Summary
The nuclear receptor CAR is a xenobiotic responsive transcription factor that plays a central role in the clearance of drugs and bilirubin while promoting cocaine and acetaminophen toxicity. In addition, CAR has established a “reverse” paradigm of nuclear receptor action where the receptor is active in the absence of ligand and inactive when bound to inverse agonists. We now report the crystal structure of murine CAR bound to the inverse agonist androstenol. Androstenol binds within the ligand binding pocket, but unlike many nuclear receptor ligands, it makes no contacts with helix H12/AF2. The transition from constitutive to basal activity (androstenol bound) appears to be associated with a ligand-induced kink between helices H10 and H11. This disrupts the previously predicted salt bridge that locks H12 in the transcriptionally active conformation. This mechanism of inverse agonism is distinct from traditional nuclear receptor antagonists thereby offering a new approach to receptor modulation.
Introduction
CAR, a 358 amino acid, 41 kDa protein from the nuclear receptor superfamily, has been directly linked to the clearance of both xenobiotics (Wei et al., 2000; Zhang et al., 2002) and endogenous toxins such as bilirubin (Huang et al., 2003). CAR is most abundantly expressed in the liver and intestine where it controls the transcription of genes involved in xenobiotic and bilirubin clearance. These target genes include certain P450 family monooxygenases, phase II conjugating enzymes, and xenobiotic transporters. Thus, CAR serves as a master regulator of xenobiotic clearance, and its activation can be considered a form of chemical immunity.
Activation of CAR is not always beneficial, because hepatic metabolism can convert certain drugs to potent toxins. For example, CAR-mediated metabolism converts acetaminophen into a reactive quinone metabolite (N-acetyl-p-benzoquinone imine). This metabolic byproduct promotes acute liver failure by binding to cellular macromolecules and by generating reactive oxygen species (Gill and Sterling, 2001; Zhang et al., 2002). Similarly, the hepatotoxic effects of cocaine are also mediated via a CAR-dependent pathway (Wei et al., 2000). Thus, the ability to either activate or repress CAR can have either protective or deleterious consequences depending on the particular chemical challenges faced by the organism.
As with other nuclear receptors, CAR is a modular protein with distinct functional domains. There is an N-terminal DNA binding domain (DBD) which determines targetgene selectivity. The ligand binding domain (LBD) is a multifunctional module that contains the ligand binding pocket, a dimerization interface that associates with the retinoid X receptor (RXR), and, importantly, a C-terminal ligand-dependent transactivation domain (AF2). Previous biochemical and structural studies on a variety of nuclear receptors have demonstrated that ligand binding results in conformation changes that are associated with a transcriptionally active state. These conformation changes are characterized by a realignment of AF2 along the receptor surface, thereby creating a new interface that recruits transcriptional coactivator proteins. These changes are required for transcriptional activity and appear to be conserved among all ligand-activated receptors that have been studied to date.
Unlike classical nuclear receptors that are ligand activated, the discovery of CAR has established a new paradigm of a constitutively active, ligand-repressible receptor (Baes et al., 1994; Choi et al., 1997; Forman et al., 1998). It is now known that CAR is nuclear localized in heterologous cell types but is cytosolic in its native hepatocyte environment (Kawamoto et al., 1999; Kobayashi et al., 2003). There are two mechanisms by which CAR can be translocated to the nucleus. First, compounds such as phenobarbital (Kawamoto et al., 1999), bilirubin (Huang et al., 2003), or 6,7-dimethylesculetin (Huang et al., 2004a) can promote translocation via an indirect mechanism that may involve activation and/or recruitment of protein phosphatase 2A (Yoshinari et al., 2003). Once in the nucleus, unliganded CAR interacts with RXR to form a heterodimer that recognizes its specific target genes. It has previously been shown that the CAR/RXR heterodimer is capable of recruiting coactivator proteins via the CAR AF2 transactivation domain (Dussault et al., 2002). However, unlike classical nuclear receptors which require ligand for activation, the transcriptional activity of CAR is induced simply by association with RXR; there is no apparent need for a CAR ligand. Although ligand is not required for activation, constitutive CAR activity is mediated through the same conserved functional domains as those utilized by ligand-activated receptors. By using a combination of homology modeling and mutagenesis, it has also been demonstrated that CAR AF2 is maintained in the active conformation by virtue of a short loop between the predicted helix H11 and AF2 (helix H12) and through a charge-charge interaction that links AF2 with Lys205 of helix H4 (Dussault et al., 2002).
In addition to ligand-independent translocation, CAR can also be transported to the nucleus via agonist ligands such as 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP) (Tzameli et al., 2000) and 6-(4-cholorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime (CITCO) (Maglich et al., 2003), which are selective for mouse and human CAR, respectively. In both cases, these agonists potentiate constitutive activity by stabilizing the constitutive AF2-coactivator interaction (Dussault et al., 2002; Maglich et al., 2003). The antiemetic, meclizine, also modulates the activity of both mouse and human CAR (Huang et al., 2004b). The functional domains required for ligand-dependent activation are similar to those utilized by the constitutively active receptor (Dussault et al., 2002).
Interestingly, the constitutive activity of apo-CAR can be reversed by a class of compounds known as inverse agonists. The prototypic members of this class are androstane metabolites, androstanol (5α-androstan-3α-ol), and androstenol (5α-androst-16-en-3α-ol) (Forman et al., 1998), which reverse constitutive activity both by displacing coactivator and by promoting a weak association with corepressor proteins (Dussault et al., 2002). This inverse agonist activity has been useful in blocking CAR activity in vivo thereby minimizing the hepatoxic effects of acetominophen.
The paradigm of a constitutively active ligand-repressible receptor raises interesting questions. The precise structural events that promote constitutive activity are of considerable interest as is the role of ligand in reversing activation. We now report the structure of the mouse CAR LBD complexed to the inverse agonist, androstenol. Unlike classical steroid receptor antagonists, androstenol does not directly displace the AF2 transactivation domain and is completely confined within the ligand binding pocket. Instead, binding to androstenol produces an H10-H11 kink, which repositions AF2 into an inactive configuration. This mechanism is reminiscent of a “toggle bolt”-like structure and has important implications for the design of inverse agonists for CAR and for antagonists of other nuclear receptors.
Results and Discussion
Structure of the mCAR LBD/Androstenol Complex
We expressed and isolated residues 109–358 (numbering convention as in Dussault et al. [2002]) of mCAR and crystallized the LBD/androstenol complex. The structure of the mCAR LBD/inverse agonist complex was determined by X-ray crystallography with the molecular replacement method to a resolution of 2.9Å (Table 1 and Figure 1).
Table 1.
Statistics for Data Collection and Refinement
| Spacegroup | C2221 |
| Resolution (Å) | 2.9 |
| Reflections, total | 157834 |
| Reflections, unique | 14399 |
| Data completeness (outer shell) (%) | 99.8 (100.0) |
| Rsym (outer shell) (%)a | 10.8 (45.1) |
| Refinement | |
| Resolution range (Å) | 30.0–2.9 R /R |
| Rwork/Rfree (%)b | 22.5/28.8 |
| Number of water molecules | 33 |
| Rmsd bond lengths (Å) | 0.007 |
| Rmsd bond angles (°) | 1.259 |
| Rmsd dihedrals (°) | 20.54 |
| Rmsd impropers (°) | 0.91 |
| Ramachandran Plot | |
| Most favored (%) | 88.8 |
| Additionally allowed (%) | 11.0 |
| Generously allowed (%) | 0.2 |
| Disallowed (%) | 0.0 |
Rsym = ΣhΣi|Ih,i — Ih|/ΣhΣiIh,i, where Ih is the mean intensity of the I observations of symmetry-related reflections of h.
Rwork = Σ∥Fobs| — |Fcalc∥/Σ|Fobs|, where Fobs = Fp, and Fcalc is the calculated protein structure factor from the atomic model (Rfree was calculated with 5% of the reflections). Rmsd in bond lengths and angles are the deviations from ideal values.
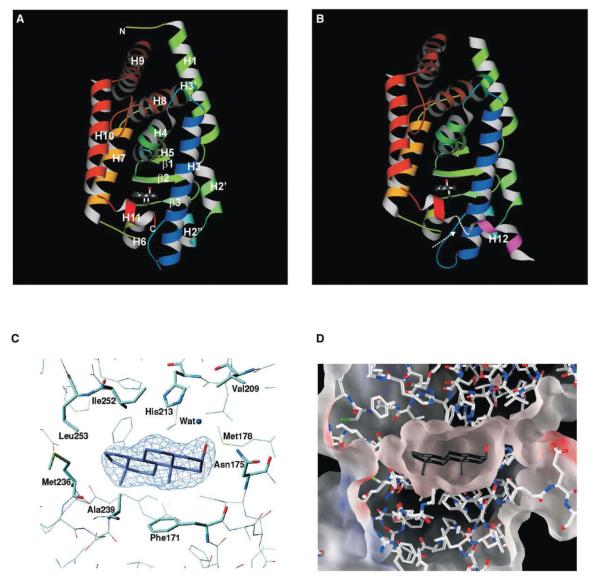
Figure 1.
Structure of mCAR LBD/Androstenol Complex
(A and B) Ribbon representation of each CAR molecule within the asymmetric unit. The arrow indicates the region of missing density between helices H11 and H12 in molecule B. The bound ligand is also displayed in each molecule as ball-and-stick representations.
(C) Fo-Fc omit map contoured at 5σ for androstenol within the binding pocket. The model is superimposed on the electron density, and the androstenol is colored blue (carbon) and red (hydroxyl).
(D) Electrostatic surface representation of the ligand binding pocket. Androstenol is shown in black (carbon) and red (hydroxyl).
There are two molecules in the crystallographic asymmetric unit. There is continuous electron density for molecule A which extends from amino acid residues 114–345, whereas density for molecule B extends from residues 120–343 and 350–357. The two molecules have a root mean square difference (rmsd) of approximately 1.07Å calculated over Cα atoms. The major conformational difference between the molecules is within the region connecting helix H1 and helix H3 (156–163), which adopts different conformations in the two molecules. Omitting these residues from the calculation results in a smaller rmsd (0.59Å), suggesting that the two molecules are nearly identical. The different conformation between these regions is because in one molecule (Figure 1A), residues within this loop are involved in an intramolecular interaction, specifically a hydrogen bond between Arg161 and Asp238. In the second molecule (Figure 1B), Arg161 makes a salt bridge with Glu339 from a symmetry-related neighbor. The CAR structure shares the canonical nuclear receptor LBD helical sand-wich fold with minor variations (Figures 1A and 1B). This model has 12 α helices and three short β strands. As with RARγ (Renaud et al., 1995), there is no helix H2. Instead, there are two 310 helices H2′ and H2″ between helices H1 and H3 and an additional helix H3′ between helices H3 and H4, similar to the mCAR/TCPOBOP structure (Suino et al., 2004 [this issue of Molecular Cell]). At the C terminus, there is no electron density for helix H12 for molecule A, but there is visible density for helix H12 in molecule B. In the modeled orientation, helix H12 has a relatively high mobility (average B factor = 115Å2). The CAR LBD is most closely related in structure to hPXR (rmsd = 0.76Å for 155 Cα atoms, sequence identity = 35.1%, 1ILH) and VDR (rmsd = 0.97Å for 155 Cα atoms, sequence identity = 30.8%, 1KKQ). An unusual characteristic of the CAR structure are the two 310 helices between helices H1 and H3. This region spans residues 136–163 and has the appearance of a cap at the “bottom” of the structure. However, this region is also quite rigid (average B factor = 33Å2). The two helices also contribute to the ligand binding pocket as Phe142 (H2′) is within the binding cavity, and although they do not appear to make any interactions with the ligand, they may contribute to the general hydrophobicity of the cavity. The conformation of this region suggests that this may be an entry point for the ligand. The corresponding region in PPARα too has been postulated to be the entry point to the ligand binding pocket, and in PPARα, this region too is α helical (H2) (Nolte et al., 1998; Gampe et al., 2000). A second distinctive feature is the orientation of helix H11 relative to that of agonist bound PXR (Watkins et al., 2001) and VDR (Rochel et al., 2000). Helix H11 in CAR is oriented closer to the main body of the protein. The second charge clamp observed in the mCAR/TCPOBOP structure (Suino et al., 2004) is absent within this structure. It is likely that this feature is induced upon binding to the TIF-2 peptide.
The mCAR ligand binding pocket is approximately 570Å3, which is nearly half the size of the PXR cavity (1130Å3) (Watkins et al., 2001). In fact, the CAR binding pocket is closest in size to that of hRXRα (Egea et al., 2000). There are twenty-seven residues that contribute to this pocket, 74% of which are apolar (Figure 1D). This highly apolar nature of the binding pocket comes from Phe142, Phe171, Ala172, Ile174, Met178, Val209, Leu212, Leu216, Phe220, Phe227, Cys229, Tyr234, Met236, Ala239, Phe244, Phe248, Leu249, Ile252, Leu253, and Leu340. Also included within this pocket are Asn175, His213, Glu225, Asn226, Lys235, Asp238, and His256. However, the aspartic acid (Asp238) forms a salt bridge with Arg156, which effectively neutralizes the charge on the aspartate. A similar interaction between Glu321 and Arg410 was observed within the PXR binding pocket (Watkins et al., 2001). The side chains of lysine (Lys235) and glutamic acid (Glu225) are oriented away from the pocket, which also minimizes their charge contribution to the pocket.
Androstenol Makes Specific Interactions with the Largely Hydrophobic mCAR Ligand Binding Pocket
Androstenol nestles within the center of the hydrophobic binding pocket of CAR and is completely sequestered from the external environment by the protein. The electron density for the ligand is intense, and its conformation is unambiguous (Figure 1C). As it floats within this cavity, it is almost equidistant from the “top” and “bottom” of this binding site (Figure 1D). The specificity for androstenol is realized through a combination of hydrogen bonding and apolar interactions between the ligand and the binding cavity (Figure 2A). The residues within the pocket that interact with androstenol come from helices H3, H5, H6, H7, H11, and β strand β2.
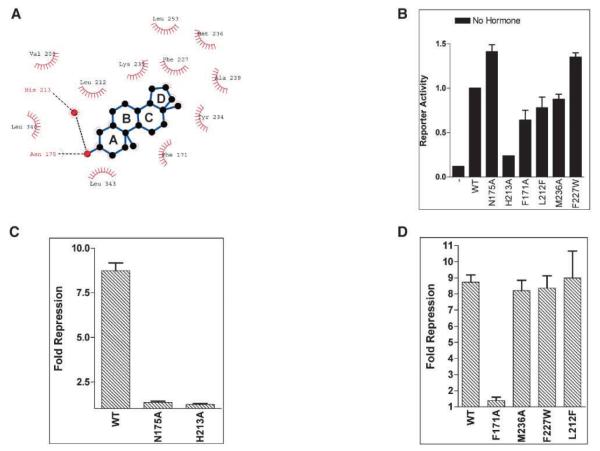
Figure 2.
Interactions between CAR and Androstenol within the Ligand Binding Pocket
(A) Schematic representation of interactions within the ligand binding pocket calculated with LIGPLOT (Laskowski et al., 1993b). The dotted lines indicated hydrogen bonds between androstenol, Asn175, His213, and a molecule of water. Residues enclosed in semicircles are in van der Waals contact with the ligand.
(B) A comparison of the constitutive transcriptional activities of wild-type CAR and mutants performed in CV-1 cells with an LXRE×3-TK-Luc reporter construct. No ligands were added to the transfected cells. Error bars represent the SD.
(C) A comparison of relative fold repression (fold repression = apo-CAR / CAR + androstenol) of wild-type and N175A and H213A mutants by androstenol with an LXRE×3-TK-Luc reporter construct. Transformed cells were incubated with 5 μM androstenol for 40 h before luciferase and galactosidase activities were measured. Error bars represent the SD.
(D) Apolar interactions and binding pocket size constraints on the ligand measured by mutagenesis and cell-based transactivation assays (LXRE×3-TK-Luc reporter construct). Error bars represent the SD.
The only polar interactions between androstenol and CAR are between the 3α-hydroxyl moiety of androstenol and the residues Asn175 (helix H3) and His213 (helix H5). These interactions are similar in both CAR molecules. The two hydrogen bonding interactions that secure the ligand within the pocket are not identical. The first is a direct hydrogen bond between the 3α-hydroxyl (acceptor) of the inverse agonist and Asn175 Nδ2 (donor). The second hydrogen bonding interaction is with His213 and is mediated by a single water molecule that bridges the distance between the 3α-hydroxyl and His213 Nϵ2 (Figure 2A). The presence of this water molecule within the ligand binding pocket is reminiscent of the interactions between ERβ and genistein, where the water molecule within ERβ also appears to mediate interactions with the ligand (Pike et al., 1999).
Asn175 and His213 were individually mutated to alanine, and the resulting mutants were analyzed in a cell-based reporter assay system to determine the role of hydrogen bonding interactions in androstenol binding. Although the N175A and H213A mutants display complete or partial constitutive activity, respectively, neither mutant displays any response to androstenol when tested in transformed CV-1 cells (Figures 2B and 2C). This indicates that hydrogen bonding with both Asn175 and His213 are critical for recognition of the inverse agonist. Next, we examined the apolar contribution to androstenol recognition and binding. Phe171 appears to stack against rings A and B of the four-membered androstenol molecule (Figures 1D and 2A). When examined, the F171A mutant was found to retain wild-type levels of constitutive activity, but the inhibitory effect of androstenol was significantly impaired (Figure 2D). This confirms an earlier study in which a F171L/I174A double mutant was completely resistant to the inhibitory effect of the inverse agonist, androstanol (Tzameli et al., 2000). Thus, it seems that the apolar contributions of Phe171 and Ile174 are additive toward androstenol binding. We also mutated Met236, but the M236A mutation displays no observable difference with wild-type CAR activity and androstenol binding, suggesting that this methionine is not critical for CAR activity (Figure 2D).
A unique feature of the CAR/androstenol interaction is the considerable difference in volume of the binding pocket (570Å3) and of the ligand (270Å3). As a result, the ligand appears to hover within the center of the pocket (Figure 1D). To examine if androstenol would tolerate minor incursions into this binding pocket, we generated two mutants designed to have bulkier amino acid side chains. Both the F227W and the L212F CAR mutants showed similar activity profiles to the wild-type protein (Figure 2D), suggesting that the binding pocket may accommodate larger ligands that make the correct contacts within the pocket. These data collectively suggest that specific apolar interactions are essential for binding androstenol. Moreover, the extra room within the binding pocket may accommodate ligand structures that have additional apolar groups.
From this structure, one can rationalize the stereo specificity displayed by CAR for various compounds. The specificity for the androstanes is limited to the 5α-reduced, 3α-hydroxy stereoisomer (Forman et al., 1998). For instance, the 5β-reduced compound is a non-planar molecule in which the 3α-hydroxy group would be oriented further away from the critical hydrogen-bonding Asn175, precluding interactions with this residue. Moreover, there is the potential for steric clashes with Leu212 and Leu216. Conversely, although the 5α-reduced, 3β-hydroxy compound is within hydrogen-bonding distance of the Asn175, this compound would not interact efficiently with His213. Molecules with 17β-hydroxy and 17-keto (androsterone) substitutions are also inactive (Forman et al., 1998), presumably because of clash with residues located in the ligand binding pocket, particularly Phe227.
The Orientation of AF2 Is Random in the mCAR/Inverse Agonist Bound Conformation
The AF2 domain functions as a critical regulatory element, the conformation of which is regulated by ligand (Moras and Gronemeyer, 1998). In the “active” conformation, this domain, which is contained within helix H12, is positioned to generate a binding site for coactivator proteins such as SRC-1 (Onate et al., 1995). In this orientation, helix H12 is folded against the body of the protein where it forms part of the ligand binding pocket and even interacts with the bound agonist (Renaud et al., 1995; Moras and Gronemeyer, 1998). The orientation of helix H12 in the “inactive” form is difficult to establish. In this inactive state, the coactivator protein is replaced by a corepressor molecule whose binding site overlaps with the binding site of the coactivator (Xu et al., 2002). However, in contrast to the nuclear receptor/coactivator complex, the corepressor molecule does not appear to require the presence of the AF2 domain (Dussault et al., 2002). What orientation does helix H12 adopt when the receptor is in the inactive conformation? Structural studies have indicated several different conformations of helix H12, such as those in the classic apo-RXRα LBD structure (Bourguet et al., 1995), the ERβ/antagonist complexes (Pike et al., 1999), and the PPARα/GW6471/SMRT complex (Xu et al., 2002). The orientation of helix H12 in each of these structures is different and, intriguingly, may be artifactually involved in a network of crystal contacts brought about by the arrangement of the protein molecules within the crystal lattice. Additionally, the mobility or B factor associated with these AF2 domains is relatively high (>100Å2) in each of the reported structures, suggesting that this domain may adopt multiple conformations.
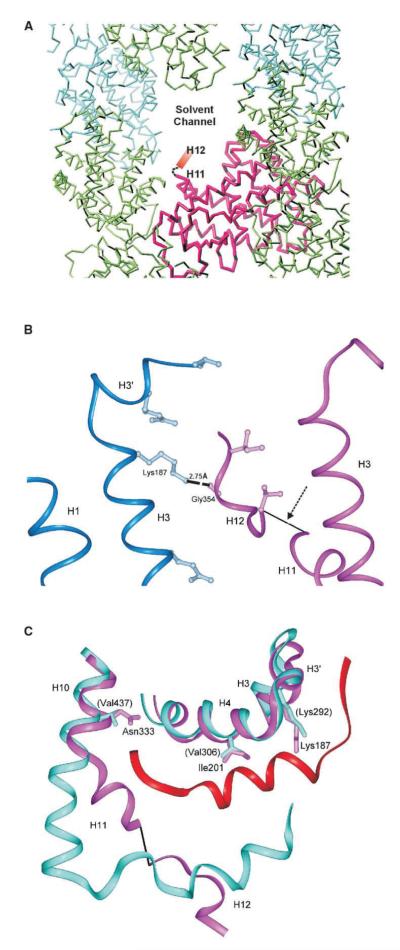
In the androstenol bound CAR structure, we see two different arrangements within the crystal lattice. One CAR molecule is oriented such that the AF2 domain is exposed to solvent channels within the crystal (Figure 3A). In this molecule, no electron density is observed for helix H12. The second CAR molecule in the crystallographic asymmetric unit is oriented such that helix H12 can interact with neighboring symmetry-related molecules, specifically through Gly354-Lys187 (symmetry molecule) (Figure 3B). These interactions stabilize helix H12 in this molecule, and there is sufficient electron density to determine the orientation of this helical domain. This helix H12, however, has an average B factor of 115Å2, also suggesting a high level of mobility.
Figure 3.
Analysis of the AF2, Helix H12, Conformation in CAR/Androstenol
(A) Crystal packing of the CAR/androstenol complex. In one CAR molecule (purple), helix H12 is exposed to solvent channels within the crystal. Symmetry partners are shown in green and cyan.
(B) Helix H12 of the second molecule (purple) is stabilized through crystal contacts with a neighboring molecule (blue) through a Gly354-Lys187 hydrogen bond. The arrow indicates missing density.
(C) Model of CAR (purple) superimposed on the PPARα/SMRT complex (cyan/red) indicates that predicted SMRT/CAR interactions are retained, particularly from the CAR Lys187. The residues in parentheses are from PPARα.
Because the recruitment of corepressor proteins accompanies the transcriptional repression of CAR, we utilized molecular docking techniques to evaluate the fit of SMRT on this CAR structure. By superimposing the structure of the PPARα/GW6471/SMRT complex (Xu et al., 2002) on the CAR/androstenol complex structure, we were able to compare the interactions made between PPARα and SMRT with those that could potentially be made by CAR and the corepressor (Figure 3C). The minimal receptor-interacting domain of SMRT encompasses the motif LXXXIXXXL, which lies across helices H3, H4, and H5. Specific interactions occur between the SMRT peptide and Lys292 of helix H3 in PPARα and are conserved between CAR (Lys187) and the docked SMRT peptide (Dussault et al., 2002). Other interactions include Val309 (Tyr314), Asn333 (Val437), Ile201 (Val306), and Thr176 (Val281), where the residues in parenthesis are from PPARα. Thus, in this structure, the CAR helix H12 is positioned to provide ample docking space for the SMRT peptide.
Structural Model for CAR Activity
There are specific structural and consequently functional changes that are initiated upon ligand binding to the nuclear receptor LBD (Moras and Gronemeyer, 1998). The most prominent of these includes a large repositioning of the C-terminal AF2 transactivation domain (helix H12) (Nolte et al., 1998). In the active conformation, the agonist interacts with helix H12 and folds this helix on to the main body of the receptor. This creates a new interface that specifically recognizes co-activator molecules (Moras and Gronemeyer, 1998; Rochel et al., 2000; Watkins et al., 2001). Antagonist ligands that bind but fail to activate the receptor displace AF2 away from the main body of the receptor and consequently disrupt the receptor/coactivator interface. For example, in both the ERα/raloxifene (Brzozowski et al., 1997) and PPARα/GW6471 (Xu et al., 2002) complexes, the antagonist protrudes out of the binding pocket and appears to dislodge helix H12 from its active conformation. It is in this conformation that the nuclear receptor can bind to corepressor molecules (Xu et al., 2002). There are other regions besides the AF2 domain that also undergo significant ligand-induced conformational changes (Moras and Gronemeyer, 1998). In the agonist bound active conformation, helix H10 and H11 appear as a single helical domain. However, in both the inactive apo and antagonist bound states, the single helix melts into the two helices H10 and H11. In the apo state, H11 adopts a conformation that is nearly orthogonal to H10. When bound to antagonist, H11 runs parallel to H10 but is displaced slightly from the general direction of H10.
It is interesting to note that the conformation of helices H10 and H11 in the repressed mCAR/androstenol structure is similar to the inactive apo conformations of traditional ligand-activated nuclear receptors (Figure 4A). In addition to being orthogonal to H10, helix H11 is twisted in the direction of the ligand binding pocket. In effect, a “kink” exists between helices H10 and H11 in the inactive androstenol bound structure. An important feature of this “kink” is a hydrogen bond between Glu339 and the backbone amide of Gln245, which functions to restrain this “kink” in place (Figure 4B). This backbone Gln245-Glu339 interaction is likely to be significant, because it is conserved in many other receptors including ERα (His) (Brzozowski et al., 1997), LXRα (Gln) (Svensson et al., 2003), ERRγ (His) (Greschik et al., 2004), RARα (Arg) (Bourguet et al., 2000), PR (Tyr) (Williams and Sigler, 1998), VDR (Gln) (Rochel et al., 2000), and TR (Arg) (Ye et al., 2003), where the residues in parenthesis are the positions corresponding to CAR Glu339. The Glu339-Gln245 hydrogen bond places several restraints upon mCAR. First, it fixes H10-H11 to the loop that connects helix H6 to H7. Second, it stabilizes the H6-H7 loop, which contains a highly conserved and presumably flexible glycine (Gly243) residue. A predicted consequence of these restraints is that the Glu339-Gln245 interaction serves as a “pin” that maintains the rigidity of certain elements of the ligand binding pocket.
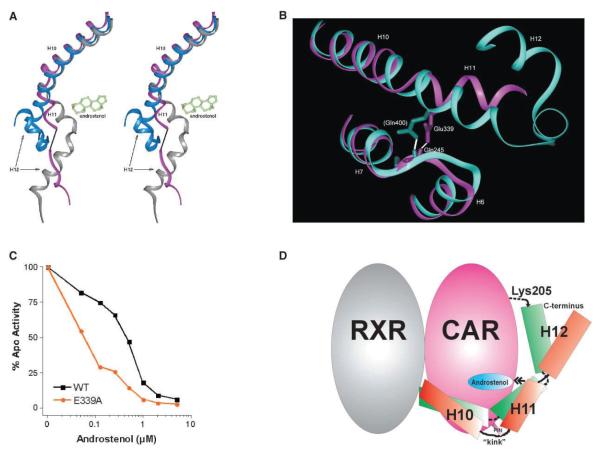
Figure 4.
Model for Androstenol-Induced Repression of CAR
(A) A stereo model to show that when bound to androstenol, the CAR helix H11 (purple) adopts a conformation that is similar to apo inactive RXR (gray), which is in contrast to the fused H10-H11 helix in the agonist bound active nuclear receptor conformations such as in PXR (blue).
(B) A Glu339-Gln245 backbone amide hydrogen bond acts as a “pin” about which helix H11 can twist toward the ligand binding pocket in CAR (purple). This interaction is conserved in most nuclear receptors even when in the active state conformation as in the VDR structure (cyan, Gln400). Residues in parentheses are from VDR.
(C) The Glu339-Gln245 backbone amide hydrogen bond is important for maintaining the integrity of the ligand binding pocket. Relative to wild-type CAR, dose response experiments demonstrate that the E339A mutant displays a 10-fold higher EC50 for androstenol (RE2×2-TK-Luc reporter construct).
(D) A conceptual model of CAR active-to-inactive state transition. Apo CAR is in green showing the predicted K205-C terminus interaction that stabilizes AF2 in the active conformation. Binding to androstenol leads to a movement (double-headed arrow) of helix H11 toward the binding pocket, generation of the H10-H11 kink, and dissociation of H12 from the body of the protein as in the CAR/androstenol structure presented here.
As noted above, androstenol floats loosely within the ligand binding pocket. Our structure suggests that this arises in part from Glu339-Gln245-mediated restraints that prevent side chains within the ligand binding pocket from making closer contact with the ligand. Our structure would further predict that disruption of the Glu339-Gln245 “pin” would relieve these constraints and allow side chains of the binding pocket to embrace androstenol more closely. The resulting increase in van der Waals contact would be sufficient to drive a significant increase in ligand binding affinity. In order to test this structural hypothesis, we selectively disrupted the Glu339-Gln245 backbone hydrogen bond by creating an E339A substitution. As predicted, a dose-response analysis demonstrated that this mutant possesses a 10-fold higher potency (EC50) for androstenol when compared with the wild-type receptor (Figure 4C). These findings support the notion that the Glu339/Gln245 backbone hydrogen bond establishes a “pin” that imposes important structural constraints upon the receptor.
This “pin” is conserved in the mCAR/TCPOBOP (Suino et al., 2004) and the hCAR (Gln349) (Xu et al., 2004 [this issue of Molecular Cell]) structures. Although Glu339 Cβ can potentially interact with the TCPOBOP ligand (Suino et al., 2004), no such interaction is observed between androstenol and the Glu339 side chain. Likewise, His524 in ERα and Gln424 in LXRα interact with the bound ligand in addition to maintaining the “pin” interaction. This “pin” is also present in both the unliganded and 4-hydroxytamoxifen/diethylstilbestrol bound ERRγ (His434) structures (Greschik et al., 2004). Taken together, these observations suggest that the “pin” is a common structural element that is utilized by several receptors regardless of their liganded state.
Conclusion
Nuclear hormone receptors represent a superfamily of structurally and functionally conserved proteins that have evolved to regulate transcription in response to small molecule ligands. The traditional nuclear receptor is inactive in the apo state and induces transcription upon binding ligand. Previous biochemical and structural studies on a variety of receptors have demonstrated that the ligand makes direct contacts with the AF2 transactivation domain (helix H12). This contact stabilizes a conformation change that realigns AF2 alongside the coregulator groove (helices H3/H4/H5) of the receptor surface. This realignment creates a new interface, which associates with transcriptional coactivator proteins that subsequently activate gene transcription. This conformation change has been referred to as the “mouse trap” model, and it appears to be a conserved feature of all ligand-activated receptors studied to date.
In contrast to this classical paradigm of ligand activated transcription, the nuclear receptor CAR defines a reverse paradigm of receptor action. This receptor is constitutively active in the apparent absence of ligand, and constitutive activity can be modestly augmented by agonists such as TCPOBOP. More importantly, the ligand-independent constitutive activity can be reversed by androstane derivatives such as androstenol. It has been previously demonstrated that both the constitutive and agonist-augmented activity of CAR require the same functional domains (AF2, coregulator groove) as those utilized for ligand-activated transcription by traditional nuclear receptors (Dussault et al., 2002). Therefore, CAR appears to share the same continuum of active to inactive transitions as traditional receptors, yet it can achieve the active state in the absence of ligand. This raises a remarkable structural question. How does CAR adapt the conserved three-dimensional scaffold of traditional receptors to produce a reverse paradigm of receptor activity?
By using homology modeling and mutagenesis, it has been demonstrated that both the apo- and TCPOBOP-activated states are maintained by virtue of a short loop between the predicted helix H11 and AF2 (H12) and via a charge-charge interaction that links the AF2 C terminus with Lys205 of helix H4 (Dussault et al., 2002). The accompanying paper by Suino et al. clearly establishes the importance of these interactions in the TCPOBOP-activated receptor. Although these unique structural features are required for constitutive activity, they are not sufficient: constitutive activity also requires formation of the CAR-RXR heterodimer (Dussault et al., 2002). Interestingly, the AF2 and coregulator groove of RXR are not required to promote constitutive activity. These findings lead to the hypothesis that RXR functions in an allosteric manner to stabilize CAR in the active AF2 C terminus/Lys205 conformation. A comparison of the active CAR/TCPOBOP structure (Suino et al., 2004) and our inactive androstenol bound structure indicates that the helix H10-H11 kink is lost in the active structure and is replaced by a single extended and continuous helix similar to the active VDR structure (Figure 4A). Thus, helices H10 and H11 appear to exist in a continuum of conformations ranging from a single continuous helix (active) to two distinct helices that are separated by a kink (inactive). Our structure indicates that when the kink is formed, H11 is drawn in toward the ligand binding cavity. This inward placement combined with the short loop between helix H11 and AF2 forces a realignment of AF2 such that it can no longer interact with K205. In effect, the kink contributes to the dissociation of the active AF2-K205 interaction. Ligand-induced changes in helix H11 have also been reported upon binding of THC to ERα and ERβ (Nettles et al., 2004) and the antagonists diethylstilbestrol and 4-hydroxytamoxifen to ERRγ (Greschik et al., 2004). In the latter case, space requirements of the ligands diethylstilbestrol and 4-hydroxytamoxifen force the helix H11 C terminus, specifically His435, away from the ERRγ ligand binding pocket.
The above observations lead us to propose the following model to explain the reverse paradigm of CAR activity (Figure 4D). The apo receptor reaches the transcriptionally active state upon binding RXR. Previous studies have indicated that allosteric communication between dimer partners can be mediated via helix H11 (Nettles et al., 2004). We suggest that this dimerization event propagates an allosteric effect onto CAR, which is associated with a full or partial “dekinking” of H10-H11 and its extension into a single helix. When H11 is extended, it is also pushed outward thus allowing the AF2 C terminus to interact with Lys205. The precise structure of this apo receptor is unclear and will await a structural analysis of a ligand-free CAR/RXR heterodimer. However, because both the apo receptor and the TCPOBOP bound receptor share functional features and domain requirements, it is likely that the two structures will have similar and overlapping features. The reverse paradigm of CAR activity is associated with an inverse agonist-mediated stabilization of the H10-H11 kink. This, in turn, results in a drawing of H11 inward, repositioning of AF2, and subsequent disruption of the AF2 C terminus-K205 interaction that defines the active state.
The H10-H11 kink is an important feature of our model. Our structure and mutagenesis studies suggest that this kink is anchored by a “pin” that is defined by a hydrogen bond between Glu339 and the Gln245 backbone. Our data suggest that this “pin” also maintains the rigidity of the ligand binding pocket. As mentioned earlier, this “pin” is conserved in many other receptors with two exceptions to this feature, PPARα and PXR. PPARα has a leucine at the corresponding position which is not capable of hydrogen bonding (Xu et al., 2002). In PXR, the helix H6—helix H7 loop is folded away such that the arginine corresponding to CAR Glu339 cannot reach this loop (Watkins et al., 2001). Conceivably, the absence of this “pin” in PPARα and PXR allows the cavities in these proteins to expand and accommodate their wide array of ligands.
It is important to note the distinct nature of the CAR structural transition between active and inactive states. Previous structural studies with ERα and PPARα demonstrate that antagonist ligands (e.g., raloxifene and GW6471, respectively) possess a functional group that extends beyond the ligand binding pocket. This extended moiety dislodges AF2 and actively “pushes” it into an inactive position. In contrast, androstenol makes no contact with the AF2 of CAR. Instead, androstenol binding is associated with a H10-H11 kink and this proximal structural change appears to lead to the repositioning of CAR AF2. This unique “toggle bolt” mechanism, where the relative position of the “wings” (H11 and H12) can be made to vary with respect to each other, is a departure from the action of the raloxifene class of nuclear receptor antagonists and has important implications for the design of a novel class of nuclear receptor antagonists.
Experimental Procedures
Protein Expression and Purification
Residues 109–358 (accession # O35627) of mCAR LBD and a human SRC-1 peptide containing all three receptor interacting domains (RID 1–3) (Asp617—Asp769, accession # U59302) were subcloned into the pET-15b (Novagen) and the pACYAC184 vectors, respectively. The two proteins were coexpressed in E. coli BL21 (DE3) Gold cells (Novagen). The mCAR LBD/SRC-1 (RID 1–3) complex was first purified by Ni-NTA affinity (Qiagen). The protein complex was liberated from the N-terminal hexa-histidine tag with thrombin protease. The complex was subsequently purified by ion exchange on a POROS HQ anion exchange column (Perseptive Biosystems). The complex was incubated with androstenol to dissociate the CAR/SRC-1 complex, and CAR was isolated from this mixture by gel filtration on a Superdex-75 Highload 16/60 (Amersham Biosciences) chromatography column. The mCAR LBD/androstenol complex was concentrated to approximately 6.5 mg/mL and crystallized at 14°C.
Data Collection, Structure Determination, Refinement, and Structure Analysis
Data was collected on beamline DuPont-Northwestern-Dow Collaborative Access Team Sector 5ID-B utilizing a MarMosaic CCD225 detector at the Advanced Photon Source in Argonne, IL. Data extended to 2.9Å and was integrated and scaled with the XDS (Kabsch, 1993). The space group was C2221 with two mCAR LBD/androstenol molecules in the crystallographic asymmetric unit. The structure was determined by the Molecular Replacement Method with a molecular model of mCAR LBD inspired by the PXR LBD structure (Watkins et al., 2001) using AMoRe (CCP4, 1994). The two molecules were built separately with the graphics package TURBO-FRODO (Roussel and Cambillau, 1991) and refined with CNS (Brunger et al., 1998). Cavity volume was calculated with VOIDOO (Kleywegt and Jones, 1994). Model analysis was performed with PROCHECK (Laskowski et al., 1993a).
Cell Assays
Cytomegalovirus (CMV)-driven receptor pCMX-mCAR, internal control pCMV-galactosidase along with the reporter plasmids LXRE×3-TK-Luc or RE2×2-TK-Luc are described elsewhere (Dussault et al., 2002). CV-1 monkey kidney cells were grown in DMEM phenol red free medium supplemented with 10% charcoal-filtered fetal bovine serum, 100 μg/ml Streptomycin, and 100 U/ml penicillin. Cells were plated to 50%–80% confluence one day prior to transfection with DMEM phenol red free supplemented with 10% charcoal-filtered fetal bovine serum. Cells were transfected with lipofectamine (Novagen) according to the manufacturer’s protocol. Reporter constructs (300–700 ng per 105 cells), CMV-driven receptors (100–200 ng per 105 cells), and CMV-galactosidase internal control (100–200 ng per 105 cells) were added as indicated. After 2 hr incubation at 37°C/5% CO2, 5 μM androstenol was added to treated cells, and the appropriate amount of DMSO was added to untreated cells as a control. After addition of ligand, cells were incubated 40 hr at 37°C/5% CO2. Cells were harvested and assayed for luciferase. Data from luciferase assays were corrected for transfection efficiency to galactasidase expression. Mutant CAR constructs were generated with the QuikChange Site-Directed Mutagenesis Kit (Stratagene).
Acknowledgments
We thank Drs. David Brian, Chris Dealwis, Hong Guo, Michael Hodsdon, Elias Lolis, and Tim Sparer for assistance and advice. We also thank Ronald Johnson for computational resources.
Accession Numbers The coordinates for the CAR/androstenol complex have been deposited in the Protein Data Bank with the PDB ID: 1XNX.
References
- Baes M, Gulick T, Choi HS, Martinoli MG, Simha D, Moore DD. A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol. Cell. Biol. 1994;14:1544–1551. doi: 10.1128/mcb.14.3.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- Bourguet W, Andry V, Iltis C, Klaholz B, Potier N, Van Dorsselaer A, Chambon P, Gronemeyer H, Moras D. Hetero-dimeric complex of RAR and RXR nuclear receptor ligand-binding domains: purification, crystallization, and preliminary X-ray diffraction analysis. Protein Expr. Purif. 2000;19:284–288. doi: 10.1006/prep.2000.1248. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- CCP4 (Collaborative Computational Project, Number 4) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Choi HS, Chung M, Tzameli I, Simha D, Lee YK, Seol W, Moore DD. Differential transactivation by two isoforms of the orphan nuclear hormone receptor CAR. J. Biol. Chem. 1997;272:23565–23571. doi: 10.1074/jbc.272.38.23565. [DOI] [PubMed] [Google Scholar]
- Dussault I, Lin M, Hollister K, Fan M, Termini J, Sherman MA, Forman BM. A structural model of the constitutive androstane receptor defines novel interactions that mediate ligand-independent activity. Mol. Cell. Biol. 2002;22:5270–5280. doi: 10.1128/MCB.22.15.5270-5280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea PF, Mitschler A, Rochel N, Ruff M, Chambon P, Moras D. Crystal structure of the human RXRalpha ligand-binding domain bound to its natural ligand: 9-cis retinoic acid. EMBO J. 2000;19:2592–2601. doi: 10.1093/emboj/19.11.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman BM, Tzameli I, Choi HS, Chen J, Simha D, Seol W, Evans RM, Moore DD. Androstane metabolites bind to and deactivate the nuclear receptor CAR-beta. Nature. 1998;395:612–615. doi: 10.1038/26996. [DOI] [PubMed] [Google Scholar]
- Gampe RT, Jr., Montana VG, Lambert MH, Miller AB, Bledsoe RK, Milburn MV, Kliewer SA, Willson TM, Xu HE. Asymmetry in the PPARgamma/RXRalpha crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol. Cell. 2000;5:545–555. doi: 10.1016/s1097-2765(00)80448-7. [DOI] [PubMed] [Google Scholar]
- Gill RQ, Sterling RK. Acute liver failure. J. Clin. Gastroenterol. 2001;33:191–198. doi: 10.1097/00004836-200109000-00005. [DOI] [PubMed] [Google Scholar]
- Greschik H, Flaig R, Renaud JP, Moras D. Structural basis for the deactivation of the estrogen-related receptor gamma by diethylstilbestrol or 4-hydroxytamoxifen and determinants of selectivity. J. Biol. Chem. 2004;279:33639–33646. doi: 10.1074/jbc.M402195200. [DOI] [PubMed] [Google Scholar]
- Huang W, Zhang J, Chua SS, Qatanani M, Han Y, Granata R, Moore DD. Induction of bilirubin clearance by the constitutive androstane receptor (CAR) Proc. Natl. Acad. Sci. USA. 2003;100:4156–4161. doi: 10.1073/pnas.0630614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhang J, Moore DD. A traditional herbal medicine enhances bilirubin clearance by activating the nuclear receptor CAR. J. Clin. Invest. 2004a;113:137–143. doi: 10.1172/JCI200418385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhang J, Wei P, Schrader WT, Moore DD. Meclizine is an agonist ligand for mouse CAR and an inverse agonist for human CAR. Mol. Endocrinol. 2004b;18:2402–2408. doi: 10.1210/me.2004-0046. Published online July 22, 2004. 10.1210/me.2004-0046. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- Kawamoto T, Sueyoshi T, Zelko I, Moore R, Washburn K, Negishi M. Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene. Mol. Cell. Biol. 1999;19:6318–6322. doi: 10.1128/mcb.19.9.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. 1994;D50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Sueyoshi T, Inoue K, Moore R, Negishi M. Cytoplasmic accumulation of the nuclear receptor CAR by a tetratricopeptide repeat protein in HepG2 cells. Mol. Pharmacol. 2003;64:1069–1075. doi: 10.1124/mol.64.5.1069. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993a;26:283–291. [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993b;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Parks DJ, Moore LB, Collins JL, Goodwin B, Billin AN, Stoltz CA, Kliewer SA, Lambert MH, Willson TM, Moore JT. Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J. Biol. Chem. 2003;278:17277–17283. doi: 10.1074/jbc.M300138200. [DOI] [PubMed] [Google Scholar]
- Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr. Opin. Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- Nettles KW, Sun J, Radek JT, Sheng S, Rodriguez AL, Katzenellenbogen JA, Katzenellenbogen BS, Greene GL. Allosteric control of ligand selectivity between estrogen receptors alpha and beta: implications for other nuclear receptors. Mol. Cell. 2004;13:317–327. doi: 10.1016/s1097-2765(04)00054-1. [DOI] [PubMed] [Google Scholar]
- Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- Onate SA, Tsai SY, Tsai MJ, O’Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- Pike AC, Brzozowski AM, Hubbard RE, Bonn T, Thorsell AG, Engstrom O, Ljunggren J, Gustafsson JA, Carlquist M. Structure of the ligand-binding domain of oestrogen receptor beta in the presence of a partial agonist and a full antagonist. EMBO J. 1999;18:4608–4618. doi: 10.1093/emboj/18.17.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud JP, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- Rochel N, Wurtz JM, Mitschler A, Klaholz B, Moras D. The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol. Cell. 2000;5:173–179. doi: 10.1016/s1097-2765(00)80413-x. [DOI] [PubMed] [Google Scholar]
- Roussel A, Cambillau C. Turbo Frodo. Silicon Graphics; Mountain View, CA: 1991. [Google Scholar]
- Suino K, Peng L, Reynolds R, Li Y, Cha J-Y, Repa JJ, Kliewer SA, Xu HE. The nuclear xenobiotic receptor CAR: structural determinants of constitutive activation and hetero-dimerization. Mol. Cell. 2004;16(this issue):893–905. doi: 10.1016/j.molcel.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Svensson S, Ostberg T, Jacobsson M, Norstrom C, Stefansson K, Hallen D, Johansson IC, Zachrisson K, Ogg D, Jendeberg L. Crystal structure of the heterodimeric complex of LXRalpha and RXRbeta ligand-binding domains in a fully agonistic conformation. EMBO J. 2003;22:4625–4633. doi: 10.1093/emboj/cdg456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzameli I, Pissios P, Schuetz EG, Moore DD. The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol. Cell. Biol. 2000;20:2951–2958. doi: 10.1128/mcb.20.9.2951-2958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins RE, Wisely GB, Moore LB, Collins JL, Lambert MH, Williams SP, Willson TM, Kliewer SA, Redinbo MR. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science. 2001;292:2329–2333. doi: 10.1126/science.1060762. [DOI] [PubMed] [Google Scholar]
- Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature. 2000;407:920–923. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- Williams SP, Sigler PB. Atomic structure of progesterone complexed with its receptor. Nature. 1998;393:392–396. doi: 10.1038/30775. [DOI] [PubMed] [Google Scholar]
- Xu HE, Stanley TB, Montana VG, Lambert MH, Shearer BG, Cobb JE, McKee DD, Galardi CM, Plunket KD, Nolte RT, et al. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature. 2002;415:813–817. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- Xu RX, Lambert MH, Wisely BB, Warren EN, Weinert EE, Waitt GM, Williams JD, Collins JL, Moore LB, Willson TM, et al. A structural basis for constitutive activity in the human CAR/RXRalpha heterodimer. Mol. Cell. 2004;16(this issue):919–928. doi: 10.1016/j.molcel.2004.11.042. [DOI] [PubMed] [Google Scholar]
- Ye L, Li YL, Mellstrom K, Mellin C, Bladh LG, Koehler K, Garg N, Collazo A.M. Garcia, Litten C, Husman B, et al. Thyroid receptor ligands. 1. Agonist ligands selective for the thyroid receptor beta1. J. Med. Chem. 2003;46:1580–1588. doi: 10.1021/jm021080f. [DOI] [PubMed] [Google Scholar]
- Yoshinari K, Kobayashi K, Moore R, Kawamoto T, Negishi M. Identification of the nuclear receptor CAR:HSP90 complex in mouse liver and recruitment of protein phosphatase 2A in response to phenobarbital. FEBS Lett. 2003;548:17–20. doi: 10.1016/s0014-5793(03)00720-8. [DOI] [PubMed] [Google Scholar]
- Zhang J, Huang W, Chua SS, Wei P, Moore DD. Modulation of acetaminophen-induced hepatotoxicity by the xenobiotic receptor CAR. Science. 2002;298:422–424. doi: 10.1126/science.1073502. [DOI] [PubMed] [Google Scholar]