Abstract
Recently, a novel oxysterol, 5-cholesten-3β, 25-diol 3-sulfate (25HC3S) was identified in primary rat hepatocytes following overexpression of the cholesterol transport protein, StarD1. This oxysterol was also detected in human liver nuclei. In the present study, 25HC3S was chemically synthesized. Addition of 25HC3S (6 μM) to human hepatocytes markedly inhibited cholesterol biosynthesis. Quantitative RT-PCR and Western blot analysis showed that 25HC3S strongly decreased HMG-CoA reductase mRNA and protein levels. Coincidently, 25HC3S inhibited the activation of sterol regulatory element binding proteins (SREBPs), suggesting that inhibition of cholesterol biosynthesis occurred via blocking SREBP-1 activation, and subsequently inhibiting the expression of HMG CoA reductase. 25HC3S decreased SREBP-1 mRNA levels and inhibited the expression of target genes encoding acetyl CoA carboxylase-1 (ACC-1) and fatty acid synthase (FAS). In contract, 25-hydroxycholesterol increased SREBP1 and FAS mRNA levels in primary human hepatocytes. The results imply that 25HC3S is a potent regulator of SREBPs mediated lipid metabolism.
Keywords: cholesterol and lipids metabolism, HMG-CoA Reductase, bile acid synthesis, acetyl CoA carboxylase-1, fatty acid synthase, SREBPs, mitochondria, 5-cholesten-3β, 25-diol 3-sulfate
Introduction
The “acidic” pathway of bile acid biosynthesis is initiated by the mitochondrial enzyme, sterol 27-hydroxylase (CYP27A1) (1). Oxysterol intermediates of the “acidic” pathway such as 27-hydroxycholesterol and 25-hydroxycholesterol have been shown to be regulators of cholesterol homeostasis (2–8). These oxysterols regulate expression of many genes encoding enzymes involved in cholesterol and lipid metabolism (3;9;10). Increased CYP27A1 activities in peripheral tissues can down-regulate cholesterol biosynthesis (6). However, the relationship between the CYP27A1 activity and intracellular cholesterol metabolism is unknown. It is possible that the regulatory oxysterols generated by the CYP27A1 pathway inhibit cholesterol biosynthesis, and increasing the expression of cholesterol transporters, such as ABCA1 and ABCG8, subsequently enhancing the cellular efflux of cholesterol via activation of liver oxysterol receptor, LXR.
Previous reports showed that overexpression of the gene encoding the steroidogenic acute regulatory protein (StarD1), a protein which facilitates cholesterol delivery into mitochondria, dramatically increases cholesterol catabolism to bile acids both in primary hepatocytes in culture and in vivo (11;12). These findings suggested that cholesterol delivery to the mitochondria, where the enzyme CYP27A1 is localized, is the rate-determining step for bile acid synthesis via the “acidic” pathway. Subsequently, StarD1 was detected in hepatocytes (13). Overexpression of the gene encoding StarD1 in vivo not only increased bile acid synthesis to the same level as overexpression of CYP7A1, but also produced a similar composition of bile acids in bile (12). Recently, a novel sulfated oxysterol, 5-cholesten-3β, 25-diol 3-sulfate (sulfated 25-hydroxycholesterol, 25HC3S) was found and characterized in mitochondria and nuclei of primary rat hepatocytes following overexpression of StarD1. This oxysterol was subsequently found to be present in human liver nuclei (14). In addition, this oxysterol could be synthesized by hydroxysteroid sulfotransferase (SULT2B1b) during the incubation of cholesterol with mitochondrial and cytosol fractions in the presence of 3′-phosphoadenosyl 5′-phosphosulfate (PAPS) (40). These observations suggest that the presence of this compound may have a physiological significance. However, the function of 25HC3S remains unknown.
Recent report showed that overexpression of SULT2B1b inactivates oxysterol-LXR signaling in several cultured mammalian cell lines but does not alter receptor response to the nonsterol agonist (T0901317) (15). Furthermore, triple-knockout mice deficient in the biosynthesis of three oxysterol ligands of LXRs, 24S-hydroxycholesterol, 25-hydroxycholesterol, and 27-hydroxysterol, respond to dietary T0901317 by inducing LXR targeting genes in liver but show impaired responses to dietary cholesterol (15). The results suggested that oxysterols are in vivo ligands for LXR and sulfation of oxysterols inactivates the LXR signaling activity (15).
In the present study, we present evidence that 25HC3S decreases expressions of HMG-CoA reductase, acetyl CoA carboxylase-1 (ACC-1), and fatty acid synthase (FAS) via inhibiting SREBP-1 expression and activation while 25-hydroxycholesterol increases SREBP-1 and FAS expression in primary human hepatocytes (PHH). The results suggest that 25HC3S may play an important role in the maintenance of intracellular cholesterol and lipid homeostasis in hepatocytes.
Materials and Methods
Materials
Cell culture reagents and supplies were purchased from GIBCO BRL (Grand Island, NY); [14C]Cholesterol and [3H]25-hydroxycholesterol from New England Nuclear (Boston, MA). [14C]27-OH Cholesterol was prepared as previously described (16). HepG2 cells were obtained from American Type Culture Collection (Rockville, MD). The reagents for real time RT-PCR were from AB Applied Biosystems (Warrington WA1 4 SR, UK). The chemicals used in this research were obtained from Sigma Chemical Co. (St. Louis, MO) or Bio-Rad Laboratories (Hercules, CA). Polyclonal rabbit antibodies against SREBP1, SREBP-2 and HMG-CoA reductase were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). All solvents were obtained from Fisher (Fair Lawn, NJ) unless otherwise indicated. The enhanced chemiluminescence (ECL) reagents were purchased from Amersham Biosciences (Piscataway, NJ). The testosterone and 27-hydroxycholesterol were obtained from Research Plus Inc. (Bayonne, NJ). LK6 20 × 20 cm thin layer chromatography (TLC) plates were purchased from Whatman Inc. (Clifton, NJ).
Chemical synthesis of 5-cholesten-3β, 25-diol 3-sulfate
A mixture of 25-hydroxycholesterol (402 mg, 1 mmol) and triethylamine-sulfur trioxide pyridine complex (160 mg, 1 mmol) in 5 ml of chloroform was stirred at 25°C for 7 days as previously described (17) with modification. After the solvent was evaporated at reduced pressure, products were purified by HPLC using a silica gel column and methylene chloride and methanol (5%) as mobile phase. The product was purified by reverse phase HPLC using C18 column as a white powder. The structure of the product was characterized by mass spectrum (MS) and nuclear magnetic resonance (NMR) spectroscopy analysis. The purity was determined by MS and HPLC.
Mass spectral analysis
The synthesized compound was analyzed by a MDS Sciex ABI 4000 Triple Quadrapole Mass Spectrometer (MDS Sciex, Toronto, Canada) with a Turbo IonSpray ionization (ESI) source. The mass spectrometer was operated in negative ion modes and data were acquired using full scan mode as previously described (14).
Proton nuclear magnetic resonance spectroscopy
Samples were prepared for 1H NMR analysis as described previously (18). Briefly, each sample, 1 mg, was dissolved in 0.5 ml of D2O and lyophilized to remove exchangeable protons. The residue was dissolved in 0.5 ml of dimethyl sulfoxide-d6/D2O (98:2, v/v). NMR spectra were obtained on spectrometers operating at 1H frequencies of 300 MHz.
Cell culture
Hepatoma cells line (HepG2) cells were grown in MEM containing non-essential amino acids, 30 mM NaHCO3, 10% FBS, 1 mM L-glutamine, 1 mM sodium pyruvate and 1% Pen/Strep and incubated at 37°C in 5% CO2. When cells reached ~90% confluence, the oxysterols in DMSO or in ethanol (final concentration, 0.1%) and/or [1-14C]acetate for cholesterol synthesis was added. Microsomal and cytosolic fractions were isolated from broken cells as described previously (19).
Primary human hepatocytes (PHH) were purchased from an NIH-approved facility (Liver Tissue Procurement Distribution System, Univ. of Minnesota). Cells were obtained from a random sampling of males and females, 18–69 yr of age. Experiments were performed as cells became available to corroborate findings in experiments conducted in HepG2 cells as previously described (20).
For study of HMG CoA expression regulation, HepG2 or PHH were cultured in the media as described above in the presence or absence of mevinolin (50 μM) and mevalonate (0.5 μM). After cultured for 48 hrs, oxysterols were added and cultured for another 6 hrs, the cells were harvested for determining mRNA and protein levels.
Determination of cholesterol biosynthesis by TLC and HPLC
After incubation of HepG2 cells in media containing different concentrations of 25HC3S as indicated for 6 hrs, cells in 60 mm dishes were given 3 ml of the same fresh medium containing 5 μCi of [1-14C]acetate. After 2 hr incubation at 37°C, the media was removed and the cells were washed twice with phosphate-buffered saline (PBS), harvested with rubber police as described, and collected in microcentrifuge tubes. The cells were sedimented by centrifugation and the pellets were washed three times by resuspension and sedimentation. Subcellular fractions, microsomal, cytosol, and nuclear, were isolated as previously described (19). The cellular or subcellular pellets were resuspended in 0.3 ml of PBS. To each sample, 1.5 μg of testosterone was added as an internal standard. The total lipids were extracted and separated by adding 3 volumes of chloroform:methanol (1:1). [14C]Cholesterol and hydroxycholesterols were isolated into chloroform phase and separated on TLC (toluene:acetyl acetate, 2/3, v/v/). [1-14C]acetate derivatives were visualized by Image Reader, Fujifilm BAS-1800 II.
For analysis of unlabeled sterol products, the extracted lipids were incubated with 2 units of cholesterol oxidase at 37°C for 20 min. The oxidation reaction was terminated by adding 1.5 ml of methanol followed by 0.5 ml of saturated KCl. The sterols were extracted twice using 3 ml of hexane. The hexane phase was collected and evaporated under a stream of nitrogen. The residues were dissolved in mobile phase solvents for HPLC analysis as previously described (8).
[1-14C]Acetate derivatives in the chloroform phase were analyzed by HPLC on an Ultrasphere Silica column (5 μ × 4.6 mm × 25 cm; Backman, USA) using HP Series 1100 solvent delivery system (Hewlett Packard) at 1.3 ml/min flow rate. The column was equilibrated and run in a solvent system of hexane:isopropanol:glacial acetic acid (965:25:10, v/v/v), as the mobile phase. The effluents were collected every 0.5 min (0.65 ml per fraction) except as indicated. The counts in [14C]acetate derivatives were determined by Scintillation Counting. The column was calibrated with [14C]cholesterol, [3H]25-hydroxycholesterol, and [14C]27-hydroxycholesterol.
Determination of mRNA levels of SREBPs and its targeting genes by real-time RT-PCR
Total RNA was isolated from HepG2 or PHH using SV Total RNA Isolation Kit (Promega). Two μg of total RNA was used for first-strand cDNA synthesis as recommended by manufacturer (Invitrogen). Real-time PCR was performed using SYBR Green on ABI 7500 Fast Real-Time PCR System (Applied Biosystems). The final reaction mixture contained 5~10 ng of cDNA, 100 nM of each primer, 10 μl of 2×SYBR® Green PCR Master Mix (Applied Biosystems), and RNase-free water to complete the reaction mixture volume to 20 μl. All reactions were performed in triplicate. The PCR was carried out for 40 cycles of 95°C for 15 s and 60°C for 1 min. The fluorescence was read during the reaction, allowing a continuous monitoring of the amount of PCR product. The data was normalized to internal control-β-actin or GAPDH mRNA. The sequences of primers are as shown in Supplemental Materials.
Western blot analysis
Microsomal fractions were isolated as previously described (19). Microsomal or total extracted proteins from the treated cells were separated on a 7.5% SDS-polyacrylamide denaturing gel. Following SDS-PAGE proteins were electrophoretically transferred to Immobilon-P (PVDF) membranes (Millipore). The membranes were then blocked at 25°C for 60 minutes in blocking buffer (PBS, pH 7.4, 0.1% Tween, 5% non-fat dry milk). Proteins were then incubated at 4°C for overnight with a rabbit polyclonal IgG against human SREBP1, SREBP-2, or HMG-CoA reductase. After washing with PBS, pH 7.4, containing 0.05% of Tween 20, goat anti-rabbit IgG-horse-radish peroxidase conjugate, 1:2500, in washing solution was added and incubated for 60 minutes. Protein bands were detected using the Amersham ECL plus Kit. Positive bands were quantitated by the Advanced Image Data Analyzer (Aida Inc., Straubenhardt, Germany).
Statistics
Data are reported as the mean ± standard deviation. Where indicated, data were subjected to t-test analysis and determined to be significantly different if p<0.05.
Results
Chemical synthesis of the nuclear oxysterol, 5-cholesten-3β, 25-diol 3-Sulfate
To study its role in cellular lipid homeostasis, 25HC3S was chemically synthesized and purified as shown in Supplement Materials. MS analysis of the synthesized compound shows the same molecular mass ion, m/z 481 as the “authentic” nuclear oxysterol (14), and the purified product was not contaminated by the starting material, 25-hydroxycholesterol, m/z 401. 1H NMR analysis shows that the proton resonance at C3 with multiple small (1.5 Hz) splits near 3.65 ppm in the spectrum of 25-hydroxycholesterol (starting material) is shifted to 4.20 ppm in the product spectrum, which confirms that a HSO3- group is added at the C3 position of 25-hydroxycholesterol (Fig 1 of Supplement Materials). The results indicate that the synthesized molecule is 5-cholesten-3β, 25-diol 3-sulphate (25HC3S).
25HC3S inhibits cholesterol biosynthesis
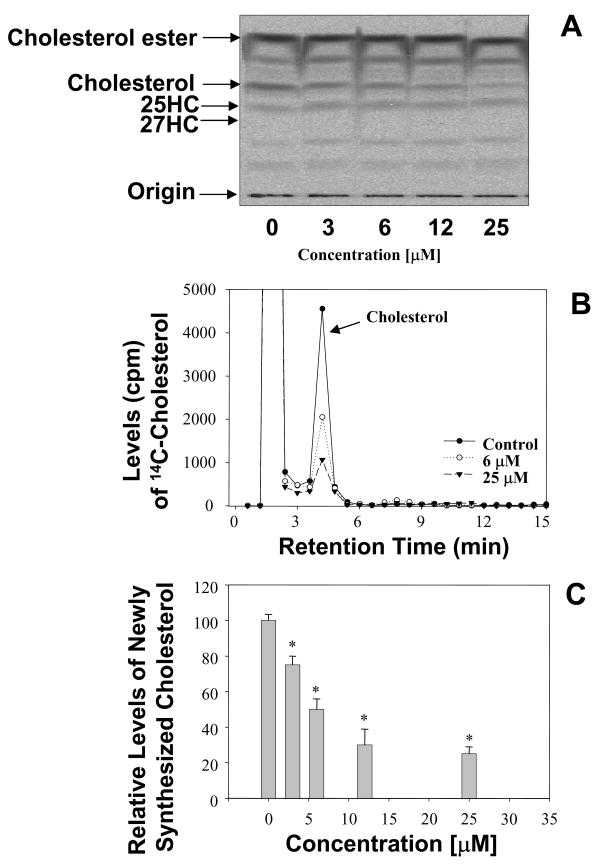
To examine the effects of 25HC3S on cholesterol biosynthesis, the rates of cholesterol synthesis were determined. The effects of 25HC3S on cholesterol biosynthesis were summarized in Fig. 1. After addition of 25HC3S, the cells were cultured for 6 hrs and with [1-14C]acetate for an additional 2 hrs, total lipids were extracted and partitioned. TLC analysis shows that [14C]cholesterol ester, [14C]cholesterol, and [14C]25-hydroxycholesterol were synthesized but [14C]27-hydroxycholesterol was not detected (Fig. 1A). Free [14C]cholesterol was found significantly decreased following addition of 25HC3S while the other labeled sterols did not significantly change in HepG2 and PHH. The decrease was concentration dependent (Fig 1A). The decreased amounts of [14C]cholesterol bands on the TLC were confirmed by HPLC analysis as shown in Fig 1B. The results are summarized in Fig. 1C.
Fig. 1. TLC and HPLC analysis of the newly synthesized [14C]cholesterol.
After incubation of the 25HC3S-treated cells with [1-14C]acetate, the total neutral lipids were extracted. TLC analysis of the [14C]-acetate derivatives extracted from the equivalent of 5 × 106 cells per each lane is shown in Panel A; HPLC analysis of the [14C]-acetate derivatives, Panel B. The column was calibrated by standards [14C]cholesterol, [14C]27-hydroxycholesterol, and [3H]25-hydroxycholesterol. A summary of three experiments of HPLC analysis, Panel C. Each bar represents the mean of three experiments ± standard deviation. The symbol * represents significant difference (p< 0.01).
It was observed that no 25-hydroxycholesterol could be detected in chloroform phase extracted from the cells treated with 25HC3S up to 25 μM (36μg) as shown Supplemental Materials. In contrast, concentration dependent levels of 25-hydroxycholesterol could be detected in the cells treated with 25-hydroxycholesterol as shown in figure 2 of Supplemental Materials.
25HC3S inhibits cholesterol biosynthesis by decreasing HMG-CoA reductase mRNA levels
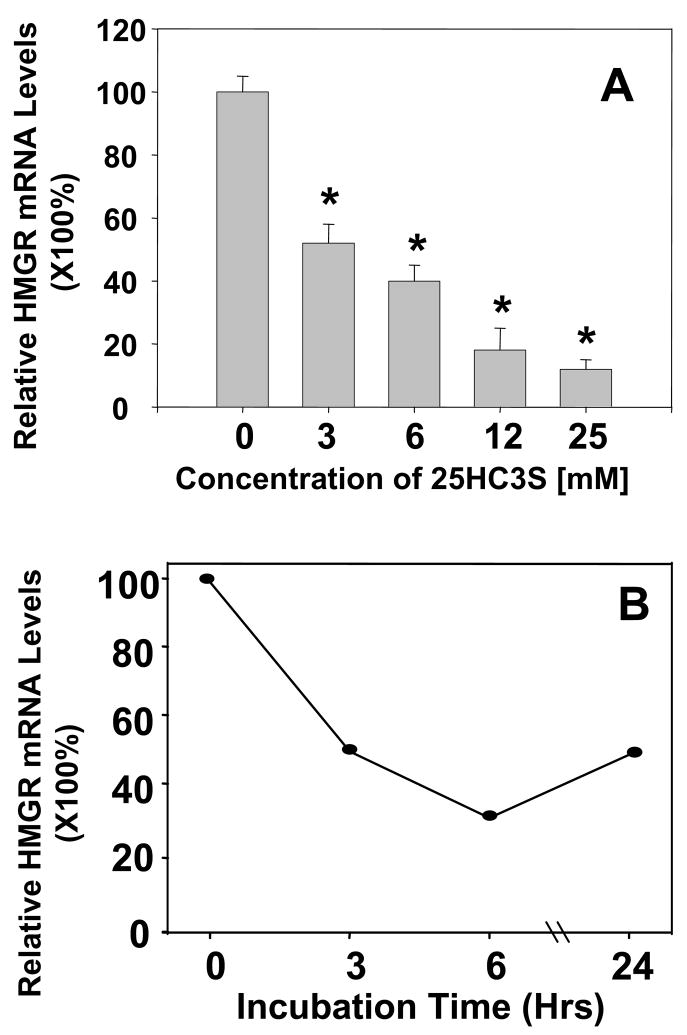
To investigate how 25HC3S inhibits cholesterol biosynthesis, total RNA were isolated from the treated HepG2 cells or PHH. The mRNA levels of HMG-CoA reductase were determined by real time RT-PCR. As shown in Fig. 2, decreases in HMG-CoA reductase mRNA levels following the addition of 25HC3S to the cells in culture were concentration (Fig. 2A) and time (Fig. 2B) dependent.
Fig. 2. 25HC3S regulates HMGR mRNA Expression.
Real time RT-PCR analysis of HMG-CoA reductase mRNA levels in HepG2 cells is shown in Panels A and B. The expression levels were normalized to GAPDH. The mRNA levels in the cells treated with 25HC3S at different concentrations as indicated are shown in Panel A (each value represents mean of three separate experiments ± standard derivation), and for the different times, Panel B. Each value represents mean of three experiments ± standard derivation. The symbol, *, represents significant difference (p< 0.05).
25HC3S inhibits HMG-CoA reductase expression by inhibition of both SREBP1 activation and expression in hepatocytes
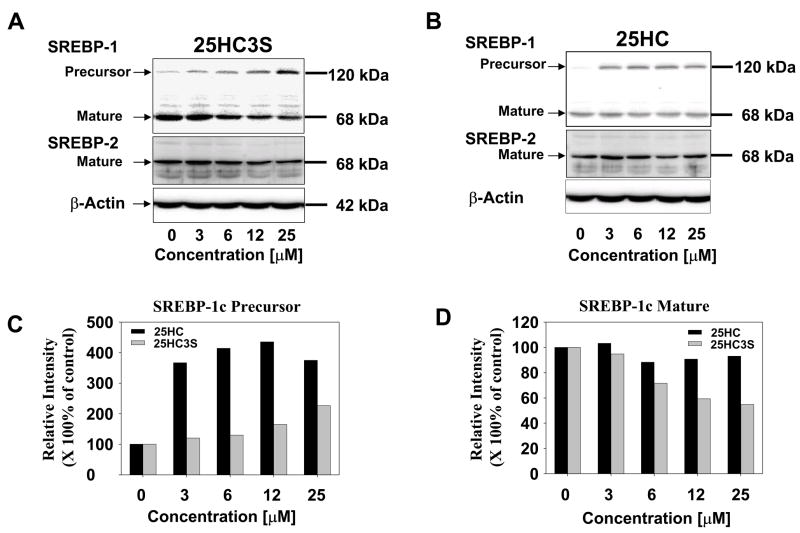
To study whether SREBP regulatory system is involved in the inhibition of HMG-CoA reductase mRNA levels and inhibition of cholesterol biosynthesis by 25HC3S, HepG2 or PHH were cultured in the presence of 50 μM of mevinolin and 0.5 μM of mevalonate. Under these conditions, HMG-CoA reductase expression is upregulated. Total cellular protein was extracted from the cells treated with 25HC3S or 25-hydroxycholesterol at different concentrations for 6 hrs. The precursor and mature forms of SREBPs were determined by Western blot analysis using specific antibodies (21;22). As expected, decreases of the mature forms of SREBP-1 (p<0.001) and SREBP-2, and increases (p<0.001) of the precursor form of SREBP-1 following addition of 25HC3S were concentration dependent (Figs. 3A, C, and D). However, 25-hydroxycholesterol increased SREBP-1 protein levels by 4-fold while mature form did not change significantly (Fig. 3B, C and D). It was observed that the activation of SREBP1 was much more sensitive to the treatment with 25HC3S than SREBP-2. The inhibition of SREBP-1 and SREBP-2 activation is consistent with the decrease in HMG-CoA reductase mRNA. Thus, 25HC3S inhibits the activation of SREBP-1 and SREBP-2, and subsequently inhibit the expression HMG-CoA reductase.
Fig. 3. Analysis of SREBPs expression and activation following 25HC3S treatment in HepG2 cells.
Western blot analysis of SREBP1 and SREBP-2 in HepG2 cells cultured in 10% of LPDS and treated with different concentrations of 25HC3S (Panels A) or 25-hydroxycholesterol (Panels B) in the presence of mevinolin (50 μM) and mevalonate (0.5 μM). The data represent a typical result from one of three independent experiments. The extracted protein (50 μg) was loaded onto each lane for each condition as indicated. Each positive band was quantified by the Image Data Analyzer. A summary of SREBP1 precursor levels is shown in Panel C and mature form levels in Panel D. The data represent mean of three independent experiments ± standard derivation. The symbol, *, represents significant difference (p< 0.05).
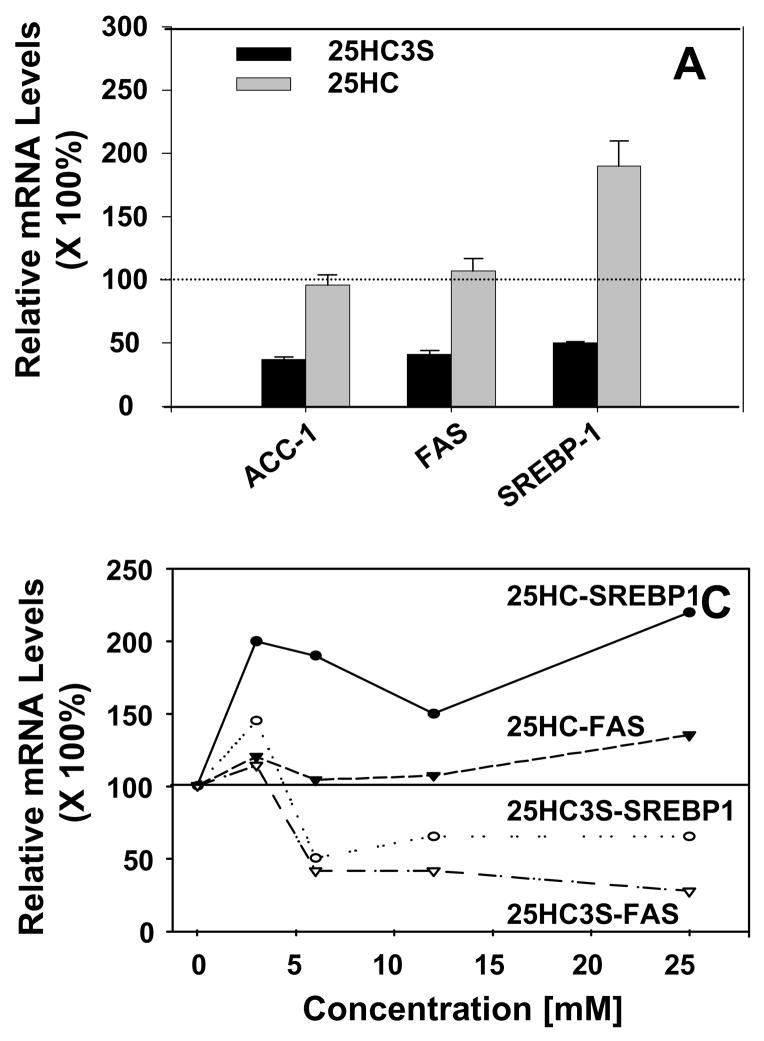
To further confirm 25HC3S functions in a different manner from 25-hydroxycholesterol, expressions of SREBP-1 and its targeting genes were determined in PHH as shown in Fig 4. At 6 uM, cultured for 6 hrs, addition of 25HC3S decreased SREBP1 mRNA by 50%, acetyl CoA carboxylase-1 (ACC-1) and fat acid synthase (FAS) by 60% (Fig. 6A). In contrast with 25HC3S and consistent with protein levels, 25-hydroxycholesterol increased SREBP1 mRNA levels by 2 folds and slightly increased FAS (Fig. 4A). The regulation of these genes was concentration dependent (Fig. 4B). Similar results were also observed in primary rat hepatocytes (data not shown).
Fig. 4. Real time RT-PCR analysis of expression of SREBP-1 targeting genes.
At 6 hr following addition of 6 μM of 25HC3S or 25-hydroxycholesterol to PHH culture (Panel A), or different concentration of 25HC3S or 25-hydroxycholesterol (Panel B), effects of 25HC3S and 25-hydroxycholesterol on FAS and SREBP1 expression were determined by real time RT-PCR. The expression levels were normalized to GAPDH. The values represent means ± SD (n =3).
Discussion
The 5-cholesten-3β, 25-diol 3-sulfate (25HC3S) unlike 25-hydroxysterol inhibits SREBP-1 expression and activation. Although there are many oxysterols reported to be LXR ligands and regulate LXR targeting gene expression (23), detail mechanism of the regulation is unclear. The identification of liver oxysterol receptors (LXRs) as regulators of lipid metabolism has been taken as strong evidence that oxysterols mediate intracellular lipids homeostasis. LXRs bind with high affinity to selective oxysterols, which are generally believed to be the most important activators for these receptors (24–29). However, administration of synthetic LXR ligands triggers induction of the lipogenic pathway, and elevates plasma triglyceride levels via induction of SREBP-1, a direct target of LXR (30). From a drug development standpoint, the most desirable compound would appear to be one that is a strong inducer of the elimination of lipids, yet lacks activity on the SREBP-1 and fatty acid synthase (FAS) promoters. The present results show that this hydrophilic 25HC3S, unlike 25-hydroxycholesterol, plays unique roles in cholesterol and lipids metabolism at both transcriptional and posttranslational levels: decreased SREBP-1 mRNA levels, inhibited SREBPs activations, and inhibited expression of SREBP-1 targeting genes, HMG-CoA reductase, ACC-1, and FAS. However, 25-Hydroxycholesterol increases SREBP-1 expression. The finding of addition of 25-hydroxycholesterol increased SREBP-1 expression is consistent with the results of administration of synthetic LXR ligands in vivo, which triggers induction of the hyperlipidemia via SREBP-1 (30). After 25-hydroxycholesterol is sulfated by SULT2B1b (40), the product, 25HC3S, decreases SREBP-1 expression and inhibits its activation by acting on the opposite direction against the substrate. Thus, 25HC3S may serve as a promising molecule for the therapy of hyperlipidemia.
Both 25-hydroxycholesterol and 25HC3S inhibit cholesterol biosynthesis but through different mechanism. 25HC3S inhibits cholesterol synthesis by inhibiting SREBP-1 expression and its activation. However, 25-hydrocholesterol inhibits the biosynthesis by increasing HMG CoA reductase fast degradation (ubiquitination) and decreasing SREBP-2 activation (3;10). Meanwhile, 25-hydroxycholesterol is able to increase SREBP-1 expression as LXR ligand (31), which is consistent with the present data that 25-hydroxycholesterol increases SREBP-1 mRNA levels (Fig. 4). 25HC3S may inhibit SREBP-1 expression by inactivating 25-hydroxycholesterol-induced LXR signaling. Recent report that overexpression SULT2B1b impairs oxysterol-induced LXR signaling (15) supports this hypothesis. Thus, the pair of the molecules, 25-hydroxycholesterol and 25HC3S, may play an important role in maintenance of intracellular lipid metabolism.
The sulfated oxysterol, 25HC3S, inhibits the cholesterol biosynthesis via decreasing mRNA levels and inhibiting the activation of SREBP-1, and subsequently inhibits the expression of HMG-CoA reductase, ACC-1, and FAS. Based on the microarray data from transgenic and knockout mice, both of SREBP-1 and SREBP-2 activation stimulate HMG-CoA reductase (32). SREBP-1 also strongly stimulates fatty acid synthase but SREBP-2 does not (32). Our results show that 25HC3S is much more potent in inhibiting SREBP-1 activation than SREBP-2. We believe that 25HC3S directly inhibits both SREBP-1 activation and subsequently inhibits HMG-CoA reductase resulting in the decline of cholesterol biosynthesis. However, Acyl CoA:cholesterol acyltransferase (ACAT) and CYP27A1 for the synthesis of cholesterol ester and 25HC are not SREBP targeting genes (32), which support the result that 25HC3S does not affect the synthesis of cholesterol ester and 25HC (Fig. 1). SREBPs play important roles not only in cholesterol synthesis and fatty acid metabolism but also many important biological events (33–39). It will be interesting to investigate whether these oxysterols are also involved in these events.
Supplementary Material
Table 1.
Primer sets for real time RT-PCR
| Gene Name | Gene Bank # | Forward Sequence | Reverse Sequence |
|---|---|---|---|
| hGAPDH | NM_002046 | CAATGACCCCTTCATTGACC | TTGATTTTGGAGGGATCTCG |
| hHMGR | NM_000859 | GTCATTCCAGCCAAGGTTGT | GGGACCACTTGCTTCCATTA |
| hFAS | NM_004104 | TGTGGACATGGTCACGGAC | GGCATCAAACCTAGACAGGTC |
| hACC1 | NM_198834 | TCGCTTTGGGGGAAATAAAGTG | ACCACCTACGGATAGACCGC |
| hSREBP1 | NM_004176 | CAGCCCCACTTCATCAAGG | ACTGTTGCCAAGATGGTTCCG |
| hSREBP-2 | NM_004599 | AACGGTCATTCACCCAGGTC | GGCTGAAGAATAGGAGTTGCC |
Acknowledgments
We acknowledge excellent technical help from Elaine Studer, Dalila Marques and Kaye Redford.
This work was supported by grants from the National Institutes of Health (R01 HL078898 and P01 DK38030, and Veterans Administration). D.R-A. is the Recipient of an American Heart Association Postdoctoral Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Vlahcevic ZR, Jairath SK, Heuman DM, Stravitz RT, Hylemon PB, Avadhani NG, Pandak WM. Transcriptional regulation of hepatic sterol 27-hydroxylase by bile acids. Am J Physiol. 1996;270:G646–G652. doi: 10.1152/ajpgi.1996.270.4.G646. [DOI] [PubMed] [Google Scholar]
- 2.Dubrac S, Lear SR, Ananthanarayanan M, Balasubramaniyan N, Bollineni J, Shefer S, Hyogo H, Cohen DE, Blanche PJ, Krauss RM, et al. Role of CYP27A in cholesterol and bile acid metabolism. J Lipid Res. 2005;46:76–85. doi: 10.1194/jlr.M400219-JLR200. [DOI] [PubMed] [Google Scholar]
- 3.Adams CM, Reitz J, De Brabander JK, Feramisco JD, Li L, Brown MS, Goldstein JL. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J Biol Chem. 2004;279:52772–52780. doi: 10.1074/jbc.M410302200. [DOI] [PubMed] [Google Scholar]
- 4.Babiker A, Andersson O, Lund E, Xiu RJ, Deeb S, Reshef A, Leitersdorf E, Diczfalusy U, Bjorkhem I. Elimination of cholesterol in macrophages and endothelial cells by the sterol 27-hydroxylase mechanism. J Biol Chem. 1997;272:26253–26261. doi: 10.1074/jbc.272.42.26253. [DOI] [PubMed] [Google Scholar]
- 5.Bjorkhem I. Do oxysterols control cholesterol homeostasis? J Clin Invest. 2002;110:725–730. doi: 10.1172/JCI16388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu X, Menke JG, Chen Y, Zhou G, Macnaul KL, Wright SD, Sparrow CP, Lund EG. 27-hydroxycholesterol is an endogenous ligand for liver X receptor in cholesterol-loaded cells. J Biol Chem. 2001;276:38378–38387. doi: 10.1074/jbc.M105805200. [DOI] [PubMed] [Google Scholar]
- 7.Meir K, Kitsberg D, Alkalay I, Szafer F, Rosen H, Shpitzen S, Avi LB, Staels B, Fievet C, Meiner V, et al. Human sterol 27-hydroxylase (CYP27) overexpressor transgenic mouse model. Evidence against 27-hydroxycholesterol as a critical regulator of cholesterol homeostasis. J Biol Chem. 2002;277:34036–34041. doi: 10.1074/jbc.M201122200. [DOI] [PubMed] [Google Scholar]
- 8.Li X, Hylemon P, Pandak WM, Ren S. Enzyme activity assay for cholesterol 27-hydroxylase in mitochondria. J Lipid Res. 2006;47:1507–1512. doi: 10.1194/jlr.M600117-JLR200. [DOI] [PubMed] [Google Scholar]
- 9.Bjorkhem I, Diczfalusy U. Oxysterols: friends, foes, or just fellow passengers? Arterioscler Thromb Vasc Biol. 2002;22:734–742. doi: 10.1161/01.atv.0000013312.32196.49. [DOI] [PubMed] [Google Scholar]
- 10.Corsini A, Verri D, Raiteri M, Quarato P, Paoletti R, Fumagalli R. Effects of 26-aminocholesterol, 27-hydroxycholesterol, and 25-hydroxycholesterol on proliferation and cholesterol homeostasis in arterial myocytes. Arterioscler Thromb Vasc Biol. 1995;15:420–428. doi: 10.1161/01.atv.15.3.420. [DOI] [PubMed] [Google Scholar]
- 11.Pandak WM, Ren S, Marques D, Hall E, Redford K, Mallonee D, Bohdan P, Heuman D, Gil G, Hylemon P. Transport of cholesterol into mitochondria is rate-limiting for bile acid synthesis via the alternative pathway in primary rat hepatocytes. J Biol Chem. 2002;277:48158–48164. doi: 10.1074/jbc.M205244200. [DOI] [PubMed] [Google Scholar]
- 12.Ren S, Hylemon PB, Marques D, Gurley E, Bodhan P, Hall E, Redford K, Gil G, Pandak WM. Overexpression of cholesterol transporter StAR increases in vivo rates of bile acid synthesis in the rat and mouse. Hepatology. 2004;40:910–917. doi: 10.1002/hep.20382. [DOI] [PubMed] [Google Scholar]
- 13.Hall EA, Ren S, Hylemon PB, Rodriguez-Agudo D, Redford K, Marques D, Kang D, Gil G, Pandak WM. Detection of the steroidogenic acute regulatory protein, StAR, in human liver cells. Biochim Biophys Acta. 2005;1733:111–119. doi: 10.1016/j.bbalip.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Ren S, Hylemon P, Zhang ZP, Rodriguez-Agudo D, Marques D, Li X, Zhou H, Gil G, Pandak WM. Identification of a novel sulfonated oxysterol, 5-cholesten-3beta, 25-diol 3-sulfonate, in hepatocyte nuclei and mitochondria. J Lipid Res. 2006;47:1081–1090. doi: 10.1194/jlr.M600019-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Chen W, Chen G, Head DL, Mangelsdorf DJ, Russell DW. Enzymatic reduction of oxysterols impairs LXR signaling in cultured cells and the livers of mice. Cell Metab. 2007;5:73–79. doi: 10.1016/j.cmet.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez-Agudo D, Ren S, Hylemon PB, Redford K, Natarajan R, Del Castillo A, Gil G, Pandak WM. Human StarD5, a cytosolic StAR-related lipid binding protein. J Lipid Res. 2005;46:1615–1623. doi: 10.1194/jlr.M400501-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Dusza JP, Joseph JP, Bernstein S. The preparation of estradiol-17 beta sulfates with triethylamine-sulfur trioxide. Steroids. 1985;45:303–315. doi: 10.1016/0039-128x(85)90079-0. [DOI] [PubMed] [Google Scholar]
- 18.Ren S, Scarsdale JN, Ariga T, Zhang Y, Klein RA, Hartmann R, Kushi Y, Egge H, Yu RK. O-acetylated gangliosides in bovine buttermilk. Characterization of 7-O-acetyl, 9-O-acetyl, and 7, 9-di-O-acetyl GD3. J Biol Chem. 1992;267:12632–12638. [PubMed] [Google Scholar]
- 19.Ren S, Hylemon P, Marques D, Hall E, Redford K, Gil G, Pandak WM. Effect of increasing the expression of cholesterol transporters (StAR, MLN64, and SCP-2) on bile acid synthesis. J Lipid Res. 2004;45:2123–2131. doi: 10.1194/jlr.M400233-JLR200. [DOI] [PubMed] [Google Scholar]
- 20.Hall E, Hylemon P, Vlahcevic Z, Mallonee D, Valerie K, Avadhani N, Pandak W. Overexpression of CYP27 in hepatic and extrahepatic cells: role in the regulation of cholesterol homeostasis. Am J Physiol Gastrointest Liver Physiol. 2001;281:G293–G301. doi: 10.1152/ajpgi.2001.281.1.G293. [DOI] [PubMed] [Google Scholar]
- 21.Yeh M, Cole AL, Choi J, Liu Y, Tulchinsky D, Qiao JH, Fishbein MC, Dooley AN, Hovnanian T, Mouilleseaux K, et al. Role for sterol regulatory element-binding protein in activation of endothelial cells by phospholipid oxidation products. Circ Res. 2004;95:780–788. doi: 10.1161/01.RES.0000146030.53089.18. [DOI] [PubMed] [Google Scholar]
- 22.Zeng L, Liao H, Liu Y, Lee TS, Zhu M, Wang X, Stemerman MB, Zhu Y, Shyy JY. Sterol-responsive element-binding protein (SREBP) 2 down-regulates ATP-binding cassette transporter A1 in vascular endothelial cells: a novel role of SREBP in regulating cholesterol metabolism. J Biol Chem. 2004;279:48801–48807. doi: 10.1074/jbc.M407817200. [DOI] [PubMed] [Google Scholar]
- 23.Schroepfer GJ., Jr Oxysterols: modulators of cholesterol metabolism and other processes. Physiol Rev. 2000;80:361–554. doi: 10.1152/physrev.2000.80.1.361. [DOI] [PubMed] [Google Scholar]
- 24.Alberti S, Steffensen KR, Gustafsson JA. Structural characterisation of the mouse nuclear oxysterol receptor genes LXRalpha and LXRbeta. Gene. 2000;243:93–103. doi: 10.1016/s0378-1119(99)00555-7. [DOI] [PubMed] [Google Scholar]
- 25.Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 26.Janowski BA, Grogan MJ, Jones SA, Wisely GB, Kliewer SA, Corey EJ, Mangelsdorf DJ. Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc Natl Acad Sci USA. 1999;96:266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landis MS, Patel HV, Capone JP. Oxysterol activators of liver X receptor and 9-cis-retinoic acid promote sequential steps in the synthesis and secretion of tumor necrosis factor-alpha from human monocytes. J Biol Chem. 2002;277:4713–4721. doi: 10.1074/jbc.M108807200. [DOI] [PubMed] [Google Scholar]
- 28.Malerod L, Juvet LK, Hanssen-Bauer A, Eskild W, Berg T. Oxysterol-activated LXRalpha/RXR induces hSR-BI-promoter activity in hepatoma cells and preadipocytes. Biochem Biophys Res Commun. 2002;299:916–923. doi: 10.1016/s0006-291x(02)02760-2. [DOI] [PubMed] [Google Scholar]
- 29.Peet DJ, Janowski BA, Mangelsdorf DJ. The LXRs: a new class of oxysterol receptors. Curr Opin Genet Dev. 1998;8:571–575. doi: 10.1016/s0959-437x(98)80013-0. [DOI] [PubMed] [Google Scholar]
- 30.Grefhorst A, Elzinga BM, Voshol PJ, Plosch T, Kok T, Bloks VW, van der Sluijs FH, Havekes LM, Romijn JA, Verkade HJ, et al. Stimulation of lipogenesis by pharmacological activation of the liver X receptor leads to production of large, triglyceride-rich very low density lipoprotein particles. J Biol Chem. 2002;277:34182–34190. doi: 10.1074/jbc.M204887200. [DOI] [PubMed] [Google Scholar]
- 31.Bocher V, Millatt LJ, Fruchart JC, Staels B. Liver X receptors: new players in atherogenesis? Curr Opin Lipidol. 2003;14:137–143. doi: 10.1097/00041433-200304000-00004. [DOI] [PubMed] [Google Scholar]
- 32.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bobard A, Hainault I, Ferre P, Foufelle F, Bossard P. Differential regulation of SREBP-1c transcriptional activity by insulin and LXR during liver development. J Biol Chem. 2004 doi: 10.1074/jbc.M406522200. [DOI] [PubMed] [Google Scholar]
- 34.Botolin D, Jump DB. Selective proteolytic processing of rat hepatic sterol regulatory element binding protein-1 (SREBP-1) and SREBP-2 during postnatal development. J Biol Chem. 2003;278:6959–6962. doi: 10.1074/jbc.M212846200. [DOI] [PubMed] [Google Scholar]
- 35.Castillo-Olivares A, Gil G. Differential effects of sterol regulatory binding proteins 1 and 2 on sterol 12 alpha-hydroxylase. SREBP-2 suppresses the sterol 12 alpha-hydroxylase promoter. J Biol Chem. 2002;277:6750–6757. doi: 10.1074/jbc.M106785200. [DOI] [PubMed] [Google Scholar]
- 36.Dif N, Euthine V, Gonnet E, Laville M, Vidal H, Lefai E. Insulin activates human sterol-regulatory-element-binding protein-1c (SREBP-1c) promoter through SRE motifs. Biochem J. 2006;400:179–188. doi: 10.1042/BJ20060499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seegmiller AC, Dobrosotskaya I, Goldstein JL, Ho YK, Brown MS, Rawson RB. The SREBP pathway in Drosophila: regulation by palmitate, not sterols. Dev Cell. 2002;2:229–238. doi: 10.1016/s1534-5807(01)00119-8. [DOI] [PubMed] [Google Scholar]
- 38.Wong J, Quinn CM, Brown AJ. SREBP-2 positively regulates transcription of the cholesterol efflux gene, ABCA1, by generating oxysterol ligands for LXR. Biochem J. 2006;400:485–491. doi: 10.1042/BJ20060914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng L, Liao H, Liu Y, Lee TS, Zhu M, Wang X, Stemerman MB, Zhu Y, Shyy JY. Sterol-responsive element-binding protein (SREBP) 2 down-regulates ATP-binding cassette transporter A1 in vascular endothelial cells: a novel role of SREBP in regulating cholesterol metabolism. J Biol Chem. 2004;279:48801–48807. doi: 10.1074/jbc.M407817200. [DOI] [PubMed] [Google Scholar]
- 40.Li X, Pandak MW, Erickson SK, Hylemon P, Ren S. A Novel Metabolic Pathway for the Synthesis of the Newly Discovered Nuclear 5-cholesten-3beta, 25-diol 3-sulphate. The FASEB J. 2007;21 (5):487–6. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.