Abstract
Regulation of neuronal gene expression is critical to establish functional connections in the mammalian nervous system. The transcription factor Sp4 regulates dendritic patterning during cerebellar granule neuron development by limiting branching and promoting activity-dependent pruning. Here, we investigate neurotrophin-3 (NT3) as a target gene important for Sp4-dependent dendritic morphogenesis. We found that Sp4 overexpression reduced NT3 promoter activity whereas knockdown of Sp4 increased NT3 promoter activity and mRNA. Moreover, Sp4 bound to the NT3 promoter in vivo, supporting a direct role for Sp4 as a repressor of NT3 expression. Addition of exogenous NT3 promoted dendritic branching in cerebellar granule neurons. Furthermore, sequestering NT3 blocked the continued addition of dendritic branches observed upon Sp4 knockdown, but had no effect on dendrite pruning. These findings demonstrate that, during cerebellar granule neuron development, Sp4-dependent repression of neurotrophin-3 is required to limit dendritic branching and thereby promote acquisition of the mature dendritic pattern.
Keywords: neurotrophin, NT3, dendrite, dendritic morphology, transcription factor, repressor
Introduction
The pattern of dendrites elaborated by a neuron determines integration of inputs. Defects in dendritic patterning are characteristic of many neurodevelopmental and mental retardation disorders (Dierssen and Ramakers, 2006; Galvez et al., 2005; Lee et al., 2003; Purpura, 1975). Dendritic development is a highly dynamic process that includes stages of addition, growth, branching, and stabilization or elimination of dendrites. Dendritic arborization patterns are regulated during development by the coordinated action of extracellular signals and gene expression programs (McAllister, 2000; Redmond and Ghosh, 2005). In addition to well-described roles in neuronal survival and differentiation, secreted polypeptide growth factors known as neurotrophins regulate growth and patterning of axons and dendrites in the mammalian nervous system (Friedman and Greene, 1999; Kaplan and Miller, 2000; Lewin and Barde, 1996; McAllister et al., 1999; Reichardt, 2006). The functions and regulated expression of neurotrophins and their receptors during dendritic development remains incompletely understood.
Neurotrophin 3 (NT3) has unique and sometimes overlapping or even opposing functions compared to the other neurotrophins: nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), and neurotrophin 4/5 (NT4/5). Among the functions described for NT3, NT3 regulates survival and differentiation of neurons in the cerebellum (Bates et al., 1999; Katoh-Semba et al., 1996; Neveu and Arenas, 1996). NT3 expression in the cerebellum is highest in the embryo and NT3 expression markedly declines with postnatal cerebellar maturation (Ernfors et al., 1990; Katoh-Semba et al., 1996; Maisonpierre et al., 1990a; Neveu and Arenas, 1996; Rocamora et al., 1993). The high affinity NT3 receptor, TrkC, is expressed in the cerebellum from the third week after birth (Neveu and Arenas, 1996; Segal et al., 1995). Thus, there is a switch in NT3 levels from high to low and in TrkC levels from low to high during postnatal development of the cerebellum. The transcriptional programs that control the balance between NT3 and TrKC gene expression during cerebellar maturation have not been fully described. The effects of NT3 on growth and branching of dendrites depends on neuronal cell type and brain region. For example, NT3 has been shown to either promote or limit dendritic growth and branching in distinct layers of pyramidal neurons (Baker et al., 1998; McAllister et al., 1997; McAllister et al., 1995). In early cerebellar granule neurons, addition of exogenous NT3 was shown to alter neurite morphology (Segal et al., 1995), however, little is known about the effects of NT3 on dendritic morphogenesis in these neurons.
We have identified an essential role for the transcription factor Sp4 during dendritic development in cerebellar granule neurons (Ramos et al., 2007). Knockdown of Sp4 in organotypic slices and dissociated cultures led to an increased number of highly branched dendrites. Total dendrite number is the summation of dendrites added less those that are removed. Time course analysis revealed that upon Sp4 knockdown, removal of dendrites was blocked and new dendritic branches continued to be added at later times. Furthermore, overexpression of Sp4 was sufficient to promote dendritic pruning in non-depolarizing conditions. These studies revealed that Sp4 is required to limit addition of new branches and promote activity-dependent pruning of dendrites. Like the related Sp1 and Sp3 proteins, Sp4 binds to specific promoter elements to regulate transcription (Black et al., 2001; Lerner et al., 2002; Philipsen and Suske, 1999; Ross et al., 2002), but the number and nature of Sp4 target genes that mediate dendritic patterning have not been described. Analysis of mutant mice expressing low levels of Sp4 revealed an age-dependent decrease of NT3 mRNA in the hippocampus (Zhou et al., 2005). When Sp4 was knocked out specifically in neural crest, NT3 levels were not affected, although a reduction in levels of TrkC mRNA was observed (St Amand et al., 2006). These studies suggest context-dependent regulation of NT3 and TrkC by Sp4.
In this study, we have investigated the hypothesis that the transcription factor Sp4 regulates NT3 expression to control dendritic development during postnatal maturation of cerebellar granule neurons. We show here that Sp4-dependent down-regulation of NT3 expression is required to suppress continued addition of dendritic branches during cerebellar granule neuron maturation. Inhibition of NT3 signaling was not observed to alter dendrite pruning, suggesting that Sp4 regulates distinct target genes to promote dendritic pruning and limit branch addition. Furthermore, we show that Sp4 activated expression of TrkC suggesting that opposing regulation by Sp4 may contribute to the dramatic change in NT3/TrKC ratios that occurs during cerebellar maturation. Our findings indicate that Sp4–dependent transcriptional regulation of Neurotrophin-3 contributes to proper dendritic morphogenesis during cerebellar development.
Results
Sp4 represses NT3 expression in developing cerebellar granule neurons
Many stages of postnatal cerebellar granule neuron development, including dendritic growth, branching and pruning, are faithfully recapitulated in vitro (Gaudilliere et al., 2004; Powell et al., 1997; Ramos et al., 2007). Sp4 protein levels are high in the post-natal cerebellum at the time of dendritic morphogenesis, whereas NT3 expression declines during cerebellar maturation (Altman, 1972; Lim et al., 2004; Maisonpierre et al., 1990a; Ramón y Cajal, 1911; Rocamora et al., 1993) (Data not shown). In order to determine whether Sp4 regulates NT3 expression in cerebellar granule neurons, we first performed luciferase reporter assays. A human NT3 promoter-driven luciferase reporter was co-transfected together with Flag-Sp4 overexpression or Sp4 RNAi knockdown plasmids in cultures of cerebellar granule neurons (Figure 1 A and B). We observed that Sp4 overexpression reduced NT3 promoter activity up to 50% while knockdown of Sp4 led to a two fold increase in NT3-promoter activity in primary granule neurons. Similar results were obtained in Neuro2A cells (Figure 1A and B). Co-transfection of NT3-luciferase with a second RNAi targeting Sp4 in Neuro2A cells also led to increased promoter activity (data not shown). These studies indicate that Sp4 represses NT3 promoter activity.
Figure 1. Sp4 transcription factor represses Neurotrophin-3 expression.
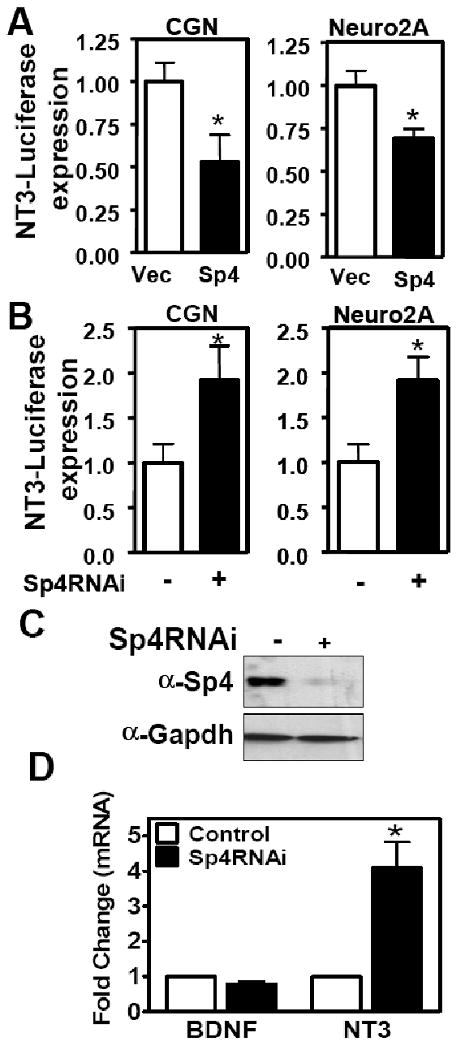
Cerebellar granule neurons or Neuro2A cells were co-transfected with NT3 (−3275/+91)-luciferase reporter and either (A) FlagSp4 (shaded bar) or empty vector (Vec) (open bar), or (B) Sp4RNAi (shaded bar) or control RNAi (open bar). Data represent the resulting luciferase activity normalized to each control condition. (Two-tailed t-test, p<0.05 (*)) (C and D) Neuro 2A cells were transfected with pMSCVpuro U6/GFP RNAi or U6/Sp4 RNAi plasmid. (C) After selection, cell lysates were immunoblotted for Sp4 and Gapdh. (D) Total RNA was subject to RT-qPCR with primers specific for NT3, BDNF and Gapdh. Values represents mean ± SEM generated from experiments performed in triplicate and normalized to Gapdh expression levels. Student's t test was used to determine the significance between groups. Asterisk denotes statistically significant value relative to Control RNAi (* p<0.05).
We also analyzed endogenous NT3 mRNA levels following depletion of Sp4 protein expression. We observed an increase in NT3 mRNA levels by RT-qPCR upon Sp4 knockdown in Neuro 2A cells (Figure 1 C and D). In contrast, Sp4 knockdown did not significantly alter BDNF mRNA levels (Figure 1D). Similar results were obtained with a second RNAi targeting Sp4 (data not shown). We further investigated whether Sp4 directly bound to endogenous NT3 promoter in granule neurons. Two transcription start sites with upstream activating and inhibitory regions have been described in the NT3 promoter (Katoh-Semba et al., 1996; Leingartner and Lindholm, 1994). We examined DNA-protein complexes by chromatin immunoprecipitation (ChIP) using primers flanking predicted Sp4 binding sites in the inhibitory region upstream of the 1B startsite (Figure 2A). ChIP assays with anti-Sp4 antisera revealed that Sp4 specifically associated with the inhibitory region of the endogenous NT3 promoter (Figure 2B). The related transcription factor Sp1 also associated with the NT3 promoter in cerebellar granule neurons. ChIP assays also revealed binding of Sp4 to the inhibitory region upstream of startsite 1A in cerebellar granule neurons as well as both inhibitory regions of the endogenous NT3 promoter in Neuro 2A (Data not shown). Taken together, these results indicate that Sp4 acts directly to repress NT3 expression.
Figure 2. Sp4 binds to the Neurotrophin-3 promoter in cerebellar granule neurons.
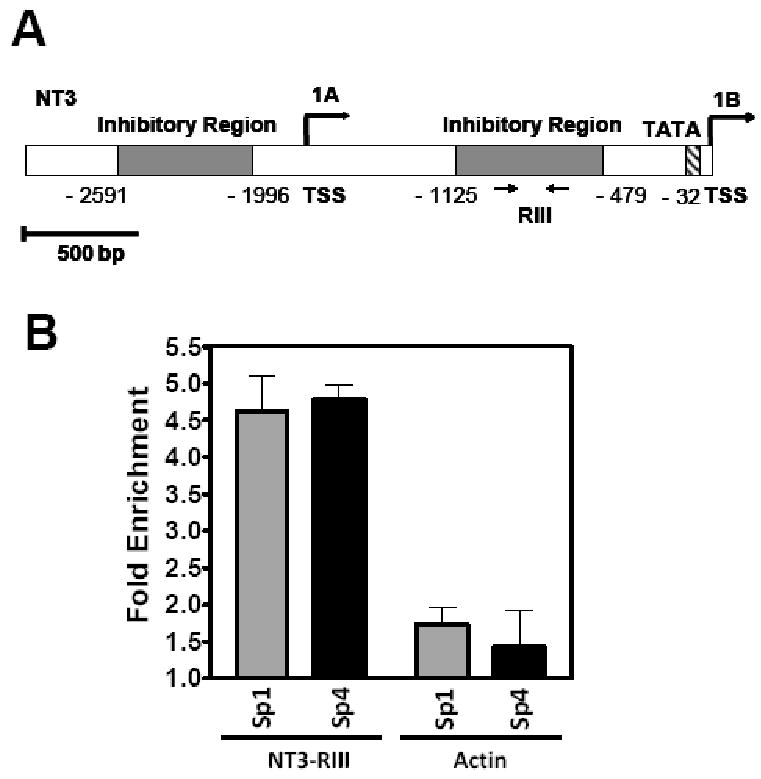
(A) Schematic of NT3 promoter in mouse. Positions of two transcription start sites (1A) and (1B), as well as a TATA box and two inhibitory regions (shaded grey) enriched in Sp4 binding sites are indicated. Arrows represent the locations of the primers used to amplify region RIII for chromatin immunoprecipitation (ChIP) assays. (B) Sp transcription factors bound to the inhibitory region of the endogenous NT3 promoter. ChIP assays were performed in cerebellar granular neurons, using antisera specific for Sp1 and Sp4 transcription factors, or Luciferase as control IgG. Immunoprecipitated DNA was analyzed by real-time PCR with oligonucleotides that amplified a fragment within the inhibitory region RIII of the NT3 promoter or a control fragment within the coding region of the beta-actin gene. Percent input normalized to control IgG is shown.
NT3 promotes dendritic branching in developing cerebellar granule neurons
We have reported that Sp4 is essential for dendritic development of cerebellar granule neurons in both organotypic slices and dissociated cultures (Ramos et al., 2007). Knockdown of Sp4 using two independent RNAi hairpins targeting Sp4 led to persistent addition of dendritic branches and blocked pruning of dendrites, with no decrease in cell viability (Ramos et al., 2007). However, Sp4 regulated target genes that control these processes are unknown. We therefore investigated whether increased levels of NT3 due to loss of Sp4-dependent repression could account for some of the morphological defects observed in Sp4 knockdown neurons. To address this issue, we first determined the effects on dendritic patterning when NT3 was added to cerebellar granule neurons. Neurons from P6 rat pups were transfected with GFP expression vector together with control RNAi or Sp4 RNAi at day in vitro (DIV) 2 and then at DIV4 neurons were treated with exogenous NT3 or control bovine serum albumin (BSA) for 48 hours (Figure 3). Dendritic arbor morphology was analyzed by counting primary dendrites emanating from the cell body and secondary dendrites emanating from the primary dendrites.
Figure 3. NT3 promotes increased dendritic branchpoints in cerebellar granule neurons.
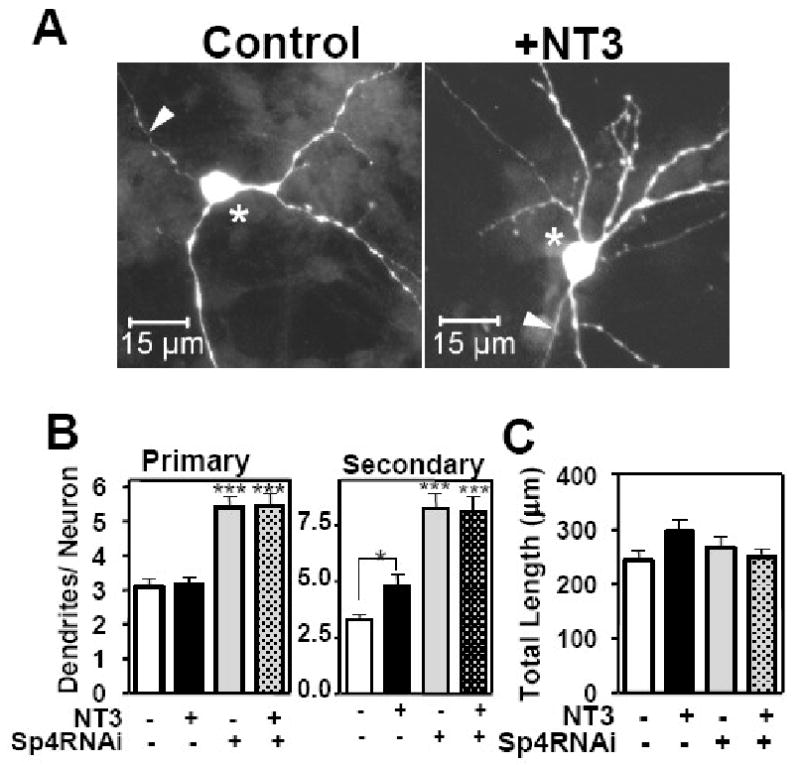
Cerebellar granule neurons were transfected with control (U6/Scr) or Sp4 RNAi together with GFP expression vector at DIV 2. At DIV4, cells were treated with exogenous NT3 or bovine serum albumin (BSA) control for 48 hours. Neurons were fixed and subjected to immunocytochemistry with anti-GFP. Dendritic trees were visualized by fluorescence microscopy. (A) Representative images of neurons transfected with control RNAi (U6/Scr) after treatment with BSA control (left) or exogenous NT3 (right). Arrowhead indicates axon and asterisk cell bodies of transfected neurons. (B) Total number of primary and secondary dendrites was quantitated. (C) Total dendritic length was quantitated. Values represent mean ± SEM of number of dendrites (B) and total dendritic length (C) per neuron. ANOVA analysis was performed by comparing each condition to control neurons p<0.001 (***) or p< 0.01 (**), n= 157 neurons were analyzed. Two-tailed t-test was also performed to compare differences plus or minus NT3 p< 0.05 (*). Quantitation was performed in 4 independent experiments with similar results.
As shown in Figure 3, in control neurons, addition of exogenous NT3 had no effect on the number of primary dendrites, whereas the number of secondary dendrites increased significantly (Figure 3A and B). These data indicate that elevated NT3 levels promote dendrite branching but have no effect on the balance of addition and elimination of primary dendrites in these neurons. As previously described, knockdown of Sp4 led to increased numbers of secondary dendrites by both inhibiting dendritic pruning and promoting new branch addition (Ramos et al., 2007) (Figure 3B). The number of primary dendrites were increased in Sp4RNAi neurons solely due to inhibition of pruning (Ramos et al., 2007). Thus, addition of NT3 recapitulates one of the phenotypes associated with Sp4 knockdown: increased numbers of secondary dendrites. Treatment with exogenous NT3 did not further increase the number of secondary dendrites seen upon Sp4 depletion, consistent with the model that NT3 levels are already elevated in the absence of Sp4-dependent repression (Figure 3B). Total dendritic length did not change after NT3 treatment (Figure 3C), indicating that dendritic growth is not regulated by either exogenous NT3 or Sp4 knockdown, as shown previously. Thus, NT3 promotes dendritic branching in cerebellar granule neurons.
Increased dendritic branching observed upon Sp4 knockdown requires NT3
We next investigated whether NT3 is required for the increased dendritic branch addition observed upon Sp4 knockdown. We transfected cerebellar granule neurons with Sp4 RNAi at DIV 2. After two days, we added a soluble form of the TrKC receptor (TrkC-Fc) to sequester NT3 in the media. TrkC-Fc is a chimera, bearing the extracellular domains of TrkC fused to the IgG heavy chain, that has the binding specificity and affinity of the NT3 receptor, TrkC, in a soluble form that can block the biological activity of NT3 (Shelton et al., 1995). We then monitored dendrite numbers. As shown in Figure 4A and B, after 48 hours treatment, TrkC-Fc reduced the number of secondary, but not primary, dendrites in Sp4 RNAi transfected cells. Addition of TrkC-Fc had no effect on dendrite numbers in control transfected neurons. This is consistent with data in Figure 3 that excess NT3 increased the number of secondary, but not primary, dendrites. Thus, the consequenes of TrKC-Fc addition was very specific only affecting the excess branching phenotype of Sp4 knockdown neurons, but not impacting the reduced pruning phenotype. TrKC-Fc treatment did not lead to changes in total dendritic length indicating again that modulation of NT3 does not affect dendritic growth in these neurons (Figure 4A).
Figure 4. Sequestering NT3 prevents excess dendritic branch addition upon Sp4 knockdown.
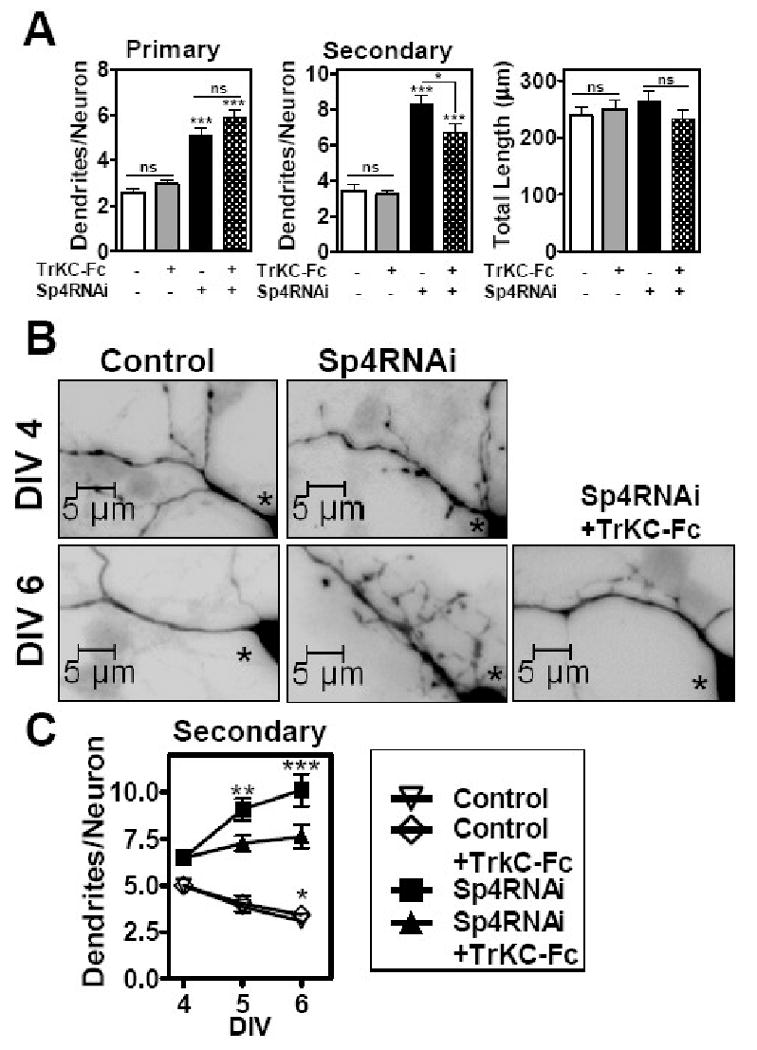
Cerebellar granule neurons were transfected with control or Sp4 RNAi as described for Figure 3. At DIV4, control bovine serum albumin or the chimeric protein TrkC-Fc was added. Neurons were fixed and subjected to immunocytochemistry with anti-GFP. Dendritic trees were visualized by fluorescence microscopy. (A) Total number of primary and secondary dendrites and total dendritic length was quantitated at DIV6. Values represent mean ± SEM of number of dendrites or total dendritic length per neuron. ANOVA analysis was performed by comparing each condition to control RNAi transfected neurons, p<0.001 (***), n= 145 neurons were analyzed. Two-tailed t-test was also performed to compare differences plus or minus addition of TrkC-Fc, p< 0.05 (*); ns= not significant. Quantitation was performed in 3 independent experiments with similar results. (B) Representative high magnification images at DIV 4 and DIV 6 of neurons transfected with control or Sp4 RNAi and treated with control BSA or TrkC-Fc (lower right panel). Asterisk indicates cell bodies of transfected neurons. (C) Total number of secondary dendrites was quantitated at DIV4, 5 and 6 as indicated. Values represent mean ± SEM of number of dendrites per neuron. ANOVA analysis was performed by comparing either Control or Sp4 RNAi transfected neurons at DIV4 with corresponding transfected neurons at DIV5 and 6, with or without TrkC-Fc treatment, p<0.001 (***) or p< 0.01 (**), n= 320 neurons were analyzed. Quantitation was performed in 3 independent experiments with similar results.
Since secondary dendrite numbers are the result of dynamic addition and elimination processes, we looked at the effect of TrkC-Fc in a time course. We reasoned that if NT3 signaling were required only for addition of new branches, then in the presence of TrkC-Fc the number of secondary dendrites in Sp4 RNAi neurons would not change during this period of dendritic maturation since no new ones would be added due to inhibition of the NT3 signal promoting branching and none would be removed due to the suppression of pruning upon Sp4 knockdownw. This is what we observed (Figure 4B and C). Control RNAi transfected neurons showed a reduction in the number of secondary dendrites during this period due to pruning (Ramos et al., 2007) which was not affected by TrkC-Fc addition (Figure 4C). In contrast, Sp4 knockdown neurons showed an increase in the number of secondary dendrites from DIV 4 to DIV6, indicating that in the absence of Sp4 these neurons not only failed to remove dendrites by pruning but continued to add new branches leading to a net gain in secondary dendrite number (Ramos et al., 2007) (Figure 4B and C). Notably, in the presence of TrkC-Fc,Sp4 RNAi neurons showed no significant change in the number of secondary dendrites during the time course examined. Treatment with TrkC-Fc blocked the continued addition of secondary dendrites in Sp4 RNAi transfected neurons. Thus, sequestering NT-3 rescued part of the Sp4 knockdown phenotype by suppressing excess dendritic branching. These results demonstrate that NT3 is required for the persistant dendritic branching observed upon knockdown of Sp4 and highlight a role for NT3 in regulating dendritic branching in cerebellar granule neurons during development.
Sp4 promotes TrKC expression in developing cerebellar granule neurons
In cerebellar granule neurons, the levels of the high affinity receptor for NT3, TrkC, are low in the early post-natal period and increase from the third week after birth (Neveu and Arenas, 1996; Segal et al., 1995). TrkC mRNA was reduced when Sp4 was knocked out in neural crest lineage of transgenic mice (St Amand et al., 2006). To explore whether Sp4 regulates TrKC expression in cerebellar granule neurons, we transfected neurons with a TrkC promoter driven luciferase reporter together with FlagSp4, Sp4RNAi, or control plasmids (Figure 5A and B). We observed that overexpression of Sp4 increased TrkC promoter activity more than two-fold while knockdown of Sp4 reduced promoter activity to 30% of levels in control transfected neurons (Figure 5A and B). Moreover, when we stably knocked down Sp4 in Neuro 2A cells and analyzed TrKC mRNA by RT-qPCR, we found that endogenous TrkC mRNA was reduced upon Sp4 depletion (Figure 5C). In addition, we analyzed the expression levels of other neurotrophin receptors in Neuro2A cells depleted of Sp4. We observed that Sp4 RNAi decreased levels of p75/NGFR and TrKA mRNA while levels of TrKB mRNA were increased (Figure 5C). Similar results were obtained with a second RNAi targeting Sp4 (data not shown). Taken together, these results suggest that Sp4 may regulate expression of several neurotrophin receptors and, notably, in maturing cerebellar granule neurons Sp4 has opposing activities on NT3 and TrkC promoter activity.
Figure 5. Transcription factor Sp4 activates TrkC expression.
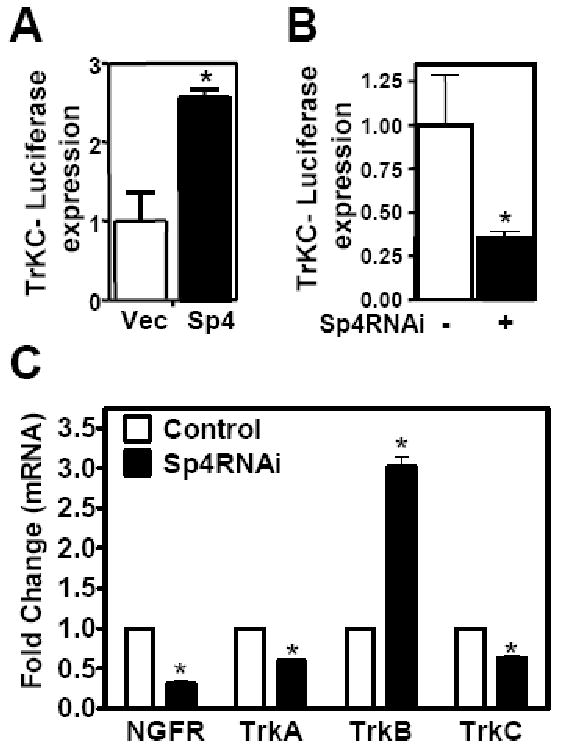
(A and B) Cerebellar granule neurons were co-transfected with TrkC-luciferase reporter and FlagSp4 expression vector (A) or Sp4RNAi (B) (shaded bar) or control vectors (open bar). The luciferase activity normalized to control is presented. (Two-tailed t-test, p<0.05 (*)). (C) Neuro 2A cells were stably transfected with control GFP or Sp4 RNAi as indicated. Total RNA was extracted and subject to RT-qPCR with primers for p75 (NGF receptor), TrKA, TrKB, TrKC, and Gapdh. Values represents mean ± SEM generated from experiments performed in triplicate and normalized to Gapdh expression levels. Student's t test was used to determine the significance between groups. Asterisk denotes statistically significant value relative to Control RNAi (* p<0.05).
Discussion
Our results indicate that the transcription factor Sp4 represses NT3 expression to control dendritic patterning during maturation of cerebellar granule neurons. Dendritic arborization patterns determine how a neuron integrates inputs and aberrant dendritic patterning is associated with mental retardation and neurological disorders (Dierssen and Ramakers, 2006; Galvez et al., 2005; Purpura, 1975). We have previously reported that the increased complexity of dendritic arbors observed upon Sp4 knockdown in cerebellar granule neurons is due to both a failure to limit new branch addition and reduced activity-dependent pruning (Ramos et al., 2007). Here we show that addition of exogenous NT3 promotes dendritic branching, and sequestering endogenous NT3 rescued one part of the Sp4 knockdown phenotype by blocking continuous branch addition (Figures 3 and 4). Together, these results support our conclusion that Sp4-dependent repression of NT3 is required to limit dendritic branching during neuronal maturation. Notably, addition of exogenous NT3 did not fully recapitulate the Sp4 RNAi phenotype, in particular, increasing or blocking NT3 signaling did not alter dendritic pruning (Figure 3 and 4), indicating that additional Sp4 target genes regulate this process. Our study therefore indicates that the pathways that control dendritic pruning and branch addition are distinct but coordinated via regulation by a common transcription factor, Sp4.
Fine timing and spatial regulation of gene expression of neurotrophins and TrK receptors suggest that there is a delicate balance between these ligands and receptors. Decrease of NT3 expression and increase of TrKC levels occurs in cerebellum shortly after birth, around the time that Sp4 levels peak (Katoh-Semba et al., 1996; Lim et al., 2004; Maisonpierre et al., 1990a; Rocamora et al., 1993; Segal et al., 1995) (data not shown). Here, we report that the transcription factor Sp4 has opposite effects on the regulation of NT3 and TrKC promoters: overexpression of Sp4 reduced NT3 promoter activity and increased TrKC promoter activity whereas knockdown of Sp4 increased expression of NT3 and reduced expression of TrKC (Figure 1 and 5). Sp4 bound the inhibitory region of the endogenous NT3 promoter in cerebellar granule neurons (Figure 2) indicating that Sp4 acts directly to repress NT3 promoter activity. The finding that Sp4 has opposing effects on NT3 and TrKC transcription raises the possibility that Sp4 contributes to differential regulation of NT3 and TrKC in the postnatal cerebellum. In fact, Sp4 regulates the expression of several neurotrophin receptors, suggesting that regulation of Sp4 activity during development contributes to maintaining the proper balance of neurotrophin and receptor expression.
Our studies have identified NT3 as an endogenous target of Sp4-dependent repression. Our findings in NT3-luciferase reporter assays are notably different from those of a previous study which suggested a role for Sp4 in activation of a small (121bp) NT3 promoter fragment in cortical neurons (Ishimaru et al., 2007). Importantly, our results in NT3 reporter assays are in agreement with the effects of Sp4 knockdown on endogenous NT3 mRNA (Figure 1). It is likely that the much larger NT3 promoter fragment used here, which includes the previously described inhibitory regions (Katoh-Semba et al., 1996; Leingartner and Lindholm, 1994) contributes to the reported differences of Sp4 activity on NT3-luciferase reporters. An age-dependent decrease in NT3 expression was observed in a hypomorphic Sp4 mouse model although NT3 expression was not affected in neural-crest derived tissue of Sp4 mutant mice (St Amand et al., 2006; Zhou et al., 2005). Taken together with our findings of Sp4-dependent repression, these studies suggest that Sp4 may have cell type-specific effects on NT3 expression. Although our data has revealed non-redundant functions of Sp4 in regulation of NT3, the finding that Sp1 also interacts with the NT3 promoter (Figure 2 and (Ishimaru et al., 2007)) suggests multiple Sp family transcription factors may contribute to fine-tuning expression of this neurotrophin. Binding sites for Sp factors have been correlated with inhibition of expression of several genes in neurons, including superoxide dismutase-2 (SOD2) and G protein receptor kinase 3 (GRK3) (Mao et al., 2006; Zhou et al., 2005), raising the possibility that Sp4-dependent repression may contribute to regulated expression of diverse genes in post-mitotic neurons.
Our data show that Sp4 has context-dependent activation and repression functions since we found that Sp4 represses NT3 and activates the TrKC promoter in cerebellar granule neurons. Sp4 has been shown to function as an activator of transcription for several genes (Hagen et al., 1995; Lerner et al., 2002; Majello et al., 1994; Ross et al., 2002). In some cases, however, Sp4 overexpression blocked activation by Sp1 (Kwon et al., 1999; Mao et al., 2002). The closely related Sp1 and Sp3 transcription factors have also been shown to function as both activators and repressors (Lee et al., 1987; Majello et al., 1997; Murata et al., 1994). One mechanism involved in the repressive activity of Sp1 and Sp3 is posttranslational modification by the small ubiquitin-related modifier (SUMO) (Ross et al., 2002; Sapetschnig et al., 2002; Spengler and Brattain, 2006). Further investigations are needed to determine the molecular mechanisms underlying the dual functions of Sp4 as a transcriptional activator or repressor.
Our study reveals that NT3 promotes dendritic branching in maturing cerebellar granule neurons. NT3 is a high affinity ligand for the TrKC receptor, however, NT3 can also stimulate TrkA and TrkB -dependent signaling in some circumstances (Davies et al., 1995; Lamballe et al., 1991; Squinto et al., 1991). This promiscuity allows NT3-dependent control of neuronal processes even in the absence of TrKC on its target cells and likely explains why the NT3 knockout phenotype is more severe than the TrkC knockout (Davies et al., 1995; Ernfors et al., 1990; Klein et al., 1994). Addition of NT3 was sufficient to increase dendritic branching in control neurons with a normal complement of neurotrophin receptors, however, it is possible that the reduced levels of TrKC, TrkA and p75/NGFR, increased levels of TrkB or other changes in Sp4 knockdown cells may sensitize these cells to the effects of NT3. The observation that TrKB expression increased in Sp4 RNAi cells makes TrKB an attractive candidate to mediate some NT3-dependent effects on dendritic branching. TrKB receptor expression is maintained in the cerebellum from postnatal stage until adulthood (Segal et al., 1995) and it has been shown that TrKB overexpression or treatment with the high affinity TrkB ligand, BDNF, promotes dendritic branching in different types of neurons (Lom et al., 2002; McAllister et al., 1997; Yacoubian and Lo, 2000). Further studies will be needed to determine the downstream signaling pathway by which NT3 promotes dendritic branching in cerebellar granule neurons.
NT3 expression is high and required in brain during embryogenesis and its levels decrease after birth becoming almost undetectable in adults (Bovolenta et al., 1996; Ernfors et al., 1990; Maisonpierre et al., 1990b). It has been unclear whether NT3 is merely not required or is actively restricted during later neuronal maturation and which transcriptional programs could be responsible for the inhibition of NT3 expression. Our results reveal that addition of new dendritic branches, a function which is restricted at later times to achieve proper dendritic patterning, is limited through an active repression of NT3 expression by the Sp4 transcription factor. Hence, our studies haveidentified a transcriptional repressor for NT3 linked to a restrictive function required during neuronal maturation. Notably, Sp4 also controls activity-dependent dendritic pruning during development, and it is likely that identification of Sp4 target genes that mediate pruning will provide further new insights into the pathways that regulate dendritic morphogenesis in the mammalian brain.
Experimental Procedures
Plasmids
Flag-Sp4 was expressed from the CMV promoter in pRC-Flag-Sp4 (Ross et al., 2002). Short hairpin RNAs were expressed from the U6 promoter in the pMSCVpuro vector (Sui et al., 2002) (Clontech Laboratories). Sequences targeted by the short hairpin RNAs were: Scrambled control (gggaattaatatgcacacaggcc); GFP control (gggcgatgccacctacggcaagc), Sp4 #1 nucleotides 1551 to 1571 (gggctccaactttaacacctt) and Sp4 #2 nucleotides 1804 to 1824 (gggtgctgcgggtgttcaagt) (Accession Number NM_003112 for Sp4) as described in (Ramos et al., 2007). The reporter plasmids containing hNT3 -3275/+91 or hTrKC – 870/+19 promoter sequence upstream of the luciferase gene have been described (Katoh-Semba et al., 1996; Nakatani et al., 2005; Shalizi et al., 2003).
Cerebellar Granule Neuron Culture and transfection
Cerebellar granule neurons were obtained from postnatal day 6 rat pups as described (Bonni et al., 1999). Animal Experimentation Protocol was approved by Harvard Medical Area Standing Committee on Animals. Briefly, neurons were maintained in Basal Minimum Essential medium supplemented with 10% fetal calf serum, 25 mM KCl, penicillin (50 units/ml), streptomycin (50 μg/ml) and 2 mM glutamine at 4.2×103 cells/mm2 density. After 18 hours, 10 μM cytosine arabinofuranoside was added. Cells were transfected by calcium phosphate precipitation at DIV 2 as described (Ramos et al., 2007). Where indicated, cerebellar granule neurons were treated with 20ng/ml of recombinant human Neurotrophin-3, 200ng/ml of recombinant human TrKC-Fc chimera (R&D Systems) or Bovine Serum Albumin 2 days after transfection. A vector expressing the antiapoptotic protein Bcl-xL was included in morphometric studies. Cerebellar granule neurons and Neuro2A cells were co-transfected with plasmids expressing Flag-Sp4 or U6/Sp4 RNAi hairpin together with luciferase reporter plasmids containing hNT3 or hTrKC promoter. Luciferase assays were performed as indicated by the manufacturer (Promega).
Immunofluorescence and Immunoblotting
At the indicated times, cells were fixed in 4% paraformaldehyde. Neurons were immunolabelled with rabbit anti-GFP (Molecular Probes), followed by a goat anti-rabbit conjugated to Cy2 (Amersham) and stained with the DNA dye bisbenzimide (Hoechst 33258). Neuro2A extracts were prepared in lysis buffer 50mM Tris pH 7.4, 150 mM NaCl containing 1% NP-40 for 30 minutes on ice, followed by sonication and centrifugation at 15000 g for 15 minutes. Lysates were resolved by SDS/PAGE and immunoblotted with polyclonal anti-Sp4 antibody (Santa Cruz) or polyclonal antibody against Gapdh (Chemicon International).
Morphometric analysis of dendrites
Images of individual transfected neurons with no overlapping processes from other transfected neurons were captured randomly in a blinded manner at 400× magnification using a Nikon eclipse TE2000 epifluorescence microscope with a CCD camera (Diagnostic instruments). Digital zoom magnification and quantitation of the length of individual primary, secondary and tertiary dendrites was performed using SPOT imaging software. Total dendritic length corresponds to the sum of the length of all individual dendrites per neuron. Statistical differences were determined by ANOVA and a post hoc Tukey test or two-tailed t-test using GraphPad Prism Software. For every condition, quantitation was performed in at least 3 independent experiments with similar results.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed following Upstate Biotechnology assay protocol with modifications. 2×107 cerebellar granule neurons were crosslinked with 1% formaldehyde for 15 min at 37°C. Chromatin was sonicated to between 500 to 1000 bp and immunoprecipitated with 3 μg of either anti-Sp1 (Upstate, anti-Sp4 (Santa Cruz), or anti-luciferase (Promega) polyclonal antibodies. Immunoprecipitated DNA was amplified by real-time PCR using specific primers as indicated. The real-time PCR was performed as described by the manufacturer using the iQ SYBR Green Supermix (BioRad). 10 % of total starting material was precipitated and used as input.
Primers for real-time PCR:
NT3RIII-F 5′- CCTCCCCCTTCATTTCTCTTGG-3′
NT3RIII-R 5′- TGAGCATTTGCCCAGCACTC-3′
actin-F 5′-TGAGAGGGAAATCGTGCGTGAC-3′
actin-R 5′-GCTCGTTGCCAATAGTGATGACC-3′
RNA extraction and Reverse transcription-qPCR analysis
Neuro2A cells were plated at 1.8 × 104 cells / cm2 24 hours before transfection with, GFP RNAi, Sp4RNAi #1 or #2, cloned into pMSCV, or pMSCV vector alone using Lipofectamine (Invitrogen). After 24 hours, cells were selected with 4μg/ml puromycin for 24 to 48 hours. RNA was extracted and purified using RNeasy Kit (QiAGEN) according to the manufacturers protocol. For quantitative RT-qPCR, first strand cDNA synthesis from 2 μg total RNA was carried out with Oligo dT primer using Superscript III reverse transcriptase (Invitrogen). cDNA were analyzed by quantitative PCR (Biorad IQ5) using the indicated primers. Expression of genes was normalized to that of Gapdh For RT-qPCR, at least 3 biological replicates were conducted and within each experiment every condition was measured in triplicate. Student's t test was used to determine the significance between groups.
NT3-F: TCACCACGGAGGAAACGCTAT
NT3-R: TCAATGGCTGAGGACTTGTCG
BDNF-F: TTGAGCACGTCATCGAAGAGC
BDNF-R: CCAAAGGCACTTGACTGCTGA
NGFR-F: AACCTCATTCCTGTCTATTGC
NGFR -R: CTGTTCCATCTCTTGAAAGC
TrkA-F: AGGTGGCTGCTGGTATGGT
TrkA-R: TCGCCTCAGTGTTGGAGAG
TrkB-F: TGTGACCCTTTCCTGCAGTGT
TrkB-R: CCCTGTGTGTGGCTTGTTTCA
TrkC-F: AGTAACCGGCTCACCACACTC
TrkC-R: AGCGGATGTCACAGCTGCAGT
Gapdh-F: CTGAGGACCAGGTTGTGTCC
Gapdh-R: CATTGTCATACCAGGAAATGAGC
Acknowledgments
We would like to thank Ary Shalizi and Azad Bonni for NT3-luciferase, Ichiro Matsuoka for TrkC-luciferase, and Maribel Rios for TrkB RT-PCR primers. These studies were supported by a Fellowship Centro de Investigación Biomedica en Red de Salud Mental to B.R and a grant from the National Institutes of Health to G.G. (HD043364).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altman J. Postnatal development of the cerebellar cortex in the rat. 3. Maturation of the components of the granular layer. J Comp Neurol. 1972;145:465–513. doi: 10.1002/cne.901450403. [DOI] [PubMed] [Google Scholar]
- Baker RE, Dijkhuizen PA, Van Pelt J, Verhaagen J. Growth of pyramidal, but not non-pyramidal, dendrites in long-term organotypic explants of neonatal rat neocortex chronically exposed to neurotrophin-3. Eur J Neurosci. 1998;10:1037–1044. doi: 10.1046/j.1460-9568.1998.00118.x. [DOI] [PubMed] [Google Scholar]
- Bates B, Rios M, Trumpp A, Chen C, Fan G, Bishop JM, Jaenisch R. Neurotrophin-3 is required for proper cerebellar development. Nat Neurosci. 1999;2:115–117. doi: 10.1038/5669. [DOI] [PubMed] [Google Scholar]
- Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- Bovolenta P, Frade JM, Marti E, Rodriguez-Pena MA, Barde YA, Rodriguez-Tebar A. Neurotrophin-3 antibodies disrupt the normal development of the chick retina. J Neurosci. 1996;16:4402–4410. doi: 10.1523/JNEUROSCI.16-14-04402.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AM, Minichiello L, Klein R. Developmental changes in NT3 signalling via TrkA and TrkB in embryonic neurons. Embo J. 1995;14:4482–4489. doi: 10.1002/j.1460-2075.1995.tb00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierssen M, Ramakers GJ. Dendritic pathology in mental retardation: from molecular genetics to neurobiology. Genes Brain Behav. 2006;5 2:48–60. doi: 10.1111/j.1601-183X.2006.00224.x. [DOI] [PubMed] [Google Scholar]
- Ernfors P, Ibanez CF, Ebendal T, Olson L, Persson H. Molecular cloning and neurotrophic activities of a protein with structural similarities to nerve growth factor: developmental and topographical expression in the brain. Proc Natl Acad Sci U S A. 1990;87:5454–5458. doi: 10.1073/pnas.87.14.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman WJ, Greene LA. Neurotrophin signaling via Trks and p75. Exp Cell Res. 1999;253:131–142. doi: 10.1006/excr.1999.4705. [DOI] [PubMed] [Google Scholar]
- Galvez R, Smith RL, Greenough WT. Olfactory bulb mitral cell dendritic pruning abnormalities in a mouse model of the Fragile-X mental retardation syndrome: further support for FMRP's involvement in dendritic development. Brain Res Dev Brain Res. 2005;157:214–216. doi: 10.1016/j.devbrainres.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Gaudilliere B, Konishi Y, de la Iglesia N, Yao G, Bonni A. A CaMKII-NeuroD signaling pathway specifies dendritic morphogenesis. Neuron. 2004;41:229–241. doi: 10.1016/s0896-6273(03)00841-9. [DOI] [PubMed] [Google Scholar]
- Hagen G, Dennig J, Preiss A, Beato M, Suske G. Functional analyses of the transcription factor Sp4 reveal properties distinct from Sp1 and Sp3. J Biol Chem. 1995;270:24989–24994. doi: 10.1074/jbc.270.42.24989. [DOI] [PubMed] [Google Scholar]
- Ishimaru N, Tabuchi A, Hara D, Hayashi H, Sugimoto T, Yasuhara M, Shiota J, Tsuda M. Regulation of neurotrophin-3 gene transcription by Sp3 and Sp4 in neurons. J Neurochem. 2007;100:520–531. doi: 10.1111/j.1471-4159.2006.04216.x. [DOI] [PubMed] [Google Scholar]
- Kaplan DR, Miller FD. Neurotrophin signal transduction in the nervous system. Curr Opin Neurobiol. 2000;10:381–391. doi: 10.1016/s0959-4388(00)00092-1. [DOI] [PubMed] [Google Scholar]
- Katoh-Semba R, Kaisho Y, Shintani A, Nagahama M, Kato K. Tissue distribution and immunocytochemical localization of neurotrophin-3 in the brain and peripheral tissues of rats. J Neurochem. 1996;66:330–337. doi: 10.1046/j.1471-4159.1996.66010330.x. [DOI] [PubMed] [Google Scholar]
- Klein R, Silos-Santiago I, Smeyne RJ, Lira SA, Brambilla R, Bryant S, Zhang L, Snider WD, Barbacid M. Disruption of the neurotrophin-3 receptor gene trkC eliminates la muscle afferents and results in abnormal movements. Nature. 1994;368:249–251. doi: 10.1038/368249a0. [DOI] [PubMed] [Google Scholar]
- Kwon HS, Kim MS, Edenberg HJ, Hur MW. Sp3 and Sp4 can repress transcription by competing with Sp1 for the core cis-elements on the human ADH5/FDH minimal promoter. J Biol Chem. 1999;274:20–28. doi: 10.1074/jbc.274.1.20. [DOI] [PubMed] [Google Scholar]
- Lamballe F, Klein R, Barbacid M. trkC, a new member of the trk family of tyrosine protein kinases, is a receptor for neurotrophin-3. Cell. 1991;66:967–979. doi: 10.1016/0092-8674(91)90442-2. [DOI] [PubMed] [Google Scholar]
- Lee A, Li W, Xu K, Bogert BA, Su K, Gao FB. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development. 2003;130:5543–5552. doi: 10.1242/dev.00792. [DOI] [PubMed] [Google Scholar]
- Lee W, Haslinger A, Karin M, Tjian R. Activation of transcription by two factors that bind promoter and enhancer sequences of the human metallothionein gene and SV40. Nature. 1987;325:368–372. doi: 10.1038/325368a0. [DOI] [PubMed] [Google Scholar]
- Leingartner A, Lindholm D. Two promoters direct transcription of the mouse NT-3 gene. Eur J Neurosci. 1994;6:1149–1159. doi: 10.1111/j.1460-9568.1994.tb00613.x. [DOI] [PubMed] [Google Scholar]
- Lerner LE, Gribanova YE, Whitaker L, Knox BE, Farber DB. The rod cGMP-phosphodiesterase beta-subunit promoter is a specific target for Sp4 and is not activated by other Sp proteins or CRX. J Biol Chem. 2002;277:25877–25883. doi: 10.1074/jbc.M201407200. [DOI] [PubMed] [Google Scholar]
- Lewin GR, Barde YA. Physiology of the neurotrophins. Annu Rev Neurosci. 1996;19:289–317. doi: 10.1146/annurev.ne.19.030196.001445. [DOI] [PubMed] [Google Scholar]
- Lim CR, Fukakusa A, Matsubara K. Gene expression profiling of mouse postnatal cerebellar development using cDNA microarrays. Gene. 2004;333:3–13. doi: 10.1016/j.gene.2004.02.026. [DOI] [PubMed] [Google Scholar]
- Lom B, Cogen J, Sanchez AL, Vu T, Cohen-Cory S. Local and target-derived brain-derived neurotrophic factor exert opposing effects on the dendritic arborization of retinal ganglion cells in vivo. J Neurosci. 2002;22:7639–7649. doi: 10.1523/JNEUROSCI.22-17-07639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonpierre PC, Belluscio L, Friedman B, Alderson RF, Wiegand SJ, Furth ME, Lindsay RM, Yancopoulos GD. NT-3, BDNF, and NGF in the developing rat nervous system: parallel as well as reciprocal patterns of expression. Neuron. 1990a;5:501–509. doi: 10.1016/0896-6273(90)90089-x. [DOI] [PubMed] [Google Scholar]
- Maisonpierre PC, Belluscio L, Squinto S, Ip NY, Furth ME, Lindsay RM, Yancopoulos GD. Neurotrophin-3: a neurotrophic factor related to NGF and BDNF. Science. 1990b;247:1446–1451. doi: 10.1126/science.247.4949.1446. [DOI] [PubMed] [Google Scholar]
- Majello B, De Luca P, Hagen G, Suske G, Lania L. Different members of the Sp1 multigene family exert opposite transcriptional regulation of the long terminal repeat of HIV-1. Nucleic Acids Res. 1994;22:4914–4921. doi: 10.1093/nar/22.23.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majello B, De Luca P, Lania L. Sp3 is a bifunctional transcription regulator with modular independent activation and repression domains. J Biol Chem. 1997;272:4021–4026. doi: 10.1074/jbc.272.7.4021. [DOI] [PubMed] [Google Scholar]
- Mao X, Moerman-Herzog AM, Wang W, Barger SW. Differential transcriptional control of the superoxide dismutase-2 kappaB element in neurons and astrocytes. J Biol Chem. 2006;281:35863–35872. doi: 10.1074/jbc.M604166200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Moerman AM, Barger SW. Neuronal kappa B-binding factors consist of Sp1-related proteins. Functional implications for autoregulation of N-methyl-D-aspartate receptor-1 expression. J Biol Chem. 2002;277:44911–44919. doi: 10.1074/jbc.M204292200. [DOI] [PubMed] [Google Scholar]
- McAllister AK. Cellular and molecular mechanisms of dendrite growth. Cereb Cortex. 2000;10:963–973. doi: 10.1093/cercor/10.10.963. [DOI] [PubMed] [Google Scholar]
- McAllister AK, Katz LC, Lo DC. Opposing roles for endogenous BDNF and NT-3 in regulating cortical dendritic growth. Neuron. 1997;18:767–778. doi: 10.1016/s0896-6273(00)80316-5. [DOI] [PubMed] [Google Scholar]
- McAllister AK, Katz LC, Lo DC. Neurotrophins and synaptic plasticity. Annu Rev Neurosci. 1999;22:295–318. doi: 10.1146/annurev.neuro.22.1.295. [DOI] [PubMed] [Google Scholar]
- McAllister AK, Lo DC, Katz LC. Neurotrophins regulate dendritic growth in developing visual cortex. Neuron. 1995;15:791–803. doi: 10.1016/0896-6273(95)90171-x. [DOI] [PubMed] [Google Scholar]
- Murata Y, Kim HG, Rogers KT, Udvadia AJ, Horowitz JM. Negative regulation of Sp1 trans-activation is correlated with the binding of cellular proteins to the amino terminus of the Sp1 trans-activation domain. J Biol Chem. 1994;269:20674–20681. [PubMed] [Google Scholar]
- Nakatani T, Ueno S, Mori N, Matsuoka I. Role of NRSF/REST in the molecular mechanisms regulating neural-specific expression of trkC/neurotrophin-3 receptor gene. Brain Res Mol Brain Res. 2005;135:249–259. doi: 10.1016/j.molbrainres.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Neveu I, Arenas E. Neurotrophins promote the survival and development of neurons in the cerebellum of hypothyroid rats in vivo. J Cell Biol. 1996;133:631–646. doi: 10.1083/jcb.133.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell SK, Rivas RJ, Rodriguez-Boulan E, Hatten ME. Development of polarity in cerebellar granule neurons. J Neurobiol. 1997;32:223–236. doi: 10.1002/(sici)1097-4695(199702)32:2<223::aid-neu7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Purpura DP. Normal and aberrant neuronal development in the cerebral cortex of human fetus and young infant. UCLA Forum Med Sci. 1975:141–169. doi: 10.1016/b978-0-12-139050-1.50014-8. [DOI] [PubMed] [Google Scholar]
- Ramón y Cajal S. Histology of the Nervous System of Man and Vertebrates. 1995. Editorial Oxford University Press; New York: 1911. [Google Scholar]
- Ramos B, Gaudilliere B, Bonni A, Gill G. Transcription factor Sp4 regulates dendritic patterning during cerebellar maturation. Proc Natl Acad Sci U S A. 2007;104:9882–9887. doi: 10.1073/pnas.0701946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond L, Ghosh A. Regulation of dendritic development by calcium signaling. Cell Calcium. 2005;37:411–416. doi: 10.1016/j.ceca.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocamora N, Garcia-Ladona FJ, Palacios JM, Mengod G. Differential expression of brain-derived neurotrophic factor, neurotrophin-3, and low-affinity nerve growth factor receptor during the postnatal development of the rat cerebellar system. Brain Res Mol Brain Res. 1993;17:1–8. doi: 10.1016/0169-328x(93)90065-w. [DOI] [PubMed] [Google Scholar]
- Ross S, Tienhaara A, Lee MS, Tsai LH, Gill G. GC box-binding transcription factors control the neuronal specific transcription of the cyclin-dependent kinase 5 regulator p35. J Biol Chem. 2002;277:4455–4464. doi: 10.1074/jbc.M110771200. [DOI] [PubMed] [Google Scholar]
- Sapetschnig A, Rischitor G, Braun H, Doll A, Schergaut M, Melchior F, Suske G. Transcription factor Sp3 is silenced through SUMO modification by PIAS1. Embo J. 2002;21:5206–5215. doi: 10.1093/emboj/cdf510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal RA, Pomeroy SL, Stiles CD. Axonal growth and fasciculation linked to differential expression of BDNF and NT3 receptors in developing cerebellar granule cells. J Neurosci. 1995;15:4970–4981. doi: 10.1523/JNEUROSCI.15-07-04970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalizi A, Lehtinen M, Gaudilliere B, Donovan N, Han J, Konishi Y, Bonni A. Characterization of a neurotrophin signaling mechanism that mediates neuron survival in a temporally specific pattern. J Neurosci. 2003;23:7326–7336. doi: 10.1523/JNEUROSCI.23-19-07326.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton DL, Sutherland J, Gripp J, Camerato T, Armanini MP, Phillips HS, Carroll K, Spencer SD, Levinson AD. Human trks: molecular cloning, tissue distribution, and expression of extracellular domain immunoadhesins. J Neurosci. 1995;15:477–491. doi: 10.1523/JNEUROSCI.15-01-00477.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler ML, Brattain MG. Sumoylation inhibits cleavage of Sp1 N-terminal negative regulatory domain and inhibits Sp1-dependent transcription. J Biol Chem. 2006;281:5567–5574. doi: 10.1074/jbc.M600035200. [DOI] [PubMed] [Google Scholar]
- Squinto SP, Stitt TN, Aldrich TH, Davis S, Bianco SM, Radziejewski C, Glass DJ, Masiakowski P, Furth ME, Valenzuela DM, et al. trkB encodes a functional receptor for brain-derived neurotrophic factor and neurotrophin-3 but not nerve growth factor. Cell. 1991;65:885–893. doi: 10.1016/0092-8674(91)90395-F. [DOI] [PubMed] [Google Scholar]
- St Amand TR, Lu JT, Zamora M, Gu Y, Stricker J, Hoshijima M, Epstein JA, Ross JJ, Jr, Ruiz-Lozano P, Chien KR. Distinct roles of HF-1b/Sp4 in ventricular and neural crest cells lineages affect cardiac conduction system development. Dev Biol. 2006;291:208–217. doi: 10.1016/j.ydbio.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Sui G, Soohoo C, Affar el B, Gay F, Shi Y, Forrester WC. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci U S A. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yacoubian TA, Lo DC. Truncated and full-length TrkB receptors regulate distinct modes of dendritic growth. Nat Neurosci. 2000;3:342–349. doi: 10.1038/73911. [DOI] [PubMed] [Google Scholar]
- Zhou X, Long JM, Geyer MA, Masliah E, Kelsoe JR, Wynshaw-Boris A, Chien KR. Reduced expression of the Sp4 gene in mice causes deficits in sensorimotor gating and memory associated with hippocampal vacuolization. Mol Psychiatry. 2005;10:393–406. doi: 10.1038/sj.mp.4001621. [DOI] [PubMed] [Google Scholar]


