Abstract
In Diffusion Weighted Magnetic Resonance Image (DW-MRI) processing, a 2nd order tensor has been commonly used to approximate the diffusivity function at each lattice point of the DW-MRI data. From this tensor approximation, one can compute useful scalar quantities (e.g. anisotropy, mean diffusivity) which have been clinically used for monitoring encephalopathy, sclerosis, ischemia and other brain disorders. It is now well known that this 2nd-order tensor approximation fails to capture complex local tissue structures, e.g. crossing fibers, and as a result, the scalar quantities derived from these tensors are grossly inaccurate at such locations. In this paper we employ a 4th order symmetric positive-definite (SPD) tensor approximation to represent the diffusivity function and present a novel technique to estimate these tensors from the DW-MRI data guaranteeing the SPD property. Several articles have been reported in literature on higher order tensor approximations of the diffusivity function but none of them guarantee the positivity of the estimates, which is a fundamental constraint since negative values of the diffusivity are not meaningful. In this paper we represent the 4th-order tensors as ternary quartics and then apply Hilbert’s theorem on ternary quartics along with the Iwasawa parametrization to guarantee an SPD 4th-order tensor approximation from the DW-MRI data. The performance of this model is depicted on synthetic data as well as real DW-MRIs from a set of excised control and injured rat spinal cords, showing accurate estimation of scalar quantities such as generalized anisotropy and trace as well as fiber orientations.
1 Introduction
Advances in medical imaging during the last decade have made it possible to collect massive amounts of data, which can be used to analyze the underlying connectivity of the tissues being scanned. More specifically, acquisition of magnetic resonance image (MRI) data that measures the apparent diffusivity of water in tissue in vivo has been extensively used for computing the diffusivity at each image lattice point.
A 2nd order tensor has commonly been used to approximate the diffusivity function from a given set of, usually seven or more, acquired DW-MR images [5]. The approximated diffusivity function is given by
| (1) |
where g = [g1 g2 g3]T is the magnetic field gradient direction and D is the estimated 2nd-order tensor. This approximation yields a diffusion tensor (DT-MRI) data set Di, which is a 3D matrix-valued image, where subscript i denotes location on a 3D lattice.
The literature is abound with techniques for estimating the diffusion tensor from DW-MRI data. Earlier techniques were restricted to the use of linearized Stejskal-Tanner equation [5, 8], until the non-linear Stejskal-Tanner equation with the positive-definite constraint on the 2nd-order tensor D to be estimated along with regularization of the tensor field was introduced by Wang et al. in [42, 43].
Geometric methods to enforce positive definiteness were introduced by Chefd’ Hotel et al. in [10]. However a linearized Stejskal-Tanner equation was used in their work to first estimate the tensor field from DW-MRI.
Mathematically, diffusion tensors are 3×3 symmetric positive-definite matrices that belong to a Riemannian symmetric space, where a Riemannian metric assigns an inner product to each point of this space. Using this metric, one can perform various tasks, e.g. interpolation, geodesic computation, principal geodesic analysis etc. [14, 30, 23, 4].
Several useful scalar quantities can be computed from the 2nd-order tensorial approximation of the diffusivity function, such as the mean diffusivity (computed as the trace of matrix Di) and various anisotropic measures of the tensor Di (fractional anisotropy [8], relative anisotropy [8] etc). The significance of these measures is that they provide an easy tool, since they are scalar quantities, for monitoring changes in the anisotropic properties of white matter fiber bundles caused either naturally by neurological disorders or artificially post-surgery or in response to a treatment. Fractional anisotropy has been successfully used clinically, and a reduction of its value has been reported in patients with trauma [33], multiple sclerosis [9, 16], Hypoxic-Ischemic Encephalopathy [44], multiple system atrophy [18], meningitis [26] and other pathologies. Similarly, reduction of the mean diffusivity, an other scalar quantity, has been also reported in the case of ischemic tissue [41].
However, the above scalar measures are computed using the 2nd-order tensorial approximation of the diffusivity, which although works well for simple tissue structures, fails to approximate more complex tissue geometry with multi-lobed diffusivity profiles. For instance, the value of fractional anisotropy drops significantly in areas of fiber crossings, although these locations are anisotropic.
In order to overcome the above limitation, higher order tensors were introduced in [27] to represent more complex diffusivity profiles which better approximate the local diffusivity function. Generalized scalar quantities such as the variance of diffusivity and the generalized anisotropy were derived as functions of the higher order tensor coefficients [13]. However, in all of these works the higher-order tensors are estimated without imposing the positivity of the diffusivity function approximation, which is significantly important since negative diffusivity values are non physical.
The use of a 4th-order covariance tensor was proposed by Basser and Pajevic in [6]. This covariance tensor is employed in defining a Normal distribution of 2nd order diffusion tensors. This distribution function has been employed in [7] for higher-order multivariate statistical analysis of DT-MRI datasets using spectral decomposition of the 4th-order covariance matrix into eigenvalues and eigentensors (2nd order). However, 2nd order tensors are used to approximate the diffusivity of each lattice point of a MR data set, failing to capture complex local tissue geometries, such as fiber crossings.
In [25], Moakher and Norris presented a way to minimize the distance between a given higher-order tensor and the closest elasticity tensor of higher symmetry, using different metrics. This work was recently extended by Moakher [24] where in he developed an interesting framework to describe the geometry of 4th-order tensors. A 4th-order symmetric positive definite tensor in three dimensions is represented by a 2nd-order symmetric positive definite tensor in six dimensions and therefore, one can use the Riemannian metric of the space of 6×6 SPD matrices for the SPD 4th-order tensor computations. However, this framework fails to parameterize the full space of SPD 4th-order tensors. For example the groups of SPD tensors and , where a, b, c are positive valued coefficients, can not be written in the form of 6×6 SPD matrices. Furthermore, the Riemannian metric of symmetric positive-definite 2nd-order tensors in six dimensions assigns infinite distance between 6×6 SPD and 6 × 6 semi-definite matrices. The SPD tensors in the aforementioned examples correspond to 6 × 6 semi-definite matrices and therefore, the Riemannian metric assigns infinite distance between them although their corresponding coefficients can be arbitrarily close. This leads to arbitrarily large errors in tensor processing and one should therefore strive to seek a more appropriate framework to characterize the full space of symmetric positive definite 4th-order tensors.
The first reported method in literature to impose the positivity constraint on 4th-order diffusion tensors was presented in Barmpoutis et al. [1] and in their work, they parameterize the full space of positive definite 4th-order tensors. They represent the 4th-order tensor as a homogeneous polynomial of degree 4 in 3 variables, the so called ternary quartic. They then invoke Hilbert’s theorem on ternary quartics [17] according to which, any non-negative 4th-order tensor can be expressed as a sum of squares of three quadratic (2nd-order) forms, which can be further expressed as an equivalent semi-definite 2nd-order tensor using a CCT parametrization, where C is a 6 × 3 coefficient matrix. Although by using this parametrization the whole space of SPD 4th-order tensors is covered, the elements on the boundary of this space are also included i.e. the positive semi-definite 4th-order tensors. Furthermore, the space of parameters is not unique since the coefficients C and their antipodal symmetric −C parameterize the same tensor. This non-uniqueness issue leads to problems when regularizing the field of coefficients C thus motivating us to seek a new way for parameterizing and estimating regularized 4th-order tensors fields.
In this paper we propose a novel parametrization of the 4th-order tensors for imposing the positivity of the estimated diffusivity. In this parametrization we express the 6 × 6 matrix G = CCT using its Iwasawa coordinates [36]. By combining the Iwasawa formulation with the Hilbert’s theorem on ternary quartics [17], we are interested in estimating Iwasawa decomposable 6×6 symmetric positive semi-definite matrices of at most rank-3. We derive formulas for uniquely computing the tensor coefficients as functions of the parameters of our model. The unknown parameters are estimated from a given DW-MRI data set by minimizing an energy function in which we add a regularization term for estimating smooth 4th-order tensor fields.
The key contribution of this work is the use of a parametrization which can decompose any 6 × 6 symmetric positive semi-definite matrix of at most rank-3. This parametrization has two main advantages: a) it minimizes the solution space by overcoming the non-uniqueness problems of the model in [1] and b) allows the simultaneous estimation and regularization of the 4th-order tensor field. The motivation for using an estimated positive higher-order tensor model is that one can derive the generalized higher-order extensions of the commonly used 2nd-order scalar measures for monitoring various brain diseases. In our experimental results we show that such scalar measures can be more accurately estimated by using the proposed framework compared to other methods. We show that our model is robust to noise in estimating fiber orientations as well as scalar measures derived from the 4th-order tensor coefficients such as the generalized trace [13]. This demonstrates that the proposed framework is a directly applicable tool that extends efficiently the clinically used measures by overcoming the limitations of the current models in literature.
The rest of the paper is organized as follows: In section 2, we present a novel parametrization of the 4th-order tensors that is used to enforce the positivity of the estimated tensors using the Iwasawa decomposition of the Gram matrix. In section 2.1, we present a functional minimization method to estimate 4th-order tensors from diffusion-weighted MR images. Furthermore, in section 2.2 we propose a distance measure for the space of 4th-order tensors, and employ it for estimating a smoothly varying tensor field as well as estimating the generalized variance of the tensors. Section 3 contains the experimental results and comparisons with other methods using simulated diffusion MRI data and real MR database of excised rat spinal cords. In section 4 we conclude.
2 Diffusion tensors of 4th order
The diffusivity function has been commonly modeled in literature by Eq. (1) using a 2nd-order tensor. Studies have shown that this approximation fails to model complex local structures of the diffusivity in real tissues [28] and higher-order tensors or other methods for multi-fiber approximation must instead be employed [40, 27, 11, 39, 38, 37, 22, 21]. In the case of 4th-order tensors, the diffusivity function can be expressed using the standard notation of quartic forms [31] as
| (2) |
where g = [g1 g2 g3]T is the magnetic field gradient direction. It should be noted that in the case of 4th-order symmetric tensors there are 15 unique coefficients Di,j,k, while in the case of 2nd-order tensors we only have 6.
In DW-MRI the diffusivity of the water is a positive quantity. This property is essential since negative diffusion coefficients are non-physical. However, in the parametrization of Eq. 2 there is no guarantee that the estimated coefficients Di,j,k form a positive tensor. Therefore, there is need for the development of a new parametrization of the 4th-order tensor, which guarantees the positivity of the estimated tensor.
Letting g1, g2 and g3 in (2) be the variables, the equivalence between symmetric tensors and homogeneous polynomials is straightforward to establish. Moreover, if a symmetric tensor is positive, then its corresponding polynomial must be positive for all real-valued variables. Hence, here we are concerned with the positive definiteness of homogenous polynomials of degree 4 in 3 variables, or the so called ternary quartics. Hilbert in 1888 proved the following theorem on ternary quartics (see [34, 32] for a modern exposition) that we will employ in this work:
Theorem 1
There exists an identity in which d is a real ternary form d = d(g1, g2, g3) of degree four which is positive semi-definite and the pi are quadratic forms with real coefficients.
By using this theorem, Eq. (2) can be expressed as a sum of 3 squares of quadratic forms as
| (3) |
where, v is a properly chosen vector of monomials, (e.g. ), C = [c1|c2|c3] is a 6 × N matrix by stacking the 6 coefficient vectors ci and G = CCT is the so called Gram matrix.
The Gram matrix G in Eq. 3 is positive semi-definite and has at most rank 3. Here we should emphasize that G being semi-definite does not imply that the 4th-order tensor d is also semi-definite, since the vector space of the former is 6-dimensional (i.e. vectors v) while the corresponding vector space of the latter is 3-dimensional (i.e. vectors g). In fact, using the parametrization of Eq. 3 the whole space of the strictly positive definite ternary quartics as well as those which are semi-definite is spanned.
Given a Gram matrix, the CCT parametrization in Eq. 3 is not unique, i.e. there exist different matrices C which parameterize the same Gram matrix. For example C parameterizes the same Gram matrix as its antipodal pair −C. Furthermore, there are infinitely many orthogonal matrices that yield the same G when appropriately applied in the aforementioned parametrization. This is due to the orthogonality property (OOT = I) of the rotation matrices O, where I is the identity matrix. Thus, in the case that C is of size 6×3, for any 3×3 orthogonal matrix O we have (CO)(CO)T = CCT. This produces an infinite solution space, which theoretically can not be handled by the optimization techniques. We should also note that in Eq. 3 there are in total 18 parameters in C, which are 3 more coefficients than the number of unique tensor coefficients in Eq. 2. The non uniqueness issues of the Gram matrix were also discussed by Powers et al. in [31], investigating how many fundamentally differnent Gram matrices parametrize the same ternary quartic.
In order to overcome the above issues, we use the Iwasawa decomposition which represents the components of a positive definite or semi-definite matrix in the Iwasawa coordinates [19, 20]. Every n × n positive definite matrix G can be uniquely expressed using its Iwasawa components as follows.
| (4) |
where W and V are SPD matrices of size k×k and m×m respectively, X ∈ ℜk×m and A[B] denotes BTAB.
In the case of n × n positive semi-definite matrices of at most rank-k, the Iwasawa coordinates are also given by Eq. 4 by setting rank(W) + rank(V) ≤ k. By computing the matrix multiplications in Eq. 4 we derive the following parametrization of positive semi-definite matrices
| (5) |
where W and V are 3×3 positive definite or positive semi-definite matrices and their ranks sum up to k. Furthermore, by using the Cholesky decomposition of W = AAT, where A is a lower triangular matrix with non-negative diagonal elements, we can establish equivalence between the parametrization in Eq. 5 and that in Eq. 3 by defining B to be the matrix that satisfies the equation BAT = XT W. By using the above we derive the following parametrization for the Gram matrix
| (6) |
where and B ∈ ℜ3×3.
If W is positive definite, then the Cholesky factor A is unique, B = XT A is also unique, V = 0 and therefore, the parametrization in Eq. 6 is unique. Note that in Eq. 6 there are in total 15 parameters, 6 in A and 9 in B, which is equal to the number of unique tensor coefficients in Eq. 2.
According to Theorem 1, Eq. 6 spans the whole space of positive semi-definite ternary quartics but can be used to define a parametrization for the case of strictly positive definite 4th-order ternary quartics as follows:
Lemma 1
For every real positive definite ternary quartic d there exists an arbitrarily small positive real number c such that d can be written as the sum of the ternary quartic c(x2 + y2 + z2)2 and three squares of quadratic forms.
Proof
Let d(g) be a strictly positive definite ternary quartic. Therefore, d(g) > 0 ∀g ∈ S2, where, S2 is the unit sphere, i.e. the space of unit vectors g. We define the following ternary quartic , where c is any real number in the interval 0 < c ≤ ming∈S2d(g). Since c > 0 this ternary quartic is also positive definite. Let us now define . Since ming∈S2f(g) = ming∈S2d(g) − c ≥ 0, f(g) is a positive semi-definite ternary quartic and therefore can be expressed as a sum of three squares of quadratic forms (by Theorem 1). Thus, every positive definite ternary quartic d(g) can be written as where c is an arbitrarily small positive real number, which proves the lemma.
The corresponding diffusivity function can be expressed using the Iwasawa coordinates as follows:
| (7) |
where C is a 3 × 3 matrix whose elements equal to the same arbitrarily small positive real number c, v is a properly chosen vector of monomials, (e.g. ), W, V and X are defined as in Eq. 5, and A and B are defined as in Eq. 6.
Here we should note that in practice due to finite precision computations c can be set to the smallest possible value of a finite precision machine and therefore, is considered as a known variable. In the double-precision floating-point IEEE 754-1985 standard the smallest positive value is approximately c =10−308.
Furthermore, we should emphasize that the semi-definite property of G does not necessarily imply semi-definiteness of d(g) since the former is semi-definite in ℜ6 while the latter may be semi-definite in ℜ3. In particular, using the proposed parametrization (Eq. 7), we compute semi-definite matrices G that correspond to strictly positive-definite d(g).
Given a Gram matrix, which in our case is parameterized using the matrices A and B in Eq. 7, we can uniquely compute the tensor coefficients Di,j,k by using the formulas in table 1. In these formulas the tensor coefficients are expressed as functions of the components of A, B and the fixed parameter c.
Table 1.
Formulas to compute the tensor coefficients Di,j,k given the components a11, a22, a33, a21, a31, a32, b11, b12, b13, b21, b22, b23, b31, b32, b33 and the fixed parameter c = 10−308.
| D400 = a112 + c |
| D040 = a212 + a222 + c |
| D004 = a312 + a322 + a332 + c |
| D220 = b112 + b122 + b132 + 2 a11 a21 + 2c |
| D202 = b212 + b222 + b232 + 2 a11 a31 + 2c |
| D022 = b312 + b322 + b332 + 2 a21 a31 + 2 a22 a32 + 2c |
| D310 = 2 a11 b11 |
| D301 = 2 a11 b21 |
| D130 = 2(a21 b11 + a22 b12) |
| D031 = 2(a21 b31 + a22 b32) |
| D103 = 2(a31 b21 + a32 b22 + a33 b23) |
| D013 = 2(a31 b31 + a32 b32 + a33 b33) |
| D211 = 2(b11 b21 + b12 b22 + b13 b23 + a11 b31) |
| D121 = 2(b11 b31 + b12 b32 + b13 b33 + a21 b21 + a22 b22) |
| D112 = 2(b21 b31 + b22 b32 + b23 b33 + a31 b11 + a32 b12 + a33 b13) |
Therefore, we can employ the parametrization in Eq. 7 for the estimation of the coefficients Di,j,k of the diffusion tensor from MR images using the following two steps: 1) first estimate the A and B matrices from the given images by using a functional minimization method (minimization of Eq. 8 using the Lavenberg-Marquardt nonlinear least-squares method), and then 2) compute the unique coefficients Di,j,k of the 4th-order tensor by using formulas in table 1.
In the following section we will employ this method to enforce the positive definite property of the estimated fourth order diffusion tensors from the diffusion weighted MR images.
2.1 Estimation from DWI
The coefficients Di,j,k of a 4th order diffusion tensor can be estimated from diffusion-weighted MR images by minimizing the following cost function:
| (8) |
where G is the Iwasawa parameterized Gram matrix given by Eq. 7, M is the number of the diffusion weighted images associated with gradient vectors gi and b-values bi; Si is the corresponding acquired signal and S0 is the zero gradient signal. Using the magnetic field gradient directions gi we construct the 6-dimensional vectors . In Eq. (8), the 4th order diffusion tensor is parameterized using the 3 × 3 matrices A and B which form together the Gram matrix G. Having estimated the matrices A and B that minimize Eq. (8), the coefficients Di,j,k can be computed directly using the formulas in Table 1. S0 can either be assumed to be known or estimated simultaneously with the coefficients Di,j,k by minimizing Eq. (8).
In the implementation of the proposed method, in order to enforce the diagonal elements a1,1, a2,2 and a3,3 of matrix A to be non negative, we should use a mapping of ℜ to the space of non-negative real numbers. In order for this mapping to be unique (one to one), the target space must be open, hence we seek a mapping to the positive part of ℜ. This does not limit the solution space in our implementation, since it has been shown that in finite precision arithmetic, open spaces are equivalent to closed spaces (Wang et al. [43]). In our experiments we used the exponential mapping and and therefore, in the minimization we solve for and instead of a1,1, a2,2 and a3,3. The total number of unknown parameters in G in Eq. 8 are 15 and they are the following: , a2,1, a3,1, a3,2 and the 9 elements of matrix B.
Starting with an initial guess for S0, A and B we can use any gradient based optimization method in order to minimize Eq. (8). We should note here that the exponent in Eq. (8) is in the form of a polynomial and therefore its gradient with respect to the unknown coefficients is easily derived analytically.
Given A and B at each iteration of the optimization algorithm we can update S0 by again minimizing Eq. (8). The derivative of this equation with respect to the unknown S0 is
| (9) |
By setting Eq. (9) equal to zero, we derive the following update formula for S0
| (10) |
In our experiments we used the well known Lavenberg-Marquardt (LM) nonlinear least-squares method, which has advantages over other optimization methods, in terms of stability and computational burden. The average execution time of our implementation on an AMD Athlon 2GHz was N ×M ×0.000545 seconds where N is the number of voxels per image and M is the number of the acquired DW-MR images used in the estimation of the 4th-order tensor field.
2.2 Distance measure
In the previous section we discussed how to estimate the positive-definite 4th-order tensors from DW-MRI data by minimizing Eq. 8. The 4th-order tensors are by definition (Eq. 2) smoothly varying functions (within a voxel). In order to impose smoothness across the image lattice we can add to the energy function the following regularization term
| (11) |
where Nj is the set of lattice points in the neighborhood of ‘j’. In the regularization term defined in Eq. (11) we need to employ an appropriate distance measure between the tensors Di and Dj. Here we use the notation D to denote the set of 15 unique coefficients Di,j,k of a 4th-order tensor.
We can define a distance measure between the 4th-order diffusion tensors D1 and D2 by computing the normalized L2 distance between the corresponding diffusivity functions d1(g) and d2(g) leading to the equation,
| (12) |
where, the integral of Eq. (12) is over all unit vectors g, i.e., the unit sphere S2 and the coefficients Δi,j,k are computed by subtracting the coefficients Di,j,k of the tensor D1 from the corresponding coefficients of the tensor D2.
As shown above, the integral of Eq. (12) can be computed analytically and the result can be expressed as a sum of squares of the terms Δi,j,k. In this simple form, this distance measure between 4th-order tensors can be implemented very efficiently. Note that this distance measure is invariant to rotations in 3-dimensional space because it is the L2 norm, which is well known for its invariance with respect to rigid motions. Furthermore, by using the formulas from Table 1 we can write the above distance as a function of the elements of the estimated matrices A and B. We should note that the obtained regularization term is in the form of polynomial and therefore its gradient with respect to the unknowns A and B of Eq. 8 can be also computed analytically.
Another property of the above distance measure is that the average element (mean tensor) D̂ of a set of N tensors Di, i = 1 … N is defined as the Euclidean average of the corresponding Di,j,k coefficients of the tensors. This property can be proved by verifying that D̂ minimizes the sum of squares of distances Σ dist(D̂, Di)2. Similarly, it can be shown that geodesics (shortest paths) between 4th-order tensors are defined as Euclidean geodesics.
Finally, the above distance measure can be used to compute the intra-voxel variance [13] of a single displacement probability function parameterized as a 4th-order tensor D. The variance is given by
| (13) |
where < D > is the generalized trace [13] given by < D >= (D4,0,0 + D0,4,0 +D0,0,4 + (D2,2,0 + D2,0,2 + D0,2,2)/3)/5. This variance has been used to define the generalized anisotropy in [13] and we use it in our experimental results as well.
3 Experimental results
In this section we present experimental results on our method applied to simulated DW-MRI data as well as real DW-MRI data from excised rat’s spinal cords.
3.1 Synthetic data experiments
In order to motivate the need for the positive-definite constraint in the 4th-order estimation process, we performed the following experiment using a synthetic data set. The synthetic data was generated by simulating the MR signal from a highly anisotropic single fiber (fractional anisotropy> 0.9) using the realistic diffusion MR simulation model in [35] (b-value= 1250s/mm2, 21 gradient directions). Then, we added different amounts of Riccian noise to the simulated data set and estimated the 4th-order tensors from the noisy data by: a) minimizing without using the proposed parametrization to enforce the SPD constraint, by employing the method in [29] and b) our method, which guarantees the SPD property of the tensors. (Si is the MR signal of the ith image and S0 is the zero-gradient signal).
It is known that the estimated 4th-order tensors are able to represent rather complicated diffusivity profiles with multiple fiber orientations which better approximate the diffusivity of the local tissue compared to the commonly used 2nd-order tensors [27]. Studies on estimating fiber orientations from the diffusivity profile have shown that the peaks of the diffusivity profile do not necessarily yield the orientations of the distinct fiber bundles [28]. One should instead employ the displacement probability functions for computing the fiber orientations. The displacement probability P(R) is given by the Fourier integral P(R) = ∫ E(q)exp(−2πiq·R)dq where q is the reciprocal space vector, E(q) is the signal value associated with vector q divided by the zero gradient signal and R is the displacement vector. In our experiments, we estimated the displacement probability profiles from the 4th-order tensors using the method in [3].
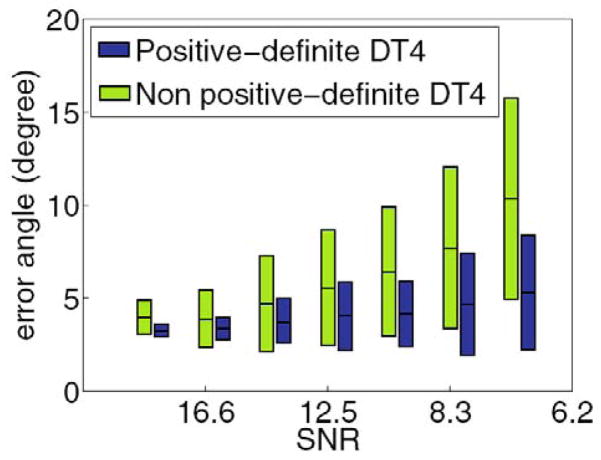
Then, we computed the displacement probability functions from the 4th-order tensors estimated earlier using the two different methods, and after that we computed the fiber orientations from the maxima of these probability functions. The error angles (mean and standard deviation) between ground truth and estimated orientations from the two methods for different amount of noise in the data are plotted in Fig. 1. Here, we should emphasize that the intensity of the simulated signal was significantly smaller when the diffusion gradient orientation formed a small angle with the fiber orientation than that corresponding to gradient orientations perpendicular to the fiber. These smaller intensity values are most likely to become negative when we corrupt the signal with noise, and as a result any method that does not impose the SPD property is affected drastically. As expected, our method yields smaller errors in comparison with the method that does not enforce the SPD property of the tensors. When we increase the amount of noise in the data, the errors observed by the latter method are significantly increased, while our method depicts less sensitivity. This demonstrates the need for enforcing the SPD property of the estimated tensors and validates the accuracy of our proposed method.
Fig. 1.
Comparison of the fiber orientation errors for different amounts of noise in the data, obtained by using: a) our parametrization to enforce positivity and b) without enforcing positivity of the estimated tensors.
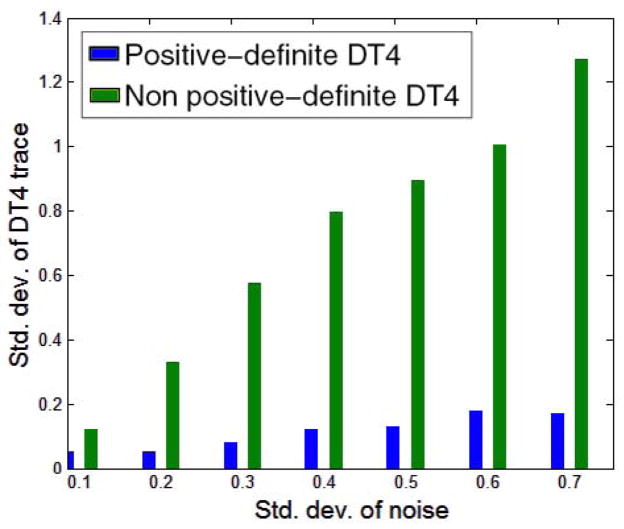
As was expected, similar results were observed by comparing the generalized trace [13] of the 4th-order diffusion tensors estimated in the previous experiment. In the case of estimating the 4th-order tensors without imposing the SPD property, 44% of the estimated tensors yielded negative generalized trace values, while the values computed using the proposed method were all positive. In both cases we computed the standard deviation of the estimated generalized trace for different amounts of noise in the data (excluding from our calculations the negative values computed by the non SPD method). Figure 2 shows the standard deviation of the generalized trace computed using both the methods. By observing the figure we conclude that the proposed method produces significantly more accurate results even in the higher noise cases.
Fig. 2.
Comparison of the standard deviation of the generalized trace [13], obtained by using: a) our parametrization that enforces positivity and b) without enforcing positivity of the estimated tensors.
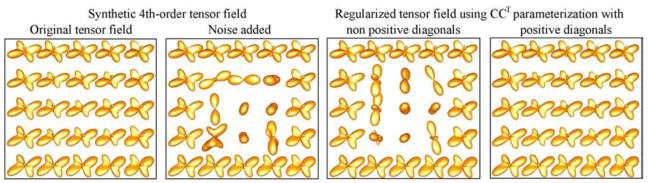
Furthermore, we compared the proposed parametrization of the SPD 4th-order tensors with the one presented in [1]. For the comparison, we synthesized a uniform tensor field (shown in Fig. 3 left) and then we corrupted the central voxels using randomly generated 4th - order tensors. The noisy field was regularized by minimizing ∫ ||∇Cx|| dx, where Cx are the parameters of each parametrization at location x (||∇Cx || was assumed to be zero at the boundary). As it was expected the non unique parametrization in [1] failed to produce a smooth result compared to the proposed parametrization.
Fig. 3.
Illustration of the advantage of the proposed parametrization, against the non unique parametrization in [1]. In this example we regularized a noisy R synthetic tensor field by minimizing ∫|| ∇ Cx || dx, where Cx are the parameters of each parametrization, and x is the lattice index.
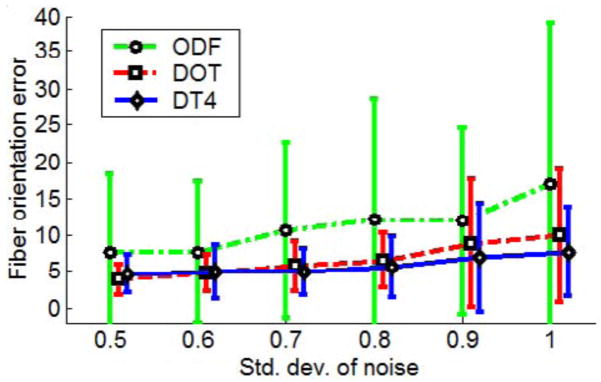
Finally, in order to compare our proposed method with other existing techniques that do not employ 4th-order tensors, we performed another experiment using synthetic data. The data were generated for different amounts of noise by following the same method as previously using the simulated MR signal of a 2-fiber crossing. We estimated SPD 4th-order tensors from the corrupted simulated MR signal using our method and then computed the fiber orientations from the corresponding probability functions. For comparison we also estimated the fiber orientations using the DOT method described in [28] and the ODF method presented in [39]. For all three methods we computed the estimated fiber orientation errors for different amounts of noise in the data (shown in Fig. 3.1). The results conclusively demonstrate the accuracy of our method, showing small fiber orientation errors (≃5°) for typical amount of noise with std. dev.≃ 0.5 – 0.8. Furthermore, by observing the plot, we also conclude that the accuracy of our proposed method is very close to that of the DOT method and is significantly better than the ODF method. For larger amounts of noise our method yielded smaller errors than all the other methods.
3.2 Real data experiments
In the following experiments, we used MR data from excised rat’s spinal cord. The protocol that we used in this experiment included acquisition of 22 images using a pulsed gradient spin echo pulse sequence with repetition time (TR) = 1.5 s, echo time (TE) = 27.2 ms, bandwidth = 30 kHz, field-of-view (FOV) = 4.3×4.3 mm. After the first image set was collected without diffusion weighting (b ~ 0 s/mm2), 21 diffusion-weighted image sets with gradient strength (G) = 664 mT/m, gradient duration (δ) = 1.5 ms, gradient separation (Δ) = 17.5 ms and diffusion time (Td) = 17 ms were collected. The image without diffusion weighting had 8 signal averages, and each diffusion-weighted image had 2 averages.
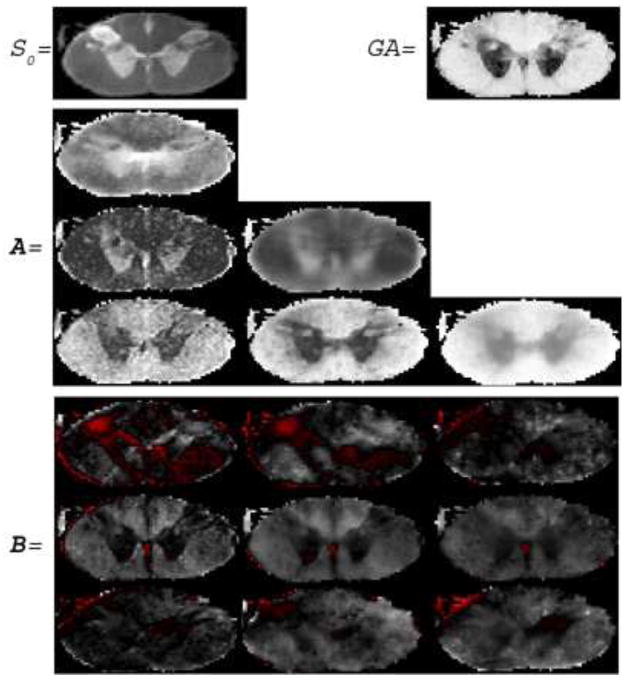
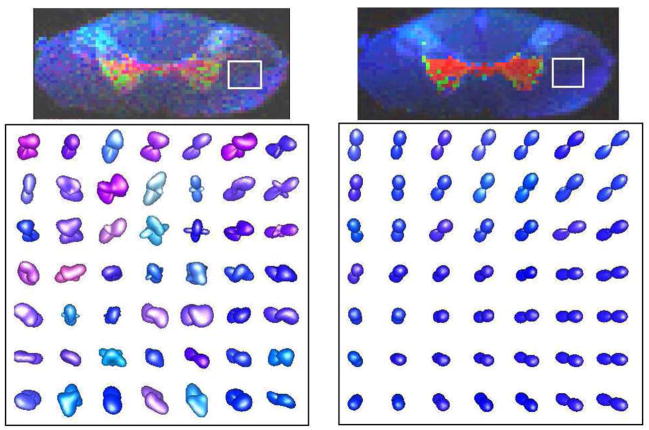
First, we estimated an SPD 4th-order tensor field by applying the proposed method to the real DW-MRI data. Figure 5 shows the estimated elements of matrices A and B, which parameterize the Gram matrix discussed in section 2. In the first row the estimated S0 and generalized anisotropy [13] are also shown. The red color in the images denotes negative values. By observing the images we can see that each coefficient shows different details of the underlying tissue.
Fig. 5.
The elements of matrices A and B estimated by the proposed method as well the estimated S0 and generalized anisotropy [13].
Furthermore, we computed the 4th-order tensor field first without imposing the positive-definite constraint. In order to compare the obtained results with that estimated by the proposed algorithm, in Fig. 6 we plot the corresponding tensors from a region of interest in the white matter. In this region of interest we expected single-lobed diffusivities with peaks predominantly in the axial direction which is represented by the blue color. The X, Y, Z components of the dominant orientation of each probability profile are assigned to R, G, B (red, green, blue) components of the color of each tensor. By observing this figure, we can say that the tensor field is incoherent if we do not enforce the SPD constraint (Fig. 6 left) and the expected single-lobed nature of this white matter region is lost. On the other hand the tensors obtained by our method (Fig. 6 right) are more coherent. Note that this is a result of enforcing the SPD constraint, since in this experiment we did not use any regularization. Similarily to the simulated data examples (Sec. 3.1), the ROI in Fig. 6 corresponds to highly anisotropic fibers in the white matter of the spinal cord, which are most likely to yield negative diffusivities when the SPD property is not imposed. This demonstrates the superior performance of our algorithm and motivates the use of the proposed SPD constraint.
Fig. 6.
4th-order tensors estimated without imposing the SPD property (left) and by using the proposed method (right). On the top the corresponding estimated S0 images are shown colored by mapping the X, Y, Z coordinates of the largest diffusivity orientation to the R, G, B color components. In this region of interest we expected single-lobed diffusivities with peaks predominantly in the axial direction (shown in blue).
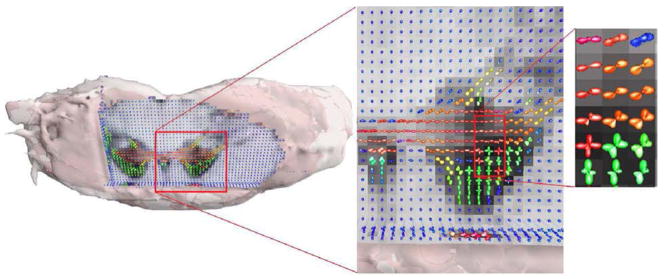
Figure 7 depicts a visualization of the tensor field estimated by the proposed technique. In the center plate multi-lobed diffusivity profiles and other complex structures can be observed. In the right image of the same figure, a zoomed in region of interest shows smooth transitions from single-lobed diffusivities to multi-lobed structures in the estimated 4th-order tensor field.
Fig. 7.
Visualization of the 4th-order tensor field estimated by applying proposed method to a real DW-MRI dataset from an excised rat’s spinal cord.
3.3 Comparison of control and injured spinal cord datasets
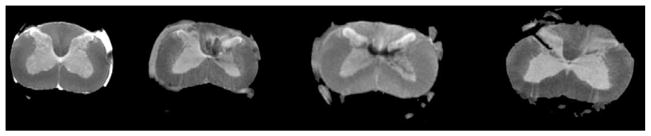
In this section we present the results obtained by performing a set of experiments using a set of 1 control and 3 injured rat spinal cords. The S0 images of the data sets are shown in Fig. 8.
Fig. 8.
The acquired S0 image of a control (left) and three injured rat spinalcords.
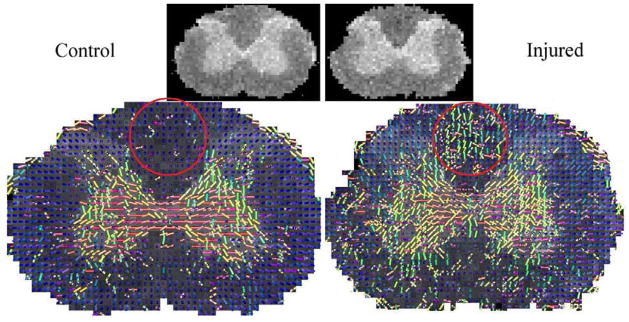
First we estimated the 4th-order diffusion tensors by applying the proposed technique to the DW-MR images. Then, we registered the estimated tensor fields using the 4th-order tensor-field registration method presented in [2]. Figure 9 shows two corresponding slices of the control and an injured cord data set. In this figure, the orientation of the peaks in the estimated 4th-order tensors are shown as tubes. By comparing the corresponding S0 images (shown on the top of Fig. 9) one cannot easily observe changes in the underlying fiber structures due to the injury. However, by comparing the corresponding fiber orientations we can observe significant changes in the white matter region between the horns (marked with a red circle).
Fig. 9.
Comparison of the fiber orientations estimated in the control and the corresponding registered injured cord dataset. The S0 images are shown on the top of the figure.
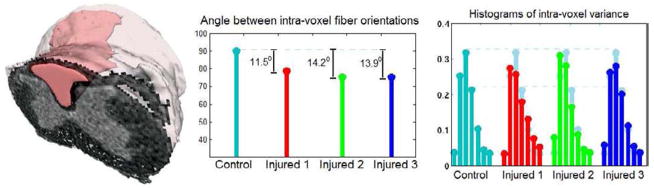
Figure 10(left) depicts a 3D visualization of the region of interest (ROI) in the white matter region between the horns (shown in pink). In order to per-from various quantitative comparisons in the ROI, we computed the average normalized diffusivity as the minimizer of the Helinger’s distance between the 4th-order tensors in the ROI. In this region, we expected to find anisotropic diffusivities with fiber orientations predominant in the axial direction. By observing the orientations in Fig. 9, one can see that the estimated diffusivities from the injured dataset are less coherent than those in the control spinal cord. In order to compare these orientation plots we computed the angle between the peaks of the average diffusivity in the ROI and the results are shown in Fig. 10(center). In this plot, there is a clear reduction of the fiber orientation angle in all injured spinal cord cases due to the changes in the underlying structures caused by the injury. The intra-voxel variances were also estimated in the ROI and the histograms of the obtained values are shown on the right plot of the same figure. This plot also shows a reduction of the variance after the injury which corresponds to a drop of the anisotropy caused by the injury.
Fig. 10.
Visualization of the ROI (shown in pink) in 3D. The plots show comparisons between the fiber orientation angle of the average diffusivity in the ROI and the histogram of variances in the ROI.
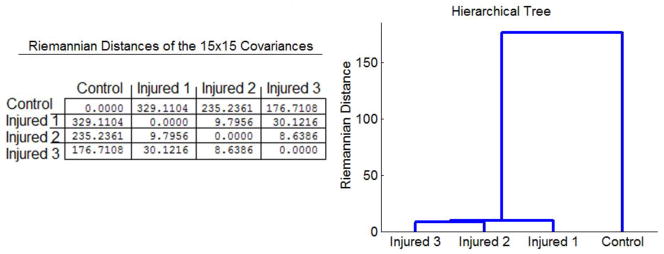
In all the above experiments we derived either a scalar or an orientational quantity in order to compare the spinal cord data sets. However, one can use all the information included in the 15 real-valued parameters of our model, which fully characterize the corresponding 4th-order tensors. In the ROI, we treated the 15 coefficients as elements of ℜ15 and we constructed a 15 × 15 covariance matrix for each data set. These matrices are symmetric and positive-definite and therefore, we can employ the Riemannian metric of P15 [30] in order to compute distances between the datasets and various statistics (e.g. Principal Geodesic analysis [14]). Figure 11 shows the table of Riemannian distances between the datasets. This table was then used in the Aglomerative clustering technique [12, 15] to construct the hierarhical tree shown on the right of the same figure. In this plot, the distance between the branches shows the affnities between the given datasets. In this case the three injured datasets were clustered together whose Riemannian distance from the control data set is significantly larger than the distance between the injured data sets.
Fig. 11.
Quantitative comparison of the rat spinal cord dataset using the Rie-mannian metric of the 15 × 15 positive definite matrices. The Riemannian distances between the covariance matrices are shown on the left. The corresponding hierarchical dendrogram computed using the Riemannian distances.
4 Conclusions
In this article a novel parametrization of symmetric positive semi-definite ten-sors has been presented using the Iwasawa decomposition and Hilbert’s theorem on ternary quartics. The advantage of Iwasawa components is that they can parametrize the Gram matrix, reducing the infinite solution space caused by the Hilbert’s sum of squares parametrization. This is the first method in literature that parametrizes the full space of the positive definite 4th-order tensors. We apply this method to estimate the diffusivity function as a field of 4th-order tensors, given a DW-MRI dataset. In the experiments presented, we used synthetic and real DW-MR images from rat spinal cords. The experimental results demonstrate the robust to noise of our method and its superiority compared with other existing techniques.
Fig. 4.
Fiber orientation errors for different SNR in the data using our method for the estimation of positive 4th-order tensors and two other existing methods: 1) DOT and 2) ODF.
Footnotes
This research was in part funded by the NIH grant EB007082 to BCV, and in part by the University of Florida Alumni Fellowship to AB.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Angelos Barmpoutis, Email: abarmpou@cise.ufl.edu.
Baba C. Vemuri, Email: vemuri@cise.ufl.edu.
Dena Howland, Email: howland@mbi.ufl.edu.
John R. Forder, Email: jforder@mbi.ufl.edu.
References
- 1.Barmpoutis A, Jian B, Vemuri BC, Shepherd TM. Symmetric positive 4th order tensors and their estimation from diffusion weighted mri. In: Karssemeijer N, Lelieveldt BPF, editors. IPMI, volume 4584 of Lecture Notes in Computer Science. Springer; 2007. pp. 308–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barmpoutis A, Vemuri BC, Forder JR. Registration of high angular resolution diffusion mri images using 4th order tensors . LNCS; Proceedings of MICCAI07: Int. Conf. on Medical Image Computing and Computer Assisted Intervention; Springer; 2007. pp. 4791pp. 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barmpoutis A, Vemuri BC, Forder JR. Fast displacement probability profile approximation from hardi using 4th-order tensors. Proceedings of ISBI08: IEEE International Symposium on Biomedical Imaging; May 2008; pp. 14–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barmpoutis A, Vemuri BC, Shepherd TM, Forder JR. Tensor splines for interpolation and approximation of DT-MRI with applications to segmentation of isolated rat hippocampi. TMI: Transactions on Medical Imaging. 2007 November;26(11):1537– 1546. doi: 10.1109/TMI.2007.903195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basser PJ, Mattiello J, Lebihan D. Estimation of the Effective Self-Diffusion Tensor from the NMR Spin Echo. J Magn Reson B. 1994;103:247–254. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- 6.Basser PJ, Pajevic S. A normal distribution for tensor-valued random variables: Applications to diffusion tensor MRI. IEEE Trans on Medical Imaging. 2003;22:785–794. doi: 10.1109/TMI.2003.815059. [DOI] [PubMed] [Google Scholar]
- 7.Basser PJ, Pajevic S. Spectral decomposition of a 4th-order covariance tensor: Applications to diffusion tensor MRI. Signal Processing. 2007;87:220–236. [Google Scholar]
- 8.Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor mri. J Magn Reson B. 1996;111(3):209–19. doi: 10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- 9.Cercignania M, Inglesea M, Pagania E, Comia G, Filippi M. Mean diffusivity and fractional anisotropy histograms of patients with multiple sclerosis. American Journal of Neuroradiology. 2001;22:952–958. [PMC free article] [PubMed] [Google Scholar]
- 10.Chefd’Hotel C, Tschumperlé D, Deriche R, Faugeras OD. Regularizing flows for constrained matrix-valued images. J Mathematical Imaging and Vision. 2004;20:147–162. [Google Scholar]
- 11.Descoteaux M, Angelino E, Fitzgibbons S, Deriche R. Regularized, fast and robust analytic q-ball imaging. MRM. 2007;58(3):497–510. doi: 10.1002/mrm.21277. [DOI] [PubMed] [Google Scholar]
- 12.Duda RO, Hart PE, Stork DG. Pattern Classification. Wiley; 2001. [Google Scholar]
- 13.Evren Ozarslan THM, Vemuri Baba C. Generalized scalar measures for diffusion mri using trace, variance, and entropy. Magn Reson Med. 2005;53(4):866–76. doi: 10.1002/mrm.20411. [DOI] [PubMed] [Google Scholar]
- 14.Fletcher P, Joshi S. Principal geodesic analysis on symmetric spaces: Statistics of diffusion tensors. Proc. of CVAMIA; 2004. pp. 87–98. [Google Scholar]
- 15.Gil-Garcia RJ, Badia-Contelles JM, Pons-Porrata A. A general framework for agglomerative hierarchical clustering algorithms. Pattern Recognition, 2006. ICPR 2006. 18th International Conference on; 2006. pp. 569–572. [Google Scholar]
- 16.Hasan KM, Gupta RK, Santos RM, Wolinsky JS, Narayana PA. Diffusion tensor fractional anisotropy of the normal-appearing seven segments of the corpus callosum in healthy adults and relapsing-remitting multiple sclerosis patients. Journal of Magnetic Resonance Imaging. 2005;21(6):735–743. doi: 10.1002/jmri.20296. [DOI] [PubMed] [Google Scholar]
- 17.Hilbert D. Über die darstellung definiter formen als summe von formenquadraten. Math Ann. 1888;32:342–350. [Google Scholar]
- 18.Ito M, Watanabe H, Kawai Y, Atsuta N, Tanaka F, Naganawa S, Fukatsu H, Sobue G. Usefulness of combined fractional anisotropy and apparent diffusion coefficient values for detection of involvement in multiple system atrophy. Journal of Neurology, Neurosurgery, and Psychiatry. 2007;78:722–728. doi: 10.1136/jnnp.2006.104075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwasawa K. On some types of topological groups. The Annals of Mathematics. 1949;50(3):507–558. [Google Scholar]
- 20.Jian B, Vemuri BC. Metric learning using iwasawa decomposition; IEEE International Conference on Computer Vision; 2007. pp. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jian B, Vemuri BC, Ozarslan E, Carney PR, Mareci TH. A novel tensor distribution model for the diffusion-weighted MR signal. NeuroImage. 2007;37(1):164–176. doi: 10.1016/j.neuroimage.2007.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jian B, et al. A unified computational framework for deconvolution to reconstruct multiple fibers from diffusion weighted MRI. TMI. 2007;26(11):1464–1471. doi: 10.1109/TMI.2007.907552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenglet C, Rousson M, Deriche R, Faugeras O. Statistics on the manifold of multivariate normal distributions: Theory and application to diffusion tensor MRI processing. Journal of Mathematical Imaging and Vision. 2006;25(3):423–444. [Google Scholar]
- 24.Moakher M. Fourth-order cartesian tensors: Old and new facts, notions and applications. Quarterly Journal of Mechanics and Applied Mathematics. 2008 [Google Scholar]
- 25.Moakher M, Norris AN. The closest elastic tensor of arbitrary symmetry to an elasticity tensor of lower symmetry. Journal of Elasticity. 2006;85(3):215–263. [Google Scholar]
- 26.Nath K, Husain M, Trivedi R, Kumar R, Prasad KN, Rathore R, Gupta R. Clinical implications of increased fractional anisotropy in meningitis associated with brain abscess. Journal of Computer Assisted Tomography. 2007;31(6):888–893. doi: 10.1097/rct.0b013e3180547118. [DOI] [PubMed] [Google Scholar]
- 27.Ozarslan E, Mareci TH. Generalized diffusion tensor imaging and analytical relationships between DTI and HARDI. MRM. 2003 Nov;50(5):955–965. doi: 10.1002/mrm.10596. [DOI] [PubMed] [Google Scholar]
- 28.Özarslan E, Shepherd TM, Vemuri BC, Blackband SJ, Mareci TH. Resolution of complex tissue microarchitecture using the diffusion orientation transform (DOT) NeuroImage. 2006;31:1086–1103. doi: 10.1016/j.neuroimage.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 29.Özarslan E, Vemuri BC, Mareci T. Fiber orientation mapping using generalized diffusion tensor imaging. ISBI. 2004:1036–1038. [Google Scholar]
- 30.Pennec X, Fillard P, Ayache N. A Riemannian framework for tensor computing. International Journal of Computer Vision. 2005;65 doi: 10.1007/11566489_116. [DOI] [PubMed] [Google Scholar]
- 31.Powers V, Reznick B. Notes towards a constructive proof of Hilbert’s theorem on ternary quartics. Quadratic Forms and Their Applications (Dublin, 1999), Contemp Math 272, Am Math Soc, Providence, RI. 2000:209–227. [Google Scholar]
- 32.Powers V, Reznick B, Scheiderer C, Sottile F. A new approach to Hilbert’s theorem on ternary quartics. C R Acad sci Paris. 2004;339:617–620. [Google Scholar]
- 33.Ptak T, Sheridan RL, Rhea JT, Gervasini AA, Yun JH, Curran MA, Borszuk P, Petrovick L, Novelline RA. Cerebral fractional anisotropy score in trauma patients: a new indicator of white matter injury after trauma. AJR Am J Roentgenol. 2003 Nov;181(5):1401–7. doi: 10.2214/ajr.181.5.1811401. [DOI] [PubMed] [Google Scholar]
- 34.Rudin W. Sums of squares of polynomials. Am Math Monthly. 2000;107:813–821. [Google Scholar]
- 35.Söderman O, Jönsson B. Restricted diffusion in cylindrical geometry. J Magn Reson, A. 1995;(117):94–97. [Google Scholar]
- 36.Terras A. Harmonic Analysis on Symmetric Spaces and Applications. Vol. 2. Springer; 1985. [Google Scholar]
- 37.Tournier J, Yeh CH, Calamante F, Cho KH, Connelly A, Lin CP. Resolving crossing fibres using constrained spherical deconvolution: Validation using diffusion-weighted imaging phantom data. NeuroImage. 2008;42(2):617–625. doi: 10.1016/j.neuroimage.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 38.Tournier JD, Calamante F, Connelly A. Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. NeuroImage. 2007;35(4):1459–1472. doi: 10.1016/j.neuroimage.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 39.Tuch D. Q-ball imaging. Magn Reson Med. 2004;52:1358–1372. doi: 10.1002/mrm.20279. [DOI] [PubMed] [Google Scholar]
- 40.Tuch DS, et al. Diffusion MRI of complex neural architecture. Neuron. 2003;40:885– 895. doi: 10.1016/s0896-6273(03)00758-x. [DOI] [PubMed] [Google Scholar]
- 41.van Gelderen P, de Vleeschouwer MHM, DesPres D, Pekar J, van Zijl PCM, Moonen CTW. Water diffusion and acute stroke. Magnetic Resonance in Medicine. 1994;31(2):154–163. doi: 10.1002/mrm.1910310209. [DOI] [PubMed] [Google Scholar]
- 42.Wang Z, Vemuri BC, Chen Y, Mareci T. A constrained variational principle for direct estimation and smoothing of the diffusion tensor field from dwi. IPMI. 2003:660–671. doi: 10.1007/978-3-540-45087-0_55. [DOI] [PubMed] [Google Scholar]
- 43.Wang Z, Vemuri BC, Chen Y, Mareci TH. A constrained variational principle for direct estimation and smoothing of the diffusion tensor field from complex dwi. IEEE Trans Med Imaging. 2004;23(8):930–939. doi: 10.1109/TMI.2004.831218. [DOI] [PubMed] [Google Scholar]
- 44.Ward P, Counsell S, Allsop J, Cowan F, Shen Y, Edwards D, Scia M, Rutherford M. Reduced fractional anisotropy on diffusion tensor magnetic resonance imaging after hypoxicischemic encephalopathy. Pediatrics. 2006;117(4):619–630. doi: 10.1542/peds.2005-0545. [DOI] [PubMed] [Google Scholar]