Abstract
Disrupted-In-Schizophrenia-1 (DISC1) is one of major susceptibility factors for a wide range of mental illnesses, including schizophrenia, bipolar disorder, major depression, and autism spectrum conditions. DISC1 is located in several subcellular domains, such as the centrosome and the nucleus, and interacts with various proteins, including NudE-like (NUDEL/NDEL1) and activating transcription factor 4 (ATF4)/CREB2. Nevertheless, a role for DISC1 in vivo remains to be elucidated. Therefore, we have generated a Drosophila model for examining normal functions of DISC1 in living organisms. DISC1 transgenic flies with preferential accumulation of exogenous human DISC1 in the nucleus display disturbance in sleep homeostasis, which has been reportedly associated with CREB signaling/CRE-mediated gene transcription. Thus, in mammalian cells, we characterized nuclear DISC1, and identified a subset of nuclear DISC1 that co-localizes with the promyelocytic leukemia (PML) bodies, a nuclear compartment for gene transcription. Furthermore, we identified three functional cis-elements that regulate the nuclear localization of DISC1. We also report that DISC1 interacts with ATF4/CREB2 and a co-repressor N-CoR, modulating CRE-mediated gene transcription.
Keywords: sleep, CREB, ATF4, schizophrenia, depression, mood disorder
Introduction
Disrupted-In-Schizophrenia-1 (DISC1) is a promising genetic factor for a wide range of mental illnesses, including schizophrenia, bipolar disorder, major depression, and autism spectrum conditions (1-5). Sleep disturbances, including deficits in sleep homeostasis and circadian rhythms, commonly accompany various types of psychiatric conditions (6, 7).
DISC1 protein has multiple functions and is located in several distinct domains in cells, including the centrosome, the postsynaptic density, the mitochondria, and the nucleus (8-11). DISC1 has specific protein interactors, corresponding to each subcellular domain, such as NudE-like (NUDEL/NDEL1) at the centrosome and activating transcription factor 4 (ATF4)/CREB2 in the nucleus (11, 12). We previously reported that a form of DISC1 enriched in the nucleus was increased in patients with sporadic schizophrenia and major depression as well as substance and alcohol abuse, suggesting that nuclear DISC1 may play a role in the pathophysiology of mental disorders (13). Understanding the mechanisms underlying DISC1 function in brains in vivo, however, remains to be elucidated.
The fruit fly, Drosophila melanogaster, has been used as an excellent invertebrate model to explore molecular functions in brains in vivo (14-17). Generation of genetically-engineered flies is much easier than generation of knockout or transgenic mice. Thus, transgenic flies expressing human disease genes have been generated to examine functions of such disease factors, with the hope that these flies can be utilized as disease models (14-19). Furthermore, a large number of genes for the biological processes or even specific factors that either enhance or suppress the phenotypes caused by expression of human disease genes can be screened. In many cases, such factors have been found to interact with disease proteins and form pathological pathways. Thus, even if disease genes by themselves are not conserved between humans and flies, the disease gene-associated phenotypes in transgenic flies have been utilized to explore unknown biological processes that are conserved between flies and humans: one successful example is shown by transgenic flies expressing α-synuclein (18, 19). Although there is no orthologue of α-synuclein in flies, the fly model exhibited adult-onset loss of specific sets of dopaminergic neurons, similar to human Parkinson's disease, and helped to identify factors that can regulate the process, including Hsp 70 (19), which was later confirmed as a modified factor or genetic risk factor for Parkinson's disease (20).
In the present study, we have generated and initially characterized a fly model expressing human DISC1. The transgenic flies with a punctate accumulation of exogenous human DISC1 in the nucleus display disturbance in sleep homeostasis, which has been reportedly associated with CREB signaling or CRE-mediated gene transcription (21). Such punctate localization of DISC1 in the nucleus is also observed in mammalian cells with co-localization with promyelocytic leukemia (PML) bodies, a nuclear compartment for gene transcription. We also identified three functional cis-elements that regulate nuclear localization of DISC1. Reporter analyses, combined with interaction assays, support the notion that DISC1 modulates CRE-mediated gene transcription by interacting with ATF4/CREB2 and recruiting a co-repressor N-CoR to the transcriptional machinery.
Materials and Methods
Constructs and antibodies
Various DISC1 constructs were prepared in the pRK5 vector containing an N-terminal HA-tag. Mutations of DISC1 were introduced by PCR-based mutagenesis (22). Constructs for fusion proteins of the C-terminus of enhanced green fluorescent protein (eGFP) with putative nuclear localization signal (NLS) and nuclear export signal (NES) were generated by subcloning with peGFP-C1 (Clontech, Mountain View, CA). Rabbit polyclonal antibody against human DISC1 was prepared as described (23). The following mouse monoclonal antibodies were used: anti-coilin antibody (Sigma, St. Louis, MO); anti-PML body antibodies (Santa Cruz Biotechnology, Santa Cruz, CA); anti-SC35 antibody (BD Pharmingen, San Diego, CA); and antibodies against HA-tag (Covance, Berkeley, CA). The following rabbit polyclonal antibodies were used: anti-CREB2 antibody (Santa Cruz); anti-mouse DISC1 antibody (Zymed, South San Francisco, CA); anti-N-CoR antibody (Upstate, Lake Placid, NY); and anti-FLAG antibody (Sigma). Mouse anti-Fasciclin II (FAS II) (mAB1D4) and anti-Dachshund (DAC) (mABdac2-3) antibodies were from Developmental Studies Hybridoma Bank (University of Iowa, IA).
Fly stocks
Canton S (CS) and a white (w) stock outcrossed with Canton S at ten times (CS10) were used as the standard. OK107 is a Gal4 enhancer trap line for the upstream regulatory modules of the eyeless/Pax6 gene, which is expressed in the majority of the MB neurons (24). Flies carrying UAS-DISC1 transgene with an HA-tag were generated by P-element-mediated transformation and outcrossed to w(CS10) at least five times before behavioral assays. Stocks were kept at 25 °C in 12 h light and 12 h dark conditions and were fed a standard diet consisting of 5.5 g agar, 40 g yeast extract, 90 g cornmeal, and 100 g glucose per liter.
Sleep assays
Flies 3-6 days old were placed individually in 65 mm × 5 mm glass tubes plugged at one end with 5% sucrose and 2% agarose gel with or without 250 μM RU486. Significant induction of Gal4 activity was confirmed with UAS-mCD8::GFP and anti-DISC1 staining. No significant alteration of sleep amount was detected for the standard stocks at 250 μM RU486. Fly activity was monitored with the Drosophila Activity Monitor System (Trikinetics, Waltham MA) at 25 °C in 12 h light and 12 h dark conditions as described (25). The fly was confined in 5 mm length by moving the cotton plug into the tube. Sleep was measured as a resting state longer than 5 min. To ensure protein induction and fly adaptation, the fly was placed in the monitor tube 48 h before starting data collection.
Walk assay
Flies were briefly sedated with CO2 and individually placed in 35-mm diameter plastic plates. After 1 h for recovery, walk behavior was monitored with a digital video camera for 2 min at 25 °C under normal light condition. To avoid fluctuation by circadian changes, all the recordings were done at the end of day when flies exhibit typical peak activity. Walk behavior was analyzed based on the digital images of the walk tracks of the flies, and various parameters were analyzed using the Dynamic Image Analysis System (DIAS 3.0, Soll Technologies, IA).
Immunofluorescent staining of fly tissues and mammalian cells
Drosophila brains were dissected and processed for staining as described (24, 26). Mouse anti-DAC and anti-FAS II were used at dilution 1:20 and 1:5, respectively. TO-PRO3 (Invitrogen, Carlsbad, CA) was used at dilution 1:200. Confocal images were taken with a Zeiss LSM510. HeLa cells were cultured in Dulbecco's modified medium as recommended by the manufacturer. About 20 h after transfection by use of PolyFect (Qiagen, Valencia, CA), cells were fixed with 4% paraformaldehyde (PFA) at 37 °C for 30 min. Coverslips were mounted with PermaFluor (Thermo electon corporation). Primary neuron cultures were prepared as described previously (11). Experimental details for cell staining were as described (27). Rabbit anti-mouse DISC1 antibody (Zymed) was used at 1: 50 for primary neurons. Cell staining was examined with a confocal microscope. Each experiment was repeated at least three times.
Cellular assays for DISC1 nuclear targeting
Cellular distribution of the C-terminus of eGFP containing putative NLS or NES was monitored by GFP fluorescence under a confocal microscope 12 to 16 h after transfection. HeLa cells with exogenously expressed HA-tagged DISC1 (wild-type, and mutants with key amino acid substitutions in the critical domains) were also examined by immunofluorescent cell staining with an anti-HA antibody, as described above. Over 100 cells were examined under each condition with three independent transfections.
Transcriptional assays
Various expression constructs were co-transfected with either Gal4 or CRE reporter construct into HeLa or IMR32 cells. IMR32 cells were cultured in the MEM medium with 10% FBS. One to 2 days after transfection, cells were suspended in the buffer provided in the kit for measurement of the luciferase activity (Luciferase Assay System (Promega, Madison, WI)) and normalized to β-galactosidase activity or total protein amount. Data are given as relative activities to the activity with Gal4 only, or that with ATF4/CREB2 only.
In vitro binding assay
Glutathione-S-transferase (GST) or GST-DISC1 protein was co-incubated with His-tagged ATF4/CREB2 at 4 °C for 2 h in 1 ml of binding buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.2 mg/ml BSA). Then, 20 μl of 50% glutathione-Sepharose beads were added to the mixture and incubated at 4 °C for 1 h. The beads were washed four times with 1 ml of washing buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1 mg/ml BSA, 0.1% CHAPS), separated by SDS-PAGE, and analyzed by Western blotting.
Immunoprecipitation
HeLa cells expressing HA-tagged DISC1 (wild-type or deletion mutant) together with ATF4/CREB2, or N-CoR, or fly CRC were lysed in RIPA buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate) containing a protease inhibitor mixture (Roche Applied Sciences, Indianapolis, IN), and the soluble proteins were subjected to immunoprecipitation by using either an anti-ATF4/CREB2 or an anti-N-CoR antibody, as described previously (13).
Statistical analysis
All data were analyzed with Student's t-test and Mann-Whitney U-test. ** P <0.01.
Results
Generation of transgenic flies expressing human DISC1
Like the successful cases of α-synuclein transgenic flies, in which human transgenes with no fly orthologues are expressed (18, 19), there is no DISC1 orthologue in flies (28). Nevertheless, many DISC1 interactors are conserved between humans and flies, including ATF4/CREB2 and NUDEL/NDEL1. Therefore, we sought to determine whether generation of transgenic flies expressing human DISC1 would be useful in exploring the in vivo function of DISC1 in association with its protein interactors.
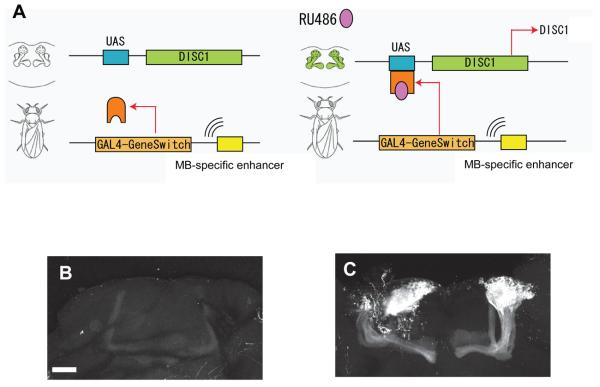
To express DISC1 in Drosophila brain, we utilized the GAL4-based GeneSwitch system, which allows inducible transgene expression in a temporally- and spatially-regulated manner by the addition of RU486 (29). As shown in Supplementary Figure 1, DISC1 transgenic flies in the conventional expression system display no robust morphological changes in the mushroom bodies (MBs) in adults. Nonetheless, in order to avoid any undetectable, but functionally critical, developmental impact due to constitutive expression of DISC1, we used GeneSwitch rather than conventional GAL4/UAS system to test functions of DISC1 in adult neurons. We induced DISC1 expression in the MBs that play pivotal roles in cognition, associative learning, attention, and sleep control in flies (Figure 1A) (30-33). Selective expression of a transgene in MB in this expression system was confirmed by the confocal assessment of adult fly brains expressing a reporter protein (Figure 1B, C). In the present study, we first generated two types of transgenic flies, those expressing full-length human DISC1 (DISC1-FL) and those expressing a C-terminal truncated (amino acids 1-597) human DISC1 [DISC1(1-597)].
Figure 1.
Expression of DISC1 in transgenic flies in a tissue- and temporal-specific manner. (A) The GAL4-GeneSwitch chimeric protein binds to a UAS-DNA sequence in the presence of RU486, which in turn induces gene transcription downstream of the UAS sequences. Inducible expression of DISC1 selectively in the mushroom bodies (MBs) is achieved by using an MB-GeneSwitch construct, in which an MB-specific enhancer is linked to the coding region of GAL4-GeneSwitch. (B, C) Confocal images of adult fly brains expressing the mCD8::GFP protein under control of the MB-GeneSwitch. In the presence of RU486 (B, 0 μM; C, 250 μM), selective expression of the protein is observed in MBs.
Behavioral alterations in transgenic flies expressing full-length DISC1 enriched in the nucleus
To address phenotypic changes in DISC1 transgenic flies, we measured their activities by using the Drosophila Activity Monitor System (DAMS), which counts the number of infrared beam crossings by an individual fly placed in a small glass tube (Figure 2A). Flies exhibit a typical circadian activity pattern with more frequent and longer rest at night than during daytime. Quantitative characteristics measured by this system are known to reflect their arousal and sleep homeostasis (25, 31, 34-38). When DISC1-FL was induced in MBs, male, but not female, flies displayed reduced arousal state as evidenced by increased sleep time per day (Figure 2B, C). This sex-specific sleep change is observed only when DISC1-FL was induced, but not DISC1(1-597) or control w(CS10) flies (Figure 2B, C). In DISC1-FL flies, the average length of each sleep (sleep bout), but not its frequency during the day (number of sleep bouts), was increased (Figure 2D). Nevertheless, the circadian cycle was not markedly changed in all types of flies (Figure 2B, C). This sleep alteration is not due to the genetic background of the transgenic flies, because there was no alteration in sleep status in heterozygous control flies carrying UAS-DISC1 or the GeneSwitch driver alone (Supplementary Figure 2). Addition of RU486 did not affect sleep amount in control flies (w(CS10)) that have no transgene (Figure 2B). The increased sleep amount was not a reflection of motor disturbance, because no change was observed in the walk speed upon induction of DISC1-FL (Figure 2E).
Figure 2.
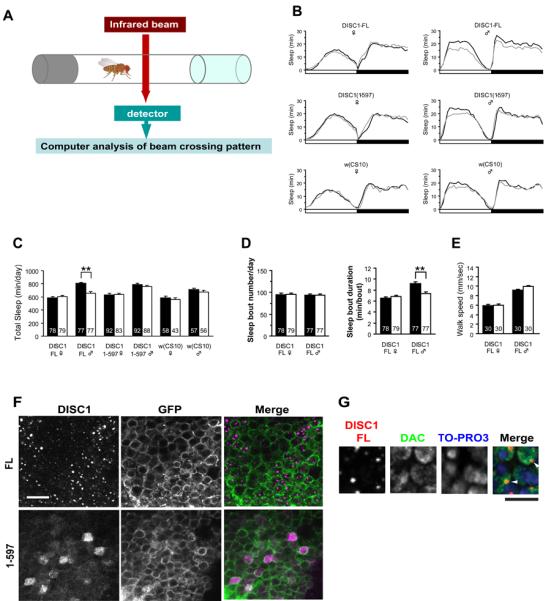
(A) Schematic illustration of activity monitoring system of flies. (B) Daily activity patterns of flies expressing DISC1-FL or DISC1(1-597) under control of the MB-GeneSwitch. Activities were measured with (+RU486, black line) or without RU486 (−RU486, gray line). Sleep was defined as a continuous rest state longer than 5 min. The results represent the average of three independent experiments. Of note, the normal circadian activity patterns are not altered by DISC1 expression. (C) Total amount of sleep per day (24 h). Expression of DISC1-FL, but not DISC1(1-597) leads to significantly longer sleep in males. Black bar, +RU486. White bar, −RU486. Total number of flies used in each group is shown in the bar. (D) Expression of DISC1-FL increases the duration, but not the number, of sleep bouts in males. Total number of flies used in each group is shown in the bar. (E) Walking speed of DISC1-FL flies assayed by the Dynamic Image Analysis System. Induction of DSC1-FL did not affect the walk speed in both sexes. Black bar, +RU486. White bar, −RU486. (F) Localization of DICS1-FL and DISC1(1-597) in MB neurons. DISC1-FL exhibited punctate nuclear localization, whereas DISC1(1-597) localized diffusely both in the nuclei and cytoplasm. DISC1 and mCD8::GFP expression were driven by OK107, a MB-Gal4 driver. Scale bar, 10 μm. (G) Co-localization of DISC1 and DAC, a nuclear transcription factor, in MB neurons. Red, DISC1; green, DAC, blue: TO-PRO3. Arrowheads indicate co-localized DISC1 and DAC. DISC1 expression was driven by OK107. Scale bar, 5 μm.
To address a molecular clue that accounts for this sleep-associated phenotypic change, we examined cellular distribution of induced DISC1 protein (Figure 2F). Interestingly, DISC1-FL displayed unique punctate staining selectively in the nucleus, whereas DISC1(1-597) did not. The punctate DISC1-FL was co-localized with a transcription factor Dachshund (DAC) in the nucleus (Figure 2G). Expression levels of DISC1(1-597) was comparable or even higher than that of DISC1-FL, but the protein distribution was diffuse in both the cytoplasm and the nucleus of the cell bodies. There was no sex difference in nuclear punctate staining of DISC1-FL (Supplementary Figure 3A). These staining patterns of DISC1-FL and DISC1(1-597) were observed also in large neurosecretary cells located at the dorsomedial region of the fly brain (Supplementary Figure 3B). No immunoreactivity of DISC1-FL and DISC1(1-597) was observed in the dendritic structures (calyx) and the extended axonal structures (lobes) in MBs (Supplementary Figure 3C). Although localization of a pool of DISC1 in the nucleus has been reported in human brains, subnuclear distribution has not yet been characterized (9, 13).
Nuclear DISC1 in mammalian cultured cells
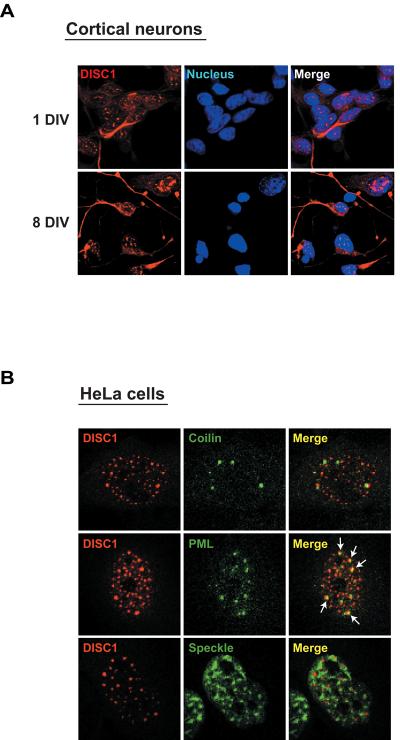
To characterize the nuclear distribution of DISC1 in greater detail, we examined the subcellular localization of DISC1 in primary cortical neurons by immunofluorescent staining. DISC1 immunoreactivity was observed in both the nucleus and the cytoplasm in both immature and mature neurons (Figure 3A). In the nucleus of primary neurons, endogenous DISC1 was present in a punctate pattern, similar to that observed in fly tissues. For further examination of the subnuclear localization of DISC1, we used HeLa cells as a model by expressing DISC1 ectopically. When cells were transfected by moderate dose of DNA (0.1-0.5 μg), DISC1 displayed predominantly a punctate nuclear distribution (Supplementary Figure 4), similar to the expression pattern observed in fly brain as well as that in primary cortical neurons (Figures 2F and 3A). We investigated the subnuclear localization of DISC1, relative to nuclear speckles (the domains for pre-mRNA splicing), coiled bodies (the domains for snRNP biogenesis and trafficking), and promyelocytic leukemia (PML) bodies (the nuclear bodies for gene transcription). DISC1 was found to be co-localized only with PML bodies (Figure 3B).
Figure 3.
Nuclear DISC1 in mammalian cells. (A) Immunofluorescent staining of primary neurons indicates that DISC1 occurs not only in the cytoplasm but also in the nucleus in a dotted pattern. (B) Exogenous expression of DISC1 at a low dose in HeLa cells depicts the nuclear pool of DISC1. A pool of DISC1 is co-localized with PML (white arrows), the main component of the nuclear body. Red, exogenous DISC1 selectively detected by a HA-antibody; green, each nuclear marker (coilin for coiled bodies; PML for PML/nuclear bodies; SC35 for speckle).
Three cis-elements that regulate nuclear distribution of DISC1
Because DISC1 exists in both the nucleus and the cytoplasm, we investigated the cis-elements that may regulate the nuclear localization of DISC1. As previously reported (39, 40), DISC1 has two putative nuclear localization signals (NLSs) (Figure 4A). First, we fused the corresponding NLS peptides with enhanced green fluorescent protein (eGFP) and examined the localization of the fused proteins. As shown in Supplementary Figure 5A, the eGFP-NLS1 fusion protein had a punctate nuclear expression pattern, but the eGFP-NLS2 or the eGFP-mNLS1 that contains a mutated NLS1 did not, suggesting that NLS1 is a functional element. To test the role of NLS1 in the context of full-length DISC1, we examined the subcellular distribution of DISC1 with deletion of NLS1 [DISC1(46-854)] or with a point mutation in the NLS1 sequences (Figure 4A). These mutants displayed cytoplasmic/perinuclear, but not nuclear, localization. In flies, expression of DISC1-mNLS1 resulted in perinuclear localization in large neurosecretary cells and MB neurons (Supplementary Figure 3B, D). Taken together, these results indicate that NLS1 is required for the nuclear localization of DISC1 in both mammalian cells and flies.
Figure 4.
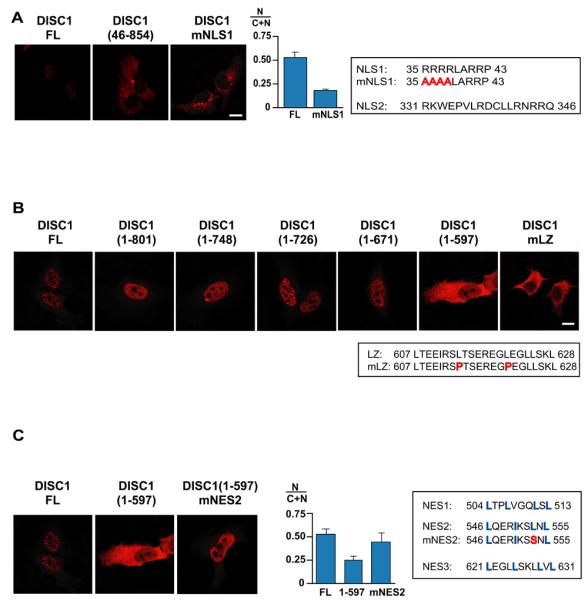
Functional cis-elements for nuclear distribution of DISC1. (A) N-terminal deletion of DISC1 (amino acids 1-45) abolishes NLS1, leading to the distribution of DISC1 mainly in the cytoplasm. Mutant DISC1 with amino acid substitutions in NLS1 (DISC1-mNLS1) occurs outside the nucleus, with enrichment at the perinuclear regions. (B) Sequential C-terminal deletion of DISC1 indicates that there is a critical domain between 598 and 671 amino acids that determines the nuclear localization of DISC1. DISC1-FL, DISC1(1-801), DISC1(1-748), DISC1(1-726), and DISC1(1-671) occur in the nucleus, but DISC1(1-597) does not. Introduction of mutations in the LZ domain (DISC1-mLZ) alters the distribution of DISC1 from the nucleus to the cytoplasm. (C) Introduction of mutations in NES2 leads to distribution of DISC(1-597) from cytoplasmic to a nuclear localization, suggesting that NES2 is functional.
Systematic deletion of DISC1 from the C-terminus indicated that another cis-element on DISC1 was also required for nuclear localization. Punctate nuclear distribution was still observed in DISC1(1-671), but not in DISC1(1-597), suggesting that an element between residues 598 and 671 plays a necessary role in the proper localization of DISC1 (Figure 4B). Inspection of the sequences in this region revealed a leucine zipper domain (LZ) at around amino acid 610. Mutation of two leucine residues in this region altered the distribution of DISC1 from the punctate nuclear pattern to the cytoplasmic localization (Figure 4B). Thus, punctate nuclear expression of DISC1 required at least two cis-elements: NLS1 and LZ. Of note, this LZ element includes a polymorphism at amino acid 607 that is associated with schizoaffective disorder (41).
Although DISC1(1-597) contains one functional NLS (NLS1), a large amount of this protein localized in the cytoplasm in a diffuse pattern (Figure 4C) as did in the Drosophila neurons (Figure 2F). This suggests a possible motif in 1-597 of DISC1 that contributes to export of the protein out of the nucleus. Treatment of HeLa cells expressing DISC1(1-597) with Leptomycin B, a potent inhibitor of CRM1/exportin-mediated export pathway (42), altered the localization of the protein to the nucleus, suggesting the existence of functional nuclear export signal(s) (NES) in the N-terminal region(1-597) (Supplementary Figure 5B). There are three sequences that fit the loose consensus for NES (Figure 4C). Expression of NES2 together with eGFP resulted in the localization of the GFP signal exclusively in the cytoplasm, in strong contrast to the ubiquitous GFP staining by eGFP-NES1, or eGFP-NES3, or eGFP alone (Supplementary Figure 5C). DISC1(1-597) with a mutation in NES2 (DISC1(1-597)-mNES2) exhibited only diffuse nuclear localization, in contrast to DISC1(1-597), suggesting that NES2 has a role to export DISC1 out of the nucleus (Figure 4C).
In all these experiments, we monitored the expression levels of the wild-type and mutant proteins by immunoblot and found comparable levels of expression for all proteins (data not shown), excluding the possibility that the changes in their subcellular distributions were due to the changes in their expression levels.
DISC1 binds to ATF4/CREB2 and N-CoR, and modulates CRE-mediated gene transcription
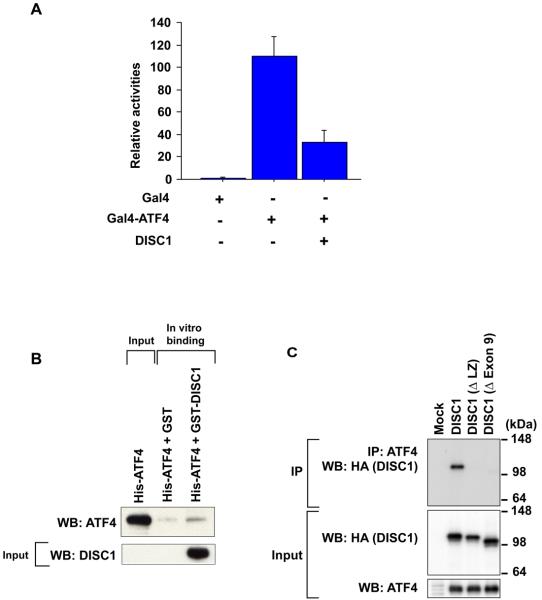
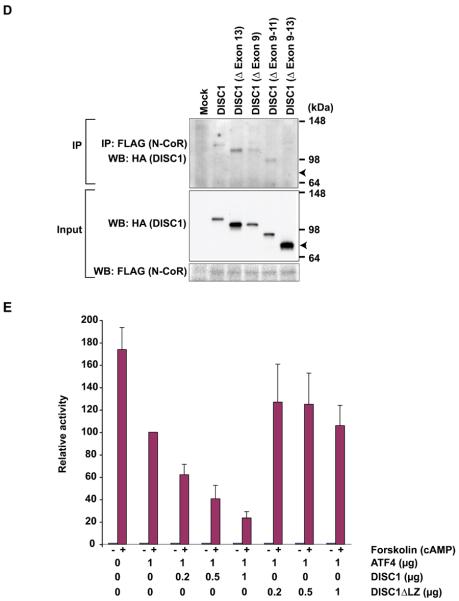
DISC1 co-localizes with the PML bodies where various gene transcriptions are controlled, and has been known to bind to a transcription factor ATF4/CREB2, which is one of the ATF/CREB family transcription factors that belong to a group of basic region-leucine zipper (bZIP) proteins originally defined by their ability to bind the consensus ATF/CRE site “TGACGTCA” (43). ATF/CREB2 mediates/modulates gene transcription, and has been implicated in regulating brain functions, including a form of long-term memory (44-47). Thus, we questioned whether DISC1 could modulate ATF4/CREB2-mediated gene transcription. We initially used a Gal4-based transcriptional assay. A reporter construct containing Gal4-binding DNA promoter sequences upstream of a luciferase gene was co-transfected with an expression construct for a fusion protein of the Gal4 DNA binding domain (Gal4-DBD) and ATF4/CREB2. Expression of Gal4-ATF4/CREB induced robust transcriptional activation as indicated by the reporter assay (Figure 5A). Additional expression of DISC1 with Gal4-ATF4/CREB2 repressed the luciferase signal, suggesting that DISC1 may recruit transcriptional repressor(s) to ATF4-containing transcriptional machinery. We further characterized the interaction of DISC1 and ATF4/CREB2, and demonstrated their direct protein interaction by in vitro binding assays with purified recombinant GST-DISC1 and His-ATF4/CREB2 (Figure 5B). Cellular interaction of human DISC1 and CRC (cryptocephal), the fly orthologue of human ATF4/CREB2, was demonstrated by co-immunoprecipitation (48) (Supplementary Figure 6). Co-immunoprecipitation with several DISC1 deletion mutants indicated that the LZ domain in exon 9 is the minimum domain of DISC1 required for binding to ATF4/CREB2 (Figure 5C). Consistent with our luciferase reporter assay, DISC1 binds with a transcriptional repressor N-CoR as indicated by co-immunoprecipitation (Figure 5D). The interaction was maintained even after the deletion of exon 9, or exons 9-11, or exon 13, but was abolished by the entire deletion from exon 9 to exon 13. These results suggest that the domain coded by exon 12 may have an important role in DISC1-N-CoR interaction. There was no overlap in the binding domains of DISC1 for ATF4/CREB2 (the coding domain in exon 9) and N-CoR, suggesting that DISC1 may interact with both ATF4/CREB2 and N-CoR simultaneously. These results suggest that, by recruiting N-CoR to an ATF4/CREB2-containing complex as a common scaffold, DISC1 might modulate gene transcription regulated by ATF4/CREB2.
Figure 5.
DISC1 binds to ATF4/CREB2 and N-CoR, and modulates CRE-mediated gene transcription. (A) Exogenous expression of DISC1 suppresses Gal4-ATF4/CREB2-mediated transcription. (B) Interaction of DISC1 and ATF4/CREB2 demonstrated by in vitro binding experiments with purified recombinant His-ATF4/CREB2 and GST-DISC1 proteins. (C) Requirement of the LZ domain of DISC1 for protein interaction of DISC1 and ATF4/CREB2 as demonstrated by co-immunoprecipitation of ATF4/CREB2 and several types of DISC1 deletion mutants with an HA-tag in HeLa cells. An anti-ATF4/CREB2 antibody was used for immunoprecipitation, and an anti-HA antibody for detection. (D) Importance of exon 12 of DISC1 for interaction of DISC1 and N-CoR is indicated by co-immunoprecipitation. An anti-N-CoR antibody is used for precipitation, and an anti-HA antibody for detection of immunoprecipitates by Western blotting. (E) Exogenous expression of DISC1, but not mutant DISC1 with deletion of LZ, augments ATF4/CREB2-mediated repression of CRE-dependent gene transcription.
ATF4/CREB2 is known to repress CRE-dependent gene transcription when the cAMP-protein kinase A (PKA) signaling pathway is activated (49). To test whether DISC1 modulates CRE-mediated gene transcription, we used a CRE-driven luciferase reporter in the absence or presence of forskolin, an agent that increases intracellular cAMP levels and activates the PKA pathway. Consistent with the literature (49), forskolin treatment greatly increased the reporter activity driven by the CRE sites, and ectopic expression of ATF4/CREB2 repressed the forskolin-induced signal (Figure 5E). Of interest, co-expression of DISC1 together with ATF4/CREB2 further decreased the reporter activity. Selective deletion of LZ in DISC1, the domain responsible for binding to ATF4/CREB2, abolished the effects, suggesting that interaction between ATF4/CREB2 and DISC1 is required for the augmented suppression by DISC1. Of note, although nuclear levels of ATF4/CREB2 are reportedly increased upon the activation of cAMP-PKA signaling (50), we did not observe changes in distribution and levels of nuclear DISC1 as a result of the addition of forskolin (data not shown).
Discussion
There are two major findings in this report. First, we generated transgenic flies expressing human DISC1. In the flies, human full-length DISC1 is expressed in a punctate pattern in the nucleus. DISC1 transgenic flies in adults display alterations in sleep homeostasis, suggesting a role for DISC1 in sleep regulation in vivo. In flies, CREB signaling or CRE-mediated gene transcription has been reported to play an essential role in sleep homeostasis (21). Second, we characterized nuclear DISC1 in mammalian cells. Consistent with the nuclear punctate pattern in flies, a pool of DISC1 was localized in the nucleus in a punctate pattern in primary neuron cultures and HeLa cells. DISC1 co-localized with PML, a main component of the nuclear body, a nuclear compartment for gene transcription. DISC1 interacted with ATF4/CREB2 and N-CoR in distinct binding domains. Consequently, DISC1 modulates CRE-mediated gene transcription in cell models.
Sleep is defined by periods of quiescence that are associated with reduced response to the external environment (51, 52). Sleep is tightly regulated, and is under the control of both a circadian clock and homeostatic mechanisms (51, 52). For the past decade, molecular mechanisms underlying the circadian clock have been substantially clarified (53), but mechanisms underlying sleep homeostasis remain to be elucidated. Fruit flies (Drosophila melanogaster) are now being used as a powerful tool to elucidate regulatory mechanisms of sleep not only for circadian rhythms but also for sleep homeostasis (25, 34). Pharmacological agents, such as caffeine and antihistamines, influence sleep in flies. A recent study has further demonstrated that methamphetamine, a psychostimulant, could suppress sleep and promote wakefulness in flies, whereas inhibitors of dopamine synthesis exhibited the opposite effects (54). Prolonged rest in flies is associated with higher arousal threshold and is homeostatically regulated independently of the circadian clock. Moreover, cAMP-CREB signaling, which leads to CRE-mediated gene transcription, has been reported to play a role in sleep homeostasis in flies (21).
In the present study, in male flies we observed that exogenous expression of full-length DISC1, but not a C-terminal truncated DISC1 [DISC1(1-597)], altered sleep time, but not the frequency of sleep bouts, or the circadian rhythm. Because full-length DISC1, but not the C-terminal truncated DISC1 [DISC1(1-597)], is targeted to the punctate nuclear bodies, this specific targeting may be associated with the phenotypic difference between these flies. Given the conservation of the fundamental mechanisms of sleep between flies and mammals, modulation of conserved endogenous nuclear factors, such as ATF/CREB family proteins, may possibly contribute to the phenotypes of flies. Two approaches will be of considerable interest in the future: 1) systematic assays with flies with various types of DISC1 containing sequential deletions; 2) screening of genetic interactors that normalize or enhance the DISC1–induced sleep alteration.
To address molecular mechanisms that target DISC1 in the punctate nuclear structures, we identified three cis-acting elements that regulate the nuclear localization of DISC (NLS1, NES2, and LZ). Mutation of either NLS1 or LZ leads to non-nuclear DISC1 staining in cells. Conceivably, each element plays a key role in nuclear or non-nuclear distribution of DISC1 by keeping subtle balances among these elements. Although regulation of DISC1 localization has been studied previously by domain mapping (8, 55), our report is the first to define the cis-elements. Porteous and colleagues (8) reported that a large deletion of the C-terminal region (598-854 amino acids), which contains the LZ identified in this study, dramatically augmented the cytoplasmic localization of DISC1 and that further deletion of the middle portion (359-597 amino acids), which contains the NES2, augmented punctate DISC1 distribution in the nucleus. Brandon and colleagues (55) used GFP-tagged DISC1 constructs at a high dose (20 μg) and reported the importance of a central coiled-coil region in regulating the subcellular distribution of DISC1, especially its targeting to the mitochondria. This region is independent of the cis-domains for the nuclear targeting (NLS1, NES2, and LZ) reported in the study. Fly models expressing mutant DISC1 selectively deficient in these cis-domains will pinpoint a nuclear role for DISC1 in sleep control in vivo. One of such models, the fly expressing mNLS1, displayed non-nuclear DISC1 staining, supporting the notion that the mutant flies generated on the basis of data from mammalian cells will be potentially useful in future studies.
Our report suggests that DISC1 modulates CRE-mediated gene transcription by interacting with ATF4/CREB2. Interaction of DISC1 with N-CoR may play a key role in this modulation, because N-CoR is responsible for recruiting histone deacetylase to targeted transcriptional machinery (56). Recent studies have suggested a role for the cascades involving histone acetyltransferase and deacetylase in psychiatric conditions, including cocaine-induced plasticity, depression, and schizophrenia (57-59). Because nuclear DISC1 is altered in brains from subjects with psychosis as well as those with substance or alcohol abuse (13), disturbance of DISC1 with respect to recruitment of the repressor complex to transcriptional machinery, such as that comprising CREB and ATF4/CREB2, may play an important role in a wide range of psychiatric disorders. Considering that sleep disturbance exists in almost all psychiatric conditions, a role for DISC1 in sleep homeostasis, at least as demonstrated in fly models would be of particular interest in mouse models in future studies (60-62). It is also intriguing to examine genetic variations that can potentially affect interaction of DISC1 with ATF4/CREB2 or N-CoR, such as non-synonymous polymorphism Leu607Phe (41), in association with sleep characteristics in human subjects, including patients with various psychiatric diseases.
Supplementary Material
Acknowledgements
We thank Dr. Pamela Talalay for critical reading of the manuscript and Ms. Y. Lema for preparing the manuscript. We thank Dr. Ron Davis for providing us with MB-GeneSwitch flies. This work was supported by U.S. Public Heath Service Grant MH-069853 (AS), grants from Stanley (AS), S-R (AS), and NARSAD (AS, NS); KD064938 (TH); Grants-in-Aid for Scientific Research from MEXT and TARA (KFT); and Establishment of Consolidated Research Institute for Advanced Science and Medical Care from MEXT (NS).
References
- 1.Sawamura N, Sawa A. Disrupted-in-schizophrenia-1 (DISC1): a key susceptibility factor for major mental illnesses. Ann N Y Acad Sci. 2006;1086:126–133. doi: 10.1196/annals.1377.018. [DOI] [PubMed] [Google Scholar]
- 2.Kilpinen H, Ylisaukko-Oja T, Hennah W, Palo OM, Varilo T, Vanhala R, et al. Association of DISC1 with autism and Asperger syndrome. Mol Psychiatry. 2008;13:187–196. doi: 10.1038/sj.mp.4002031. [DOI] [PubMed] [Google Scholar]
- 3.Ishizuka K, Paek M, Kamiya A, Sawa A. A review of Disrupted-In-Schizophrenia-1 (DISC1): neurodevelopment, cognition, and mental conditions. Biol Psychiatry. 2006;59:1189–1197. doi: 10.1016/j.biopsych.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 4.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. image 45. [DOI] [PubMed] [Google Scholar]
- 5.Porteous DJ, Millar JK. Disrupted in schizophrenia 1: building brains and memories. Trends Mol Med. 2006;12:255–261. doi: 10.1016/j.molmed.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 6.Monti JM, Monti D. Sleep disturbance in schizophrenia. Int Rev Psychiatry. 2005;17:247–253. doi: 10.1080/09540260500104516. [DOI] [PubMed] [Google Scholar]
- 7.Costa e Silva JA. Sleep disorders in psychiatry. Metabolism. 2006;55:S40–44. doi: 10.1016/j.metabol.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 8.Millar JK, James R, Christie S, Porteous DJ. Disrupted in schizophrenia 1 (DISC1): subcellular targeting and induction of ring mitochondria. Mol Cell Neurosci. 2005;30:477–484. doi: 10.1016/j.mcn.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 9.Kirkpatrick B, Xu L, Cascella N, Ozeki Y, Sawa A, Roberts RC. DISC1 immunoreactivity at the light and ultrastructural level in the human neocortex. J Comp Neurol. 2006;497:436–450. doi: 10.1002/cne.21007. [DOI] [PubMed] [Google Scholar]
- 10.Miyoshi K, Honda A, Baba K, Taniguchi M, Oono K, Fujita T, et al. Disrupted-In-Schizophrenia 1, a candidate gene for schizophrenia, participates in neurite outgrowth. Mol Psychiatry. 2003;8:685–694. doi: 10.1038/sj.mp.4001352. [DOI] [PubMed] [Google Scholar]
- 11.Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y, et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- 12.Morris JA, Kandpal G, Ma L, Austin CP. DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum Mol Genet. 2003;12:1591–1608. doi: 10.1093/hmg/ddg162. [DOI] [PubMed] [Google Scholar]
- 13.Sawamura N, Sawamura-Yamamoto T, Ozeki Y, Ross CA, Sawa A. A form of DISC1 enriched in nucleus: altered subcellular distribution in orbitofrontal cortex in psychosis and substance/alcohol abuse. Proc Natl Acad Sci U S A. 2005;102:1187–1192. doi: 10.1073/pnas.0406543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bilen J, Bonini NM. Drosophila as a model for human neurodegenerative disease. Annu Rev Genet. 2005;39:153–171. doi: 10.1146/annurev.genet.39.110304.095804. [DOI] [PubMed] [Google Scholar]
- 15.Bonini NM, Fortini ME. Human neurodegenerative disease modeling using Drosophila. Annu Rev Neurosci. 2003;26:627–656. doi: 10.1146/annurev.neuro.26.041002.131425. [DOI] [PubMed] [Google Scholar]
- 16.Karsten SL, Sang TK, Gehman LT, Chatterjee S, Liu J, Lawless GM, et al. A genomic screen for modifiers of tauopathy identifies puromycin-sensitive aminopeptidase as an inhibitor of tau-induced neurodegeneration. Neuron. 2006;51:549–560. doi: 10.1016/j.neuron.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 17.Jackson GR, Wiedau-Pazos M, Sang TK, Wagle N, Brown CA, Massachi S, et al. Human wild-type tau interacts with wingless pathway components and produces neurofibrillary pathology in Drosophila. Neuron. 2002;34:509–519. doi: 10.1016/s0896-6273(02)00706-7. [DOI] [PubMed] [Google Scholar]
- 18.Feany MB, Bender WW. A Drosophila model of Parkinson's disease. Nature. 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 19.Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson's disease. Science. 2002;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 20.Wu YR, Wang CK, Chen CM, Hsu Y, Lin SJ, Lin YY, et al. Analysis of heat-shock protein 70 gene polymorphisms and the risk of Parkinson's disease. Hum Genet. 2004;114:236–241. doi: 10.1007/s00439-003-1050-1. [DOI] [PubMed] [Google Scholar]
- 21.Hendricks JC, Williams JA, Panckeri K, Kirk D, Tello M, Yin JC, et al. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci. 2001;4:1108–1115. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- 22.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 23.Ozeki Y, Tomoda T, Kleiderlein J, Kamiya A, Bord L, Fujii K, et al. Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci U S A. 2003;100:289–294. doi: 10.1073/pnas.0136913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adachi Y, Hauck B, Clements J, Kawauchi H, Kurusu M, Totani Y, et al. Conserved cis-regulatory modules mediate complex neural expression patterns of the eyeless gene in the Drosophila brain. Mech Dev. 2003;120:1113–1126. doi: 10.1016/j.mod.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 25.Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, et al. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 26.Kurusu M, Awasaki T, Masuda-Nakagawa LM, Kawauchi H, Ito K, Furukubo-Tokunaga K. Embryonic and larval development of the Drosophila mushroom bodies: concentric layer subdivisions and the role of fasciclin II. Development. 2002;129:409–419. doi: 10.1242/dev.129.2.409. [DOI] [PubMed] [Google Scholar]
- 27.Sawa A, Khan AA, Hester LD, Snyder SH. Glyceraldehyde-3-phosphate dehydrogenase: nuclear translocation participates in neuronal and nonneuronal cell death. Proc Natl Acad Sci U S A. 1997;94:11669–11674. doi: 10.1073/pnas.94.21.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bord L, Wheeler J, Paek M, Saleh M, Lyons-Warren A, Ross CA, et al. Primate disrupted-in-schizophrenia-1 (DISC1): high divergence of a gene for major mental illnesses in recent evolutionary history. Neurosci Res. 2006;56:286–293. doi: 10.1016/j.neures.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 29.Mao Z, Roman G, Zong L, Davis RL. Pharmacogenetic rescue in time and space of the rutabaga memory impairment by using Gene-Switch. Proc Natl Acad Sci U S A. 2004;101:198–203. doi: 10.1073/pnas.0306128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heisenberg M. Mushroom body memoir: from maps to models. Nat Rev Neurosci. 2003;4:266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- 31.Swinderen B. The remote roots of consciousness in fruit-fly selective attention? Bioessays. 2005;27:321–330. doi: 10.1002/bies.20195. [DOI] [PubMed] [Google Scholar]
- 32.Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–756. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- 33.Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–760. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- 34.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 35.Nitz DA, van Swinderen B, Tononi G, Greenspan RJ. Electrophysiological correlates of rest and activity in Drosophila melanogaster. Curr Biol. 2002;12:1934–1940. doi: 10.1016/s0960-9822(02)01300-3. [DOI] [PubMed] [Google Scholar]
- 36.van Swinderen B, Nitz DA, Greenspan RJ. Uncoupling of brain activity from movement defines arousal States in Drosophila. Curr Biol. 2004;14:81–87. [PubMed] [Google Scholar]
- 37.Andretic R, Shaw PJ. Essentials of sleep recordings in Drosophila: moving beyond sleep time. Methods Enzymol. 2005;393:759–772. doi: 10.1016/S0076-6879(05)93040-1. [DOI] [PubMed] [Google Scholar]
- 38.Ho KS, Sehgal A. Drosophila melanogaster: An Insect Model for Fundamental Studies of Sleep. Methods Enzymol. 2005;393:772–793. doi: 10.1016/S0076-6879(05)93041-3. [DOI] [PubMed] [Google Scholar]
- 39.Ma L, Liu Y, Ky B, Shughrue PJ, Austin CP, Morris JA. Cloning and characterization of Disc1, the mouse ortholog of DISC1 (Disrupted-in-Schizophrenia 1) Genomics. 2002;80:662–672. doi: 10.1006/geno.2002.7012. [DOI] [PubMed] [Google Scholar]
- 40.Taylor MS, Devon RS, Millar JK, Porteous DJ. Evolutionary constraints on the Disrupted in Schizophrenia locus. Genomics. 2003;81:67–77. doi: 10.1016/s0888-7543(02)00026-5. [DOI] [PubMed] [Google Scholar]
- 41.Hodgkinson CA, Goldman D, Jaeger J, Persaud S, Kane JM, Lipsky RH, et al. Disrupted in schizophrenia 1 (DISC1): association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am J Hum Genet. 2004;75:862–872. doi: 10.1086/425586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, et al. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 43.Hai T, Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- 44.Chen A, Muzzio IA, Malleret G, Bartsch D, Verbitsky M, Pavlidis P, et al. Inducible enhancement of memory storage and synaptic plasticity in transgenic mice expressing an inhibitor of ATF4 (CREB-2) and C/EBP proteins. Neuron. 2003;39:655–669. doi: 10.1016/s0896-6273(03)00501-4. [DOI] [PubMed] [Google Scholar]
- 45.Guan Z, Giustetto M, Lomvardas S, Kim JH, Miniaci MC, Schwartz JH, et al. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111:483–493. doi: 10.1016/s0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- 46.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 47.Blais JD, Filipenko V, Bi M, Harding HP, Ron D, Koumenis C, et al. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol Cell Biol. 2004;24:7469–7482. doi: 10.1128/MCB.24.17.7469-7482.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hewes RS, Schaefer AM, Taghert PH. The cryptocephal gene (ATF4) encodes multiple basic-leucine zipper proteins controlling molting and metamorphosis in Drosophila. Genetics. 2000;155:1711–1723. doi: 10.1093/genetics/155.4.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karpinski BA, Morle GD, Huggenvik J, Uhler MD, Leiden JM. Molecular cloning of human CREB-2: an ATF/CREB transcription factor that can negatively regulate transcription from the cAMP response element. Proc Natl Acad Sci U S A. 1992;89:4820–4824. doi: 10.1073/pnas.89.11.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yukawa K, Tanaka T, Tsuji S, Akira S. Regulation of transcription factor C/ATF by the cAMP signal activation in hippocampal neurons, and molecular interaction of C/ATF with signal integrator CBP/p300. Brain Res Mol Brain Res. 1999;69:124–134. doi: 10.1016/s0169-328x(99)00086-8. [DOI] [PubMed] [Google Scholar]
- 51.Campbell SS, Tobler I. Animal sleep: a review of sleep duration across phylogeny. Neurosci Biobehav Rev. 1984;8:269–300. doi: 10.1016/0149-7634(84)90054-x. [DOI] [PubMed] [Google Scholar]
- 52.Hendricks JC, Sehgal A, Pack AI. The need for a simple animal model to understand sleep. Prog Neurobiol. 2000;61:339–351. doi: 10.1016/s0301-0082(99)00048-9. [DOI] [PubMed] [Google Scholar]
- 53.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 54.Andretic R, van Swinderen B, Greenspan RJ. Dopaminergic modulation of arousal in Drosophila. Curr Biol. 2005;15:1165–1175. doi: 10.1016/j.cub.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 55.Brandon NJ, Schurov I, Camargo LM, Handford EJ, Duran-Jimeniz B, Hunt P, et al. Subcellular targeting of DISC1 is dependent on a domain independent from the Nudel binding site. Mol Cell Neurosci. 2005;28:613–624. doi: 10.1016/j.mcn.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 56.Jepsen K, Rosenfeld MG. Biological roles and mechanistic actions of co-repressor complexes. J Cell Sci. 2002;115:689–698. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- 57.Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 58.Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 59.Simonini MV, Camargo LM, Dong E, Maloku E, Veldic M, Costa E, et al. The benzamide MS-275 is a potent, long-lasting brain region-selective inhibitor of histone deacetylases. Proc Natl Acad Sci U S A. 2006;103:1587–1592. doi: 10.1073/pnas.0510341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clapcote SJ, Lipina TV, Millar JK, Mackie S, Christie S, Ogawa F, et al. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron. 2007;54:387–402. doi: 10.1016/j.neuron.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 61.Hikida T, Jaaro-Peled H, Seshadri S, Oishi K, Hookway C, Kong S, et al. Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proc Natl Acad Sci U S A. 2007;104:14501–14506. doi: 10.1073/pnas.0704774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li W, Zhou Y, Jentsch JD, Brown RA, Tian X, Ehninger D, et al. Specific developmental disruption of disrupted-in-schizophrenia-1 function results in schizophrenia-related phenotypes in mice. Proc Natl Acad Sci U S A. 2007;104:18280–18285. doi: 10.1073/pnas.0706900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.