3´-UTR | [17–19] |
| 2 | IL1B | rs1143623 | C/G | −2023 | Promoter | [33] |
| 2 | CASP10 | rs11674246 | C/T | 93 | Intron 1 boundary | [14,17] |
| 2 | CASP10 | rs12613347 | C/T | 4811 | Intron 3 | |
| 2 | CASP10 | rs3731714 | A/G | 10319 | Intron 5 | |
| 2 | CASP10 | rs13006529 | A/T | 31958 | Exon 8/10, Intron 9 | |
| 2 | CFLAR | rs6728771 | A/C | −2182 | Intron 1 | [14] |
| 2 | CFLAR | rs2041765 | C/T | 28571 | Intron 8 | [17] |
| 2 | PRKR | rs12712526 | A/G | −8366 | Promoter | [10,14,17,19,34,35] |
| 2 | PRKR | rs2254958 | C/T | −1342 | Exon 1 | |
| 2 | PRKR | rs2270414 | C/T | 69 | Intron 1 boundary | |
| 2 | PNPT1 | rs782584 | C/G | −158 | Promoter | [36] |
| 3 | DRD3 | rs6280 | C/T | 24 | Exon 2 | [37–39] |
| 4 | CASP3 | rs2019978 | G/T | 2026 | Intron 3 | [14] |
| 5 | DUSP1 | rs881150 | A/T | −950 | Promoter | [19,40] |
| 6 | IRF4 | rs1514346 | A/G | −3523 | Promoter | [14] |
| 6 | IRF4 | rs1473037 | A/T | 5926 | Intron 6 | |
| 6 | IRF4 | rs1877175 | A/G | 17340 | 3´-UTR | |
| 7 | LEPTIN | rs7799039 | A/G | −13289 | Promoter | [41] |
| 7 | CASP2 | rs3181166 | G/T | 1349 | Intron 1 | [14] |
| 8 | IDO1 | rs3824259 | G/T | −1849 | Promoter | [42] |
| 9 | DBH | rs1611115 | C/T | −1021 | Promoter | [37–39,43] |
| 9 | JAK2 | rs1887427 | A/G | −42258 | Promoter | [14] |
| 9 | JAK2 | rs3808850 | A/T | −38677 | Promoter | |
| 9 | JAK2 | rs1887429 | G/T | −37439 | Promoter | |
| 9 | CD274 | rs822342 | C/T | −2141 | Intron 1 | [44] |
| 9 | CD274 | rs2282055 | A/C | −382 | Intron 1 | |
| 10 | CASP7 | rs12415607 | A/C | −19049 | Promoter | [14] |
| 10 | CASP7 | rs11196418 | A/G | −18787 | Promoter | |
| 10 | CASP7 | rs2227310 | C/G | 31899 | Exon 8 | |
| 11 | TH | rs10770140 | C/T | −581 | Promoter | [37–39] |
| 11 | CASP5 | rs507879 | A/G | 1739 | Exon 2 | [14] |
| 14 | IRF9 | rs2236350 | A/C | −344 | Intron 1 | [17] |
| 15 | PIAS1 | rs8029670 | C/T | 36750 | Intron 2 | |
| 15 | PIAS1 | rs10162905 | A/G | 49382 | Intron 2 | |
| 17 | SOCS3 | rs4969170 | A/G | −5362 | Promoter | [17] |
| 17 | PRKCA | rs3764402 | C/T | −1517 | Promoter | [45–47] |
| 17 | STAT3 | rs4796793 | C/G | −41676 | Promoter | [14,17] |
| 19 | TYK2 | rs280501 | C/T | −2240 | Promoter | [14] |
| 19 | TYK2 | rs280500 | A/G | −1320 | Exon 2 | |
| 19 | TYK2 | rs2304256 | G/T | 13430 | Exon 8 | |
| 19 | TYK2 | rs12720356 | A/C | 19107 | Exon 15 | |
| 21 | ITGB2 | rs760452 | A/G | 96 | Intron 1 boundary | [14,32] |
| 21 | ITGB2 | rs3788150 | G/T | −3105 | Intron 1 | |
| 21 | IFNAR2 | rs8127890‡ | A/G | 20650 | Exon 9 | [17] |
| 21 | IL10RB | rs2834167§ | A/G | 2017 | Exon 2 |
Data according to [103].
Can also be referred to as IFNAR2, 26560 (downstream SNP).
Can also be referred to as IL10RB, −3893 (promoter SNP).
Materials & methods
Patients
Samples were obtained from 255 relapsing remitting MS patients (1 male:2.5 females) from Belfast and Dublin, diagnosed with clinically definite MS according to the Poser Criteria [15]. All patients were receiving IFN-β treatment and had data available for 2 years before initiation of treatment and at least 2 years after. Response status was defined as described previously (see [9]). Briefly, response to treatment was defined as a reduction in relapse rate by one-third after the initial 6–9 months of treatment and no sustained progression (verified at 3 months) on the Expanded Disability Status Scale (EDSS) (≥1 point if baseline EDSS <5.5 or <0.5 points if baseline EDSS ≥5.5). A NR to treatment is a patient whose relapse rate remains the same or increases after the initial 6- to 9-month period of treatment. A total of 155 patients were classified as Rs (1 male:2.9 females) and 100 as NRs (1 male:1.9 females). All patients participated in the study on the basis of informed consent, and were informed about the scope and extent of the study through a detailed information sheet. Ethics approval of the study was obtained via the Queen’s University Belfast (Belfast, Northern Ireland) institutional review board.
Genotyping
SNPs were selected as described in the introduction. The primary genotyping method for this study was Matrix Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) mass spectrometry (Sequenom Inc., CA, USA). Additionally, genotypes of rs1473037, rs1877175, rs2041765 and rs4796793 were determined by Taqman™ Assay-on-Demand kits (CatNo:C_1576168_1, C_12576157_10, C_11460441_1_, C_27977213_10) (Applied Biosystems, CA, USA). The PCR conditions and subsequent detection were according to the manufacturer’s instructions. The Taqman assays were carried out in a DNA Engine Opticon® 2 system obtained from MJ Research Inc, (CA, USA). A further SNP, rs3181166, was genotyped using the template-directed dye-terminator incorporation with fluorescence polarization (FP-TDI) method.
Statistical analysis
Genotype and allele frequencies, as well as carriage rates were compared between Rs and NRs using a standard χ2 test (EpiInfo™ Statcalc, version 6 [Centers for Disease Control and Prevention, GA, USA]). Haplotype analysis was performed using Haploview software [101]. The uncorrected p-value calculated for single marker and haplotype analysis is referred to hereafter as puc. Allelic combinations, which influence treatment response, were identified using the APSampler algorithm [16] (see also [102]). The validation framework for APSampler was improved for this work. Each allelic combination was validated by a permutation test as well as by a standard Fisher’s test (uncorrected). The permutation test gathered a null-distribution of the Fisher’s test p-values for carriage of allelic combinations by a series of runs of the APSampler algorithm with randomly permuted R/NR labels. As a result, the test estimates the probability for a random allelic combination (i.e., a combination that is obtained for a random dataset) to have an exact Fisher’s p-value (pf) better than or equal to that of the combination being validated. The probability is referred to as the permutation p-value (pperm). In more simple words, the more significant (lower) is pperm, the lower the probability is to obtain the same or better pf by chance. Here, we present the patterns with both pf and pperm less than 0.0025.
Results
All individual markers were examined in terms of genotype distribution, allele frequency and carriage, in relation to response versus nonresponse (data not shown). No significant associations were found.
Haplotype analysis of individual genes did not uncover any significant results. A suggestive increase in the JAK2 rs1887427*A–rs3808850*T–rs1887429*G haplotype in Rs (40.1%) compared with NRs (32.7%) was noted (puc = 0.096). Similarly two-marker analysis suggested a greater rs1887427*A–rs3808850*T frequency in Rs (46.7%) compared with NRs (39.0%) (puc = 0.094) (data not shown).
Analysis of allelic combinations influencing treatment response using the APSampler algorithm highlighted many combinations (two to five markers), which differed significantly in carriage between the two groups (pperm < 0.025). As the number of alleles in a combination increases, the number of the combination carriers decreases. Only one pattern contained five informative alleles. In the generated dataset, combinations of six or more informative alleles were not identifiable.
The ten most significant associations are listed in Table 2. The most significant association was observed for a JAK2–IL10RB–GBP1–PIAS1 combination, followed by a JAK2–IL10–CASP3 combination. The latter and the TYK2–DUSP1–PRKCA combination were more frequent in Rs (combinations favorable for beneficial IFN-β treatment response), whereas the other eight were found mostly in NRs (unfavorable combinations). Allele G of JAK2 rs1887429 was part of the favorable JAK2–IL10–CASP3 combination, whereas the alternative allele T of this SNP was present, accordingly, in three unfavorable combinations.
Table 2.
Ten most significant allelic combinations differing in carriage between responders and nonresponders, their carriage rates among responders and nonresponders and the exact Fisher’s and permutation test p-values.
| SNP1 | SNP2 | SNP3 | SNP4 | Carriage in | Fisher’s p-value |
Odds ratio (95% CI) |
Permutation p-value |
|
|---|---|---|---|---|---|---|---|---|
| R (%) | NR (%) | |||||||
| JAK2 rs1887429*T | IL10RB rs2834167*G | GBP1 rs12089335*T | PIAS1 rs10162905*A | 4.8 | 20.7 | 0.00019 | 0.2 (0.1–0.5) | 0.0008 |
| JAK2 rs1887429*T | IL10RB rs2834167*G | GBP1 rs10493822*G | 6.3 | 21.1 | 0.0008 | 0.2 (0.1–0.6) | 0.0018 | |
| JAK2 rs1887429*T | IL10RB rs2834167*G | GBP1 rs12089335*T | 6.2 | 20.7 | 0.0009 | 0.3 (0.1–0.6) | 0.0019 | |
| JAK2 rs1887429*G | IL10 rs1800872*A | CASP3 rs2019978*G | 13.9 | 1.1 | 0.0004 | 14.3 (1.9–109.1) | 0.0012 | |
| JAK1 rs310249*G | PNPT1 rs782584*C | TYK2 rs2304256*C | 5.3 | 20.7 | 0.0007 | 0.2 (0.1–0.5) | 0.0017 | |
| JAK1 rs310249*G | PNPT1 rs782584*C | LEP rs7799039*G | 5.3 | 20.7 | 0.0007 | 0.2 (0.1–0.5) | 0.0017 | |
| JAK1 rs310249*G | PNPT1 rs782584*C | CASP7 rs12415607*C | 6.2 | 22.0 | 0.0008 | 0.2 (0.1–0.6) | 0.0018 | |
| JAK1 rs310249*G | PNPT1 rs782584*C | PRKR rs2270414*T | 3.8 | 17.1 | 0.0013 | 0.2 (0.1-.06) | 0.0023 | |
| JAK1 rs310249*G | IFNAR2 rs8127890*A | LEP rs7799039*A | 1.5 | 12.5 | 0.0009 | 0.1 (0.02–00.5) | 0.0019 | |
| TYK2 rs280501*C | DUSP1 rs881150*T | PRKCA rs3764402*A | 90.7 | 73.1 | 0.0012 | 3.6 (1.6–8.0) | 0.0023 | |
NR: Nonresponder; R: Responder
Discussion
In general, APSampler prefers to identify the minimal allelic combinations [16] associated with the trait of interest (e.g., the treatment response), rather than the redundant ones. However, the stochastic nature of the algorithm allows it to find nonoptimal combinations as well as the optimal one. The output of the sampler is, therefore, a ranked list of allelic combinations. Thus, additional validation of the initial APSampler findings was suggested for this study. The substance of the validation was to test how probable each allelic combination would be, given the dataset characteristics (number of patients, number of alleles and so on) independent of the data itself. To evaluate the probability, we introduced and implemented the permutation test. Those patterns that had a low probability in the permutation test (pperm < 0.025) were considered as associated with response.
No significant findings emerged from single marker analysis. Although haplotype analysis did not draw out any significant findings for response status, it is interesting that a trend in JAK2 rs1887427–rs3808850 haplotypes was observed. Analysis of multi-allelic combinations in Rs and NRs identified a JAK2–IL10RB–GBP1–PIAS1 combination to be most significant (pf = 0.00019, pperm = 0.0008). Combinations of JAK2, IL10RB and GBP1 alleles were prominent throughout these ten most significant results. GBP1 rs10493822*G and GBP1 rs12089335*T are in strong linkage disequilibrium (D´ = 0.993, r2 = 0.979). JAK2 rs1887429*T–IL10RB rs2834167*G–GBP1 rs10493822*G and JAK2 rs1887429*T–IL10RB rs2834167*G–GBP1 rs12089335*T are therefore equivalent combinations. Examination of the potential underlying biallelic patterns in this gene triplet identified JAK2–IL10RB (pf = 0.08, pperm = 0.14) as the core element but the triplet including a GBP1 allele (pf = 0.0008, pperm = 0.0018) or quartet with an additional PIAS1 allele (pf = 0.00019, pperm = 0.0008), proved to be more influential. The GBP1 rs10493822 polymorphism is located in the 5´-UTR region, nucleotide position −763 (relative to ATG). IFN-α has been shown to upregulate GBP1 in peripheral blood mononuclear cells of hepatitis C patients [17]. In MS patients two distinct expression patterns of this gene have been detected [18], and evidence suggests it may be upregulated to a greater extent in patients responding to IFN-β treatment than in NRs [19].
The second most significant gene triplet was a JAK2–IL10–CASP3 (pf = 0.0004, pperm = 0.0012) combination. CASP3 appeared in the best scoring gene triplets predicting IFN-β response in MS reported by Baranzini et al., and in the same study JAK2 expression appeared to modestly differ between Rs and NRs [14]. It is worth noting that IL-10 appears to regulate CASP3 activation, though somewhat contradictory results have been reported. IL-10-mediated CASP3 activation has been implicated in apoptosis of mast cells and macrophages [20]. However, IL-10 inhibits glutamate-mediated apoptosis of cerebellar granule cells by blocking caspase-3 induction [21] and lipopolysaccharide-treated IL-10 knockout mice have increased hepatocellular production of caspase-3 [22].
The third group of significant tri-allelic combinations were underpinned by a JAK1-PNPT1 combination. However the ability of this pair to differentiate between Rs and NRs (pf = 0.0051, pperm = 0.03) was not as strong as in the associated triplets. JAK1 forms part of the type I IFN receptor complex. PNPT1 is a type I IFN-inducible early response gene that seems to impart the pro-apoptotic and growth inhibitory effects of IFN-β in melanoma cells [23]. Further triplets were detected in our top scoring combinations, including JAK1–IFNAR2–LEP (pf = 0.0009, pperm = 0.0019), and TYK2–DUSP1–PRKCA (pf = 0.0012, pperm = 0.0023). The first of these is particularly interesting as JAK1 and IFNAR2 are in direct contact as part of the IFN type I receptor complex [24]. Overall examination of the top ten scoring combinations indicated that JAK2, JAK1, IL10RB, GBP1 and PNPT1 were the most frequently encountered genes.
Conclusion
Response to IFN-β is thought to constitute a polygenic trait [9,11,12]. The present study has uncovered a number of allele combinations significantly associated with (non)response to IFN-β. Care should be exerted when interpreting this data as the relative importance of these combinations will have to be considered in the context of whole-genome polygenic elucidation of IFN-β response. Nevertheless, this study shows the benefit of using an advanced algorithmic tool in determining pharmacogenomics of polygenic traits, as illustrated by its ability to extract more information than single-marker analysis alone. The functional and biological relationship between combinations of genes uncovered in this study as putative MS IFN-β pharmacogenomic biomarkers is represented in Figure 1. While we have tried to screen a relatively large number of potentially response-altering polymorphisms in our MS treatment group, there is an obvious likelihood of overlooking an important marker. In addition to the limited number of candidate genes investigated to this point, and only one genome-wide screen published to date [11], a further hurdle for investigators in this field is the difficulty in obtaining large, well-powered cohorts [12]. The present study, though in strict sense is still underpowered, includes one of the largest numbers of Rs and NRs to IFN-β analyzed to date. Replication in independent datasets will be required.
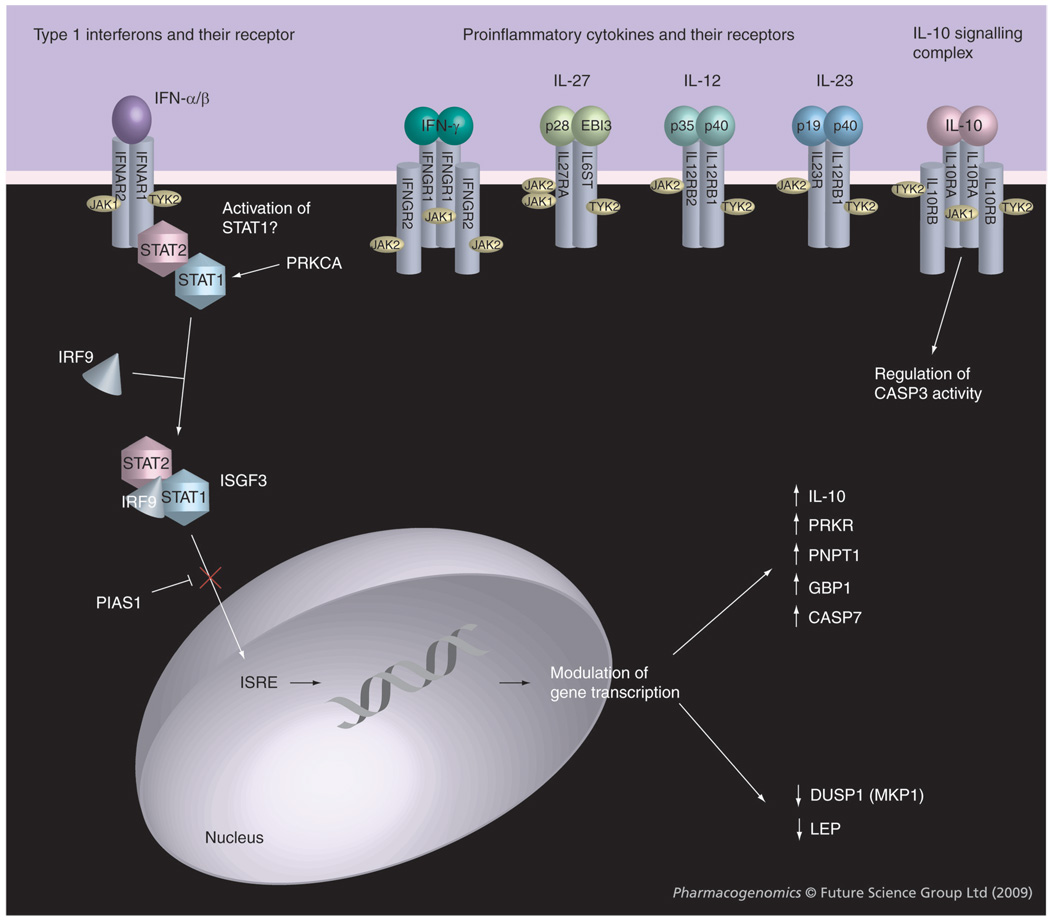
Figure 1. Potential biological relationships between the most significant gene combinations differing in carriage between responders and nonresponders to IFN-β in the present study.
Signaling via the JAK-STAT pathway by IFN-β (IFNAR2, JAK1, TYK2, PRKCA and PIAS1) [46,48,49] causes upregulation (GBP1, PRKR, PNPT1, IL10) [17,19,36,50,51] or downregulation (DUSP1 and LEP) [19,40,41] of various genes. Also indicated are proinflammatory molecules and their receptors involved in the pathogenesis of multiple sclerosis, which may be indirectly affected by IFN-β treatment and that use JAK1, JAK2 and TYK2. [48,52]. Other genes highlighted in the analysis include those encoding IL10RB (part of the IL10 receptor complex [53]) and proteins involved in apoptosis such as CASP3 [14,21,22] and CASP7 [50]. ISRE: Interferon stimulated response element.
Future perspective
The next 5 years is likely to see a shift away from candidate gene studies toward whole-genome screens, both for first- and second-line MS treatments. The polygenic nature of medication treatment efficacy in MS, as well as of the diagnosis and clinical course of this disease, will increasingly be addressed using genomic, transcriptomic and proteomic methods. While IFN-β is thought to act predominantly through direct induction or repression of IFN-stimulated genes, a major challenge will be to separate redundant from response-involved genes. Polymorphisms in IFN-β regulated genes may affect transcription, mRNA stability and splicing, as well as the final protein sequence and its associated biological behavior. Integration of systems biology approaches will be crucial to model the regulatory networks underlying treatment outcome, and to identify the primary molecular mediators of drug action. Application of bioinformatic methods that provide complex simultaneous analysis for the contribution of numerous genes to the medication response, such as APSampler [16], will allow both validation and identification of new results. In conclusion, the simultaneous implementation of genetic, transcriptomic and proteomic methods and corresponding network analysis [2] may guarantee the highest chance for successful identification of biomarker combinations predictive for response.
Executive summary
A significant proportion of multiple sclerosis (MS) patients do not respond to IFN-β drug treatment with clear-cut clinical amelioration.
The pharmacogenomics of IFN-β therapy in MS is as yet poorly understood, but is thought to constitute a polygenic trait.
Patients & methods
A total of 61 SNPs in 34 candidate genes were analyzed as possible determinants of IFN-β response in 255 Irish MS patients.
Allelic combinations associated with treatment response were identified using the APSampler algorithm.
Results
No significant associations were found for individual SNPs.
Analysis of allelic combinations influencing treatment response using the APSampler algorithm highlighted many combinations (two to five markers), which differed significantly in carriage between responders and nonresponders (pperm < 0.05).
The most significant association was a JAK2–IL10RB–GBP1–PIAS1 combination, followed by a JAK2–IL10–CASP3 combination.
Discussion
Overall examination of the top ten scoring combinations indicated that JAK2, JAK1, IL10RB, GBP1 and PNPT1 were the most frequently encountered genes.
Conclusion
This study shows the benefit of using an advanced Markov chain Monte Carlo-based approach for identifying gene combinations potentially involved in phenotypic appearance of polygenic pharmacogenomic traits. This is best illustrated by its ability to extract much more information from the dataset than single-marker or, wherever applicable, haplotype analysis alone.
Footnotes
Financial & competing interests disclosure
Genotyping was performed with grants from NI HPSS R&D Office (RSG/1726) and MS Ireland to KV. We thank NLM/NIH for support to Michael Ochs, Olga Favorova, and Alexander Favorov (LM008932) and RFBR (Russian Foundation for Basic Research) for support to OF and AF (08-04-01834-a). Stanley Hawkins declares that he received an unrestricted educational grant of 40,000 GBP from Serono in 2004–2005 via the Irish Institute of Clinical Neuroscience, and that he has performed consultancy work for Bayer/Schering. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Contributor Information
Catherine O’Doherty, University of South Australia, Adelaide, Australia.
Alexander Favorov, Laboratory for Bioinformatics, GosNIIGenetika, Moscow, Russia and Johns Hopkins School of Medicine, MD, USA.
Shirley Heggarty, Queens University, Belfast, UK.
Colin Graham, Queens University, Belfast, UK.
Olga Favorova, Russian State Medical University, Moscow, Russia.
Michael Ochs, Johns Hopkins School of Medicine, MD, USA.
Stanley Hawkins, Royal Victoria Hospital, Belfast, UK.
Michael Hutchinson, St Vincent’s Hospital, Dublin, Ireland.
Killian O’Rourke, St Vincent’s Hospital, Dublin, Ireland.
Koen Vandenbroeck, Neurogenomiks Laboratory, Ikerbasque and Universidad Del País Vasco (UPV-EHU), Parque Tecnológico de Bizkaia, 48170 Zamudio, Spain.
Bibliography
Papers of special note have been highlighted as:
•of interest
•• of considerable interest
- 1.Prevention of Relapses and Disability by Interferon β-1a Subcutaneously in Multiple Sclerosis (PRISMS) Study Group. Randomised double-blind placebo-controlled study of interferon β-1α in relapsing/remitting multiple sclerosis. Lancet. 1998;352(9139):1498–1504. [PubMed] [Google Scholar]
- 2. Martinez-Forero I, Pelaez A, Villoslada P. Pharmacogenomics of multiple sclerosis: in search for a personalized therapy. Expert Opin. Pharmacother. 2008;9(17):3053–3067. doi: 10.1517/14656560802515553. • Discussion of current state and challenges ahead in pharmacogenomics research in multiple sclerosis (MS)
- 3.Rudick RA, Goodkin DE, Jacobs LD, et al. Impact of interferon β-1α on neurologic disability in relapsing multiple sclerosis. Neurology. 1997;49(2):358–363. doi: 10.1212/wnl.49.2.358. [DOI] [PubMed] [Google Scholar]
- 4.The IFNB Multiple Sclerosis Study Group. Interferon β-1b is effective in relapsing-remitting multiple sclerosis. I. Clinical results of a multicenter, randomized, double-blind, placebo-controlled trial. Neurology. 1993;43(4):655–661. doi: 10.1212/wnl.43.4.655. [DOI] [PubMed] [Google Scholar]
- 5.Villoslada P, Barcellos LF, Rio J, et al. The HLA locus and multiple sclerosis in Spain. Role in disease susceptibility, clinical course and response to interferon-β. J. Neuroimmunol. 2002;130(1–2):194–201. doi: 10.1016/s0165-5728(02)00215-1. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez O, Fernandez V, Mayorga C, et al. HLA class II and response to interferon-β in multiple sclerosis. Acta Neurol. Scand. 2005;112(6):391–394. doi: 10.1111/j.1600-0404.2005.00415.x. [DOI] [PubMed] [Google Scholar]
- 7.Sriram U, Barcellos LF, Villoslada P, et al. Pharmacogenomic analysis of interferon receptor polymorphisms in multiple sclerosis. Genes Immun. 2003;4(2):147–152. doi: 10.1038/sj.gene.6363946. [DOI] [PubMed] [Google Scholar]
- 8.Leyva L, Fernandez O, Fedetz M, et al. IFNAR1 and IFNAR2 polymorphisms confer susceptibility to multiple sclerosis but not to interferon-β treatment response. J. Neuroimmunol. 2005;163(1–2):165–171. doi: 10.1016/j.jneuroim.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 9. Cunningham S, Graham C, Hutchinson M, et al. Pharmacogenomics of responsiveness to interferon IFN-β treatment in multiple sclerosis: a genetic screen of 100 type I interferon-inducible genes. Clin. Pharmacol. Ther. 2005;78(6):635–646. doi: 10.1016/j.clpt.2005.08.018. •• First large-scale study of the pharmacogenomics of IFN-β in MS based on identification of polymorphisms in 100 type I IFN-inducible genes
- 10.Weinstock-Guttman B, Tamano-Blanco M, Bhasi K, Zivadinov R, Ramanathan M. Pharmacogenetics of MXA SNPs in interferon-β treated multiple sclerosis patients. J. Neuroimmunol. 2007;182(1–2):236–239. doi: 10.1016/j.jneuroim.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 11. Byun E, Caillier SJ, Montalban X, et al. Genome-wide pharmacogenomic analysis of the response to interferon β therapy in multiple sclerosis. Arch. Neurol. 2008;65(3):337–344. doi: 10.1001/archneurol.2008.47. •• First genome-wide screen for identification of genetic markers for response to IFN-β in MS based on genotyping of DNA pools for 100K SNPs
- 12. Vandenbroeck K, Matute C. Pharmacogenomics of the response to IFN-β in multiple sclerosis: ramifications from the first genome-wide screen. Pharmacogenomics. 2008;9(5):639–645. doi: 10.2217/14622416.9.5.639. • Discussion of current status of MS pharmacogenomics research based on polymorphic markers.
- 13.O’Doherty C, Villoslada P, Vandenbroeck K. Pharmacogenomics of type I interferon therapy: a survey of response-modifying genes. Cytokine Growth Factor Rev. 2007;18(3–4):211–222. doi: 10.1016/j.cytogfr.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 14. Baranzini SE, Mousavi P, Rio J, et al. Transcription-based prediction of response to IFNβ using supervised computational methods. PLoS Biology. 2005;3(1):E2. doi: 10.1371/journal.pbio.0030002. •• Pioneering study identifying transcriptional markers of response to IFN-β. Design and use of advanced data-mining and predictive modelling approaches.
- 15.Poser CM, Paty DW, Scheinberg L, et al. New diagnostic criteria for multiple sclerosis: guidelines for research protocols. Ann. Neurol. 1983;13(3):227–231. doi: 10.1002/ana.410130302. [DOI] [PubMed] [Google Scholar]
- 16. Favorov AV, Andreewski TV, Sudomoina MA, Favorova OO, Parmigiani G, Ochs MF. A Markov chain Monte Carlo technique for identification of combinations of allelic variants underlying complex diseases in humans. Genetics. 2005;171(4):2113–2121. doi: 10.1534/genetics.105.048090. •• Description of the APSampler algorithm for exploration of allelic combinations associated with outcome. APSampler was applied in the present study
- 17.Ji X, Cheung R, Cooper S, Li Q, Greenberg HB, He XS. Interferon α regulated gene expression in patients initiating interferon treatment for chronic hepatitis C. Hepatology. 2003;37(3):610–621. doi: 10.1053/jhep.2003.50105. [DOI] [PubMed] [Google Scholar]
- 18.Weinstock-Guttman B, Badgett D, Patrick K, et al. Genomic effects of IFN-β in multiple sclerosis patients. J. Immunol. 2003;171(5):2694–2702. doi: 10.4049/jimmunol.171.5.2694. [DOI] [PubMed] [Google Scholar]
- 19.Sturzebecher S, Wandinger KP, Rosenwald A, et al. Expression profiling identifies responder and nonresponder phenotypes to interferon-β in multiple sclerosis. Brain. 2003;126(Pt 6):1419–1429. doi: 10.1093/brain/awg147. [DOI] [PubMed] [Google Scholar]
- 20.Bailey DP, Kashyap M, Bouton LA, Murray PJ, Ryan JJ. Interleukin-10 induces apoptosis in developing mast cells and macrophages. J. Leukoc. Biol. 2006;80(3):581–589. doi: 10.1189/jlb.0405201. [DOI] [PubMed] [Google Scholar]
- 21.Bachis A, Colangelo AM, Vicini S, et al. Interleukin-10 prevents glutamate-mediated cerebellar granule cell death by blocking caspase-3-like activity. J. Neurosci. 2001;21(9):3104–3112. doi: 10.1523/JNEUROSCI.21-09-03104.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong J, Deaciuc IV, Burikhanov R, de Villiers WJ. Lipopolysaccharide-induced liver apoptosis is increased in interleukin-10 knockout mice. Biochim. Biophys. Acta. 2006;1762(4):468–477. doi: 10.1016/j.bbadis.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 23.Sarkar D, Leszczyniecka M, Kang DC, et al. Down-regulation of Myc as a potential target for growth arrest induced by human polynucleotide phosphorylase (hPNPaseold-35) in human melanoma cells. J. Biol. Chem. 2003;278(27):24542–24551. doi: 10.1074/jbc.M302421200. [DOI] [PubMed] [Google Scholar]
- 24.Novick D, Cohen B, Rubinstein M. The human interferon α/β receptor: characterization and molecular cloning. Cell. 1994;77(3):391–400. doi: 10.1016/0092-8674(94)90154-6. [DOI] [PubMed] [Google Scholar]
- 25.Soilu-Hanninen M, Laaksonen M, Hanninen A, Eralinna JP, Panelius M. Downregulation of VLA-4 on T cells as a marker of long term treatment response to interferon β–1 α in MS. J. Neuroimmunol. 2005;167(1–2):175–182. doi: 10.1016/j.jneuroim.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 26.Graber J, Zhan M, Ford D, et al. Interferon-β–1 α induces increases in vascular cell adhesion molecule: implications for its mode of action in multiple sclerosis. J. Neuroimmunol. 2005;161(1–2):169–176. doi: 10.1016/j.jneuroim.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 27.Edwards-Smith CJ, Jonsson JR, Purdie DM, Bansal A, Shorthouse C, Powell EE. Interleukin-10 promoter polymorphism predicts initial response of chronic hepatitis C to interferon α. Hepatology. 1999;30(2):526–530. doi: 10.1002/hep.510300207. [DOI] [PubMed] [Google Scholar]
- 28.Wergeland S, Beiske A, Nyland H, et al. IL-10 promoter haplotype influence on interferon treatment response in multiple sclerosis. Eur. J. Neurol. 2005;12(3):171–175. doi: 10.1111/j.1468-1331.2004.01102.x. [DOI] [PubMed] [Google Scholar]
- 29.Abbas Z, Moatter T, Hussainy A, Jafri W. Effect of cytokine gene polymorphism on histological activity index, viral load and response to treatment in patients with chronic hepatitis C genotype 3. World J. Gastroenterol. 2005;11(42):6656–6661. doi: 10.3748/wjg.v11.i42.6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yee LJ, Tang J, Gibson AW, Kimberly R, van Leeuwen DJ, Kaslow RA. Interleukin 10 polymorphisms as predictors of sustained response in antiviral therapy for chronic hepatitis C infection. Hepatology. 2001;33(3):708–712. doi: 10.1053/jhep.2001.22347. [DOI] [PubMed] [Google Scholar]
- 31.Bartosik-Psujek H, Stelmasiak Z. The interleukin-10 levels as a potential indicator of positive response to interferon β treatment of multiple sclerosis patients. Clin. Neurol. Neurosurg. 2006;108(7):644–647. doi: 10.1016/j.clineuro.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 32.Muraro PA, Liberati L, Bonanni L, et al. Decreased integrin gene expression in patients with MS responding to interferon-β treatment. J. Neuroimmunol. 2004;150(1–2):123–131. doi: 10.1016/j.jneuroim.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Kawanokuchi J, Mizuno T, Kato H, Mitsuma N, Suzumura A. Effects of interferon-β on microglial functions as inflammatory and antigen presenting cells in the central nervous system. Neuropharmacology. 2004;46(5):734–742. doi: 10.1016/j.neuropharm.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 34.Asahina Y, Izumi N, Uchihara M, et al. Interferon-stimulated gene expression and hepatitis C viral dynamics during different interferon regimens. J. Hepatol. 2003;39(3):421–427. doi: 10.1016/s0168-8278(03)00287-3. [DOI] [PubMed] [Google Scholar]
- 35.Knapp S, Yee LJ, Frodsham AJ, et al. Polymorphisms in interferon-induced genes and the outcome of hepatitis C virus infection: roles of MxA, OAS-1 and PKR. Genes Immun. 2003;4(6):411–419. doi: 10.1038/sj.gene.6363984. [DOI] [PubMed] [Google Scholar]
- 36.Leszczyniecka M, Su ZZ, Kang DC, Sarkar D, Fisher PB. Expression regulation and genomic organization of human polynucleotide phosphorylase, hPNPase(old-35), a type I interferon inducible early response gene. Gene. 2003;316:143–156. doi: 10.1016/s0378-1119(03)00752-2. [DOI] [PubMed] [Google Scholar]
- 37.Cosentino M, Zaffaroni M, Marino F, et al. Catecholamine production and tyrosine hydroxylase expression in peripheral blood mononuclear cells from multiple sclerosis patients: effect of cell stimulation and possible relevance for activation-induced apoptosis. J. Neuroimmunol. 2002;133(1–2):233–240. doi: 10.1016/s0165-5728(02)00372-7. [DOI] [PubMed] [Google Scholar]
- 38.Cosentino M, Zaffaroni M, Ferrari M, et al. Interferon-γ and interferon-β affect endogenous catecholamines in human peripheral blood mononuclear cells: implications for multiple sclerosis. J. Neuroimmunol. 2005;162(1–2):112–121. doi: 10.1016/j.jneuroim.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 39.Giorelli M, Livrea P, Trojano M. Dopamine fails to regulate activation of peripheral blood lymphocytes from multiple sclerosis patients: effects of IFN-β. J. Interferon Cytokine Res. 2005;25(7):395–406. doi: 10.1089/jir.2005.25.395. [DOI] [PubMed] [Google Scholar]
- 40.Chen L, Borozan I, Feld J, et al. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 2005;128(5):1437–1444. doi: 10.1053/j.gastro.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 41.Batocchi AP, Rotondi M, Caggiula M, et al. Leptin as a marker of multiple sclerosis activity in patients treated with interferon-β. J. Neuroimmunol. 2003;139(1–2):150–154. doi: 10.1016/s0165-5728(03)00154-1. [DOI] [PubMed] [Google Scholar]
- 42.Jansen M, Reinhard JF., Jr Interferon response heterogeneity: activation of a pro-inflammatory response by interferon α and β. A possible basis for diverse responses to interferon β in MS. J. Leukoc. Biol. 1999;65(4):439–443. doi: 10.1002/jlb.65.4.439. [DOI] [PubMed] [Google Scholar]
- 43.Zabetian CP, Anderson GM, Buxbaum SG, et al. A quantitative-trait analysis of human plasma-dopamine β-hydroxylase activity: evidence for a major functional polymorphism at the DBH locus. Am. J. Hum. Genet. 2001;68(2):515–522. doi: 10.1086/318198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schreiner B, Mitsdoerffer M, Kieseier BC, et al. Interferon-β enhances monocyte and dendritic cell expression of B7-H1 (PD-L1), a strong inhibitor of autologous T-cell activation: relevance for the immune modulatory effect in multiple sclerosis. J. Neuroimmunol. 2004;155(1–2):172–182. doi: 10.1016/j.jneuroim.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 45.Johnson J, Albarani V, Nguyen M, Goldman M, Willems F, Aksoy E. Protein kinase C α is involved in interferon regulatory factor 3 activation and type I interferon-β synthesis. J. Biol. Chem. 2007;282(20):15022–15032. doi: 10.1074/jbc.M700421200. [DOI] [PubMed] [Google Scholar]
- 46.Fimia GM, Evangelisti C, Alonzi T, et al. Conventional protein kinase C inhibition prevents a interferon-mediated hepatitis C virus replicon clearance by impairing STAT activation. J. Virol. 2004;78(23):12809–12816. doi: 10.1128/JVI.78.23.12809-12816.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reich NC, Pfeffer LM. Evidence for involvement of protein kinase C in the cellular response to interferon α. Proc. Natl Acad. Sci. 1990;87(22):8761–8765. doi: 10.1073/pnas.87.22.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taniguchi T, Takaoka A. A weak signal for strong responses: interferon-α/β revisited. Nat. Rev. Mol. Cell Biol. 2001;2(5):378–386. doi: 10.1038/35073080. [DOI] [PubMed] [Google Scholar]
- 49.Liu B, Mink S, Wong KA, et al. PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity. Nat. Immunol. 2004;5(9):891–898. doi: 10.1038/ni1104. [DOI] [PubMed] [Google Scholar]
- 50.Natsume A, Ishii D, Wakabayashi T, et al. IFN-β down-regulates the expression of DNA repair gene MGMT and sensitizes resistant glioma cells to temozolomide. Cancer Res. 2005;65(17):7573–7579. doi: 10.1158/0008-5472.CAN-05-0036. [DOI] [PubMed] [Google Scholar]
- 51.Krakauer M, Sorensen P, Khademi M, Olsson T, Sellebjerg F. Increased IL-10 mRNA and IL-23 mRNA expression in multiple sclerosis: interferon-β treatment increases IL-10 mRNA expression while reducing IL-23 mRNA expression. Mult. Scler. 2008;14(5):622–630. doi: 10.1177/1352458507087136. [DOI] [PubMed] [Google Scholar]
- 52.Paunovic V, Carroll HP, Vandenbroeck K, Gadina M. Signalling, inflammation and arthritis: crossed signals: the role of interleukin (IL)-12, -17, -23 and -27 in autoimmunity. Rheumatology. 2008;47(6):771–776. doi: 10.1093/rheumatology/kem352. [DOI] [PubMed] [Google Scholar]
- 53.Donnelly RP, Sheikh F, Kotenko SV, Dickensheets H. The expanded family of class II cytokines that share the IL-10 receptor-2 (IL-10R2) chain. J. Leukoc. Biol. 2004;76(2):314–321. doi: 10.1189/jlb.0204117. [DOI] [PubMed] [Google Scholar]
Websites
- 101.Haploview. www.broad.mit.edu/mpg/haploview.
- 102.APSampler. http://astor.com.jhmi.edu/~sasha/APSampler/
- 103.SNPper. http://snpper.chip.org.