Abstract
Cancer is a pathology that is associated with aberrant gene expression and an altered metabolism. While changes in gene expression have historically been attributed to mutations, it has become apparent that epigenetic processes also play a critical role in controlling gene expression during carcinogenesis. Global changes in epigenetic processes including DNA methylation and histone modifications have been observed in cancer. These epigenetic alterations can aberrantly silence or activate gene expression during the formation of cancer; however, the process leading to this epigenetic switch in cancer remains unknown. Carcinogenesis is also associated with metabolic defects that increase mitochondrially derived reactive oxygen species, create an atypical redox state, and change the fundamental means by which cells produce energy. Here, we summarize the influence of these metabolic defects on epigenetic processes. Metabolic defects affect epigenetic enzymes by limiting availability of the cofactors like S-adenosylmethionine. Increased production of reactive oxygen species alters DNA methylation and histone modifications in tumor cells by oxidizing DNMTs and HMTs, or through direct oxidation of nucleotide bases. Lastly, the Warburg effect and increased glutamine consumption in cancer influences histone acetylation and methylation by affecting the activity of sirtuins and histone demethylases.
Introduction
Carcinogenesis is operationally divided into three discreet steps; initiation, an irreversible genetic alteration or mutation that predisposes a clonogenic cell to cancer formation; promotion, the clonal expansion of an initiated clonogenic cell that increases the likelihood of additional events occurring on the background of the initiating mutation; and progression, the acquisition of additional genetic and epigenetic changes that lead to the generation of diverse phenotypes within a solid tumor during its evolution. During the progression stage in particular, gene expression is globally altered in cancers cells compared to the tissues from which they arise. These changes in gene expression have classically been attributed to the increased rate of mutation and genomic instability seen in cancer. However, over the past decade numerous studies have suggested that epigenetic alterations can be just as effectively alter gene expression in cancer. Epigenetics is managed by two major processes: cytosine methylation, and the post-translational modification of histone tails. Wide-ranging changes are observed for both processes in most types of cancer, and these changes constitute an epigenetic switch. Characterization of this epigenetic switch has clearly established epigenetic dysfunction as an intrinsic mechanism of carcinogenesis; however, while the effect of the epigenetic switch in cancer is well characterized, its cause remains elusive.
Another hallmark of tumor cells is a metabolic defect which is responsible for altering how tumor cells produce and utilize metabolites. Such metabolic changes lead to increased glycolysis, dysfunctional mitochondrial electron transport, aberrant production of oxidants, and the formation of an atypical redox state. Roles for each of these have been hypothesized to be causal in the initiation, promotion, and progression of the malignant phenotype [1-4]. These hypotheses have centered on the ability of these metabolic changes to elicit genetic alterations during carcinogenesis; however, these alterations are also concomitant with the epigenetic switch in cancer mentioned above. A connection between the epigenetic switch and metabolic defects of cancer was first suggested by Peter Cerutti in 1985. Cerutti aptly proposed that epigenetic processes were disrupted by metabolic defects to causally change gene expression in cancer [5]. However, since Cerutti the depth and breadth of our knowledge regarding the mechanisms of epigenetics and their complexity has grown significantly. We have previously reviewed the ability metabolic changes to elicit epigenetic changes during development [6]. The central theme for this review will be to discuss the novel relationship between metabolic defects and altered epigenetic processes in cancer. We will first discuss how aberrant production of mitochondrial oxidants influences the epigenetic cofactor SAM. Next we will discuss the relationship between the altered redox status of cancer cells and changes in epigenetic processes. Lastly we will introduce the novel concept of how defects in oxidative metabolism might directly influence epigenetics.
The metabolic defect of cancer
As early as the 1920's Otto Warburg and others were measuring fundamental changes in tumor cell metabolism [7, 8]. Today, gross metabolic alterations are found in all forms of cancer and center around two major changes: the Warburg effect, and alterations in mitochondrial electron transport. The Warburg effect describes the increased glucose consumption and glycolytic activity of tumor cells (for a recent review see [9]). Increased glucose consumption and glycolytic activity are common in rapidly dividing normal and tumor cells, however the Warburg effect of cancer is accompanied by increased lactate dehydrogenase activity to recycle NADH back into NAD+ and remove pyruvate [10, 11]. Lactic acid fermentation, or negative regulation of pyruvate dehydrogenase disrupts the use of pyruvate as a carbon entry source into the Krebs cycle [12, 13]. Combined, these defects shift the NAD+/NADH ratio. It is this aspect of the Warburg effect that we will relate to epigenetic processes in carcinogenesis. Even with limited amounts of pyruvate entering the Krebs cycle, tumor cells continue to produce limited energy via mitochondrial electron transport. Instead of pyruvate, tumor cells use amino acids, like glutamine, as carbon sources for the Krebs cycle. Beginning in the 1970's several groups reported increased glutamine oxidation in cancer [14-16]. Amino acids such as glutamine are great carbon sources for the Krebs cycle because their high concentrations facilitate their passive diffusion in cells, and they can easily be converted into Krebs cycle intermediates [10, 17]. Before glutamine can be used as a carbon source for the Krebs cycle it is first converted to glutamate by phosphate-activated glutaminase (PAG) then into α-ketoglutarate by glutamate dehydrogenase (GDH) [18, 19]. Modern reports suggest that aberrant glutamine consumption can be attributed to increased expression of both PAG and GDH [18]. Because of lactate fermentation, α-ketoglutarate derived from glutamine, not pyruvate from glucose, becomes the metabolite of entry into the Krebs cycle. This phenomenon has been termed a “truncated Krebs cycle” by Loris Baggetto in which the flux of carbon from α-ketoglutarate to oxaloacetate are much higher than that of citrate to α-ketoglutarate, suggesting carbon is entering the cycle by a means other than pyruvate (i.e. α-ketoglutarate) [17]. The most intriguing aspect of the metabolic defects mentioned above is that their severity increases with cancer progression. The Warburg effect and glutamine consumption are both higher in poorly differentiated advanced stages of cancer [17-19]. This observation suggests metabolic defects may have a causal role in carcinogenesis.
Free Radicals and Cancer
Tumor cells have increased production of reactive oxygen species (ROS) and an atypical redox balance. The fundamental change in tumor cells that increases their ROS production are defects in mitochondrial electron transport. Tumor cell mitochondrial are rife with structural and functional defects, and may either be the cause or result of increased ROS levels [20]. Confounding this relationship between ROS and mitochondrial defects is that these same cells often exhibit altered expression of antioxidant enzymes. Nevertheless, there is considerable evidence to suggest that the aberrant production of reactive oxygen species in the mitochondria leads to the accumulation of damage that drives carcinogenesis [1, 3, 4, 21]. The expression of antioxidant enzymes like MnSOD, GPx and catalase are often altered in cancer, thus leaving these cells more susceptible to damage from ROS [22, 23]. To remedy this, tumor cells decrease their use of mitochondrial electron transport and utilize pyruvate produced during glycolysis as a scavenger of peroxides [24]. But how can aberrant production of ROS drive carcinogenesis? It was first hypothesized by Oberley and Buettner in 1979 that the pro-oxidant state of cancer, created by decreased antioxidant capacity, generates mutations that drive the initiation and progression of cancer [1]. These ideas have since been expanded by others to include metabolic and redox changes in cancer [1, 3, 21]. The aforementioned hypotheses all centered on the ability of the pro-oxidant state to generate mutations that create the common phenotypic changes associated with cancer. However epigenetic changes are just as capable at inducing phenotypic changes in cancer.
Epigenetics
Epigenetics was a term first coined by Conrad Waddington in 1938. Waddington defined it as “the science concerned with the causal analysis of development” [25]. A more modern understanding of the mechanisms and principles of epigenetics has led to the unified definition of epigenetics proposed by Adrian Bird, where he describes it as “the structural adaptation of chromosomal regions so as to register, signal or perpetuate altered activity states” [26]. Several epigenetic processes govern the structural adaptation of chromosomes, but paramount among these are cytosine methylation and the post-translational modification of histones. These two mechanisms can work separately, or in concert to control the function of the genome. Epigenetics is most appreciated for its role in regulating transcription; however it has additional roles in DNA repair, DNA replication, and cell division [27, 28]. Loss of epigenetic control of transcription, DNA repair, replication, and cell division becomes relevant in many diseases, especially cancer.
DNA methylation
DNA is methylated almost exclusively at cytosines that are part of a CpG di-nucleotides [29]. CpG di-nucleotides are unusual for two reasons. First, they occur at approximately one fifth their expected frequency in the genome compared to the other possible nucleotide doublets, and second they are unevenly distributed towards gene regulatory elements and highly repetitive sequences [30]. CpG dense regions in gene regulatory regions are referred as “CpG islands” by epigeneticists and these have been characterized and operationally defined [31]. As a general rule, methylation of these CpGs is associated with transcriptional repression and condensed chromatin. CpG islands are generally unmethylated in normal cells; however smaller regions, called CpG clusters, can be hypermethylated and silence gene expression [32, 33]. Approximately half of all human genes have CpG dense regions located within their regulatory elements, hinting that this mechanism might play a role to regulate gene expression [34]. Methylated CpGs have also been observed in repetitive elements and centromeric repeats [35]. Loss of DNA methylation in these regions leads to gross changes in chromosome structure and function as observed in ICF syndrome and possibly cancer [36]. Methylated CpG di-nucleotides influence DNA in two ways, by inhibiting protein binding, and by serving as a substrate for new proteins to bind DNA. The addition of the methyl group to cytosine has little effect on the overall structure of DNA, however it can inhibit the binding of several “methylation sensitive” proteins such as CREB, NFkB, and AP2 [37-39]. Conversely, methylated CpGs can serve as specific binding sites that recruit methyl-CpG binding domain proteins (MBPs). With MBP binding comes histone modifying enzymes that influence chromatin structure at the site of CpG methylation. Thus, these proteins can serve as a linker between CpG methylation and histone modifications.
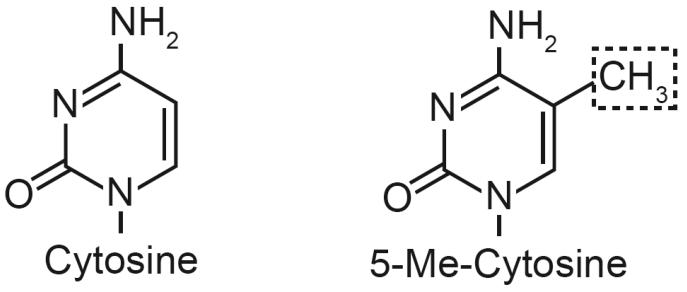
The methylation of cytosine is a post-replicative event, meaning it occurs after cytosine is incorporated into double stranded DNA. Catalyzing the methylation of cytosine at CpG di-nucleotide are several DNA methyltransferases (DNMTs). These enzymes specifically recognize CpGs as targets for methylation. DNMTs catalyze the transmethylation of cytosine by transferring methyl groups from S-adenosylmethionine (SAM) to position 5 of the pyrimidine ring to create 5-methylcytosine (Fig. 1). Humans have three DNA methyltransferases: DNMT1, DNMT3a and DMNT3b. These enzymes are subdivided into two classes based on their functionality in vivo: maintenance methylation, and de novo methylation [40]. Maintenance methylation is catalyzed by DNMT1 and occurs rapidly following DNA replication [41]. DNMT1 has high affinity for hemimethylated CpGs (CpG doublets where only one DNA strand is methylated), which it quickly converts into fully methylated CpG di-nucleotides [42]. DNMT1 is expressed in all cell types during S phase of the cell cycle, when its activity is highest [43]. It is through DNMT1 that the pattern of DNA methylation is recapitulated in daughter cells [44]. The de novo methyltransferases, DNMT3a and DNMT3b, are so named because they equally methylate hemimethylated and unmethylated CpGs [45]. Different genes encode these enzymes, but both exist as multiple splice variants that give these enzymes functional flexibility [45-49]. De novo methylation creates new epigenetic events, meaning these enzymes could possibly initiate gene silencing.
Fig. 1. Pyrimidine ring structures for cytosine and 5-methyl-cytosine (5-Me-C).
The addition of methyl group to position 5 of the cytosine nucleobase (dashed box) creates 5-Me-C in genomic DNA.
Modification of the nucleosome
Genetic information is packaged into higher order structures by nucleosomes. Each nucleosome encompasses ∼146 base pairs of DNA wrapped around an octamer of histone proteins. This octamer contains two H2A, H2B, H3, and H4 histone proteins. Another structural characteristic of the nucleosome are the “histone tails” that extend from the core octamer [50]. These tails consist of the N-termini of the histone proteins and are the main site for their post-translational modification. Modifying histones allows the nucleosome to have dynamic roles in transcription, DNA repair, DNA replication, and the cell cycle. The list of potential modifications includes: acetylation, methylation, phosphorylation, ADP-ribosylation, ubiquitination, and sumoylation (For review of these topics see [51-53]. A majority of these modifications take place at lysines, arginines and serines within histone tails (Fig. 2). Given the breadth of literature on this topic it is impossible to sufficiently cover all modifications in depth here. Therefore, for the sake of brevity, we will focus on lysine acetylation and methylation citing specific examples of how they control nucleosome function. Lysine acetylation is associated with active gene expression and open chromatin. H3K9ac and H4K16ac are two histone modifications often associated with euchromatin [54]. Histone methylation is more complex because the ε-nitrogen on lysine can be modified by up to three methyl groups, thus creating multiple degrees of lysine methylation. For example, lysine 4 of histone H3 can be umethylated, monomethylated (H3K4me1), dimethylated (H3K4me2), or trimethylated (H3K4me3). The complexity of histone methylation is further increased by the possibility that each degree may have a unique function. Histone methylation is associated with both active and silent genes, or as a fundamental mark of all histones. The recent development of Illumina® chromatin immunoprecipitation sequencing (ChIP Seq) has identified where these modifications reside and hint at their potential roles [55, 56]. Active genes have high levels of H3ac, H4ac, and H3K4me3 at their promoters and abundant H3K36me3 and H3K4me1 within coding regions [57]. Having specific modifications at defined tracks along the axis of expressed genes has led to the hypothesis that histone modifications may regulate both transcriptional initiation and elongation [58]. Modifications such as H3K9me3, H3K27me3, and H4K20me3 at promoters are indicative of transcriptionally silent heterochromatin [59-61]. By what means do these modifications influence DNA? Histone modifications actuate DNA/nucleosome interactions in to ways. First they alter the charge of histone tail to influence the contact between negatively charged DNA and the lysine-rich positively charged nucleosome. Second, modified histone tails can serve as binding sites for effector proteins that manipulate DNA. This facet of nucleosome function forms the central tenet of the “histone code” hypothesis, which proposes that the covalent modification of amino acids, and the interaction with one another, generates a language that is read by a series of effector proteins that act upon DNA [62]. Loss of the either the histone modification or the effector proteins would influence DNA function in similar ways. Two common examples of proteins that read the histone code are transcriptional repressors such as heterochromatin protein 1 (HP1) that binds H3K9me3, and lethal 3 malignant brain tumor 1 (L3MBTL1) which interacts with H4K20me1, both of which organize DNA into heterochromatin and silence gene expression [63-67].
Fig. 2. The sites of lysine acetylation and methylation in histone tails.
Histone tails protrude from the central globular domain of histone proteins and can be modified by acetylation and methylation in various ways. (* denotes lysine can be mono, di, or tri methylated)
Histone Acetylation
Establishment and maintenance of the histone code is accomplished by a multitude of enzymes that target specific amino acids for modification. Histone acetyltranferases (HATs) use Acetyl-CoA to add acetyl groups to lysines within histone tails. Mammalian histone acetyltransferases are divided into five distinct families: GNAT (Gcn5-related N-Acetyltransferase) superfamily, MYST (MOZ, YBF2/Sas3, Sas2, Tip60) family, p300/CBP, TAFII 250, and nuclear receptors [68]. Each of these families have different roles in regulating chromatin structure, however they share a common enzymatic activity. HATs transfer acetyl groups from acetyl CoA to lysine to form ε-Nacetyl lysine in histones and CoA [69, 70]. Recruitment of HAT activity to a gene regulatory element is generally associated with active transcription and open chromatin. Histone acetylation occurs at lysine residues in histones H2B, H3 and H4 (Fig. 2). Once acetylated, histone tails become sites for bromodomain containing effector proteins to bind and act upon the DNA or nucleosome [71]. Acetylation of histone tails is not a permanent modification. Histone deacetylases (HDACs) remove acetyl groups from histones, thus providing a certain degree of plasticity to the epigenetic control of gene expression [72, 73]. The presence of HDACs within gene regulatory regions is consistent with epigenetic gene silencing and closed chromatin. The HDACs are divided into three groups (Classes I-III) [74]. Combined, there are a total of ten Class I and II HDACs that utilize hydrolysis to remove acetyl groups, while class III HDACs include the sirtuin family of protein deacetylases. Sirtuins are a class of NAD+ dependent protein deacetylases that are commonly known for their role in increasing lifespan of yeast and C. elegans during caloric restriction [75]. The HDAC activity of sirtuins is intimately linked to metabolism by the NAD+/NADH ratio. With increased NAD+, sirtuins can more readily deacetylate histones and other proteins [76, 77].
Methylation of histone tails is carried out by several histone methyltransferases (HMTs). Methylation requires concerted activity among several protein complexes and HMTs whose full description go beyond the scope of this review (for reviews of these topics see [51, 78, 79]). In the context of this review we will focus on the relationship of HMTs with redox and metabolism. Members of HMT family can methylate lysine or arginines in histone tails (Fig. 2). Like DNMTs, HMTs use SAM as a cofactor during transmethylation and produce SAH as a byproduct. HMT substrate specificity and activity is centralized in SET domains and surrounding motifs [80, 81]. This specificity is responsible for creating the progressive methylation of lysines mentioned above. Some HMTs are exclusively monomethyltransferases (i.e. create Kme1), while others progressively methylate lysines to di and tri methylated forms. For example, PR-Set7 is the H4K20 monomethyltransferase that creates a substrate for SUV4-20H1/H2 to progressive methylate K20 to higher degrees of methylation [82, 83]. Until recently it was believed that histone methylation was a terminal event. This has changed with the discovery of lysine specific demethylase 1 (LSD1) and the jumonji C (JmjC) family of histone demethylases, collectively known as KDMs [84-86]. LSD1 specifically demethylates H3K4me2 and is a member of several transcriptional repression complexes [84, 87-89]. Histone demethylation by LSD1 uses oxygen as an electron acceptor to reduce methylated lysine to form lysine, formaldehyde, and hydrogen peroxide [84]. JmjC demethylases have a different mechanism than LSD1 that allows them to demethylate trimethylated lysine [85]. Like HMTs, the KDMs have target lysines. The most intriguing aspect of these proteins is the cofactors they require. JmjC demethylases all need molecular oxygen (O2), α-ketoglutarate, Fe2+, and ascorbate to demethylate mono, di or tri methyl lysines [85, 90].
The epigenetic switch in cancer
Cancer cells have an altered epigenotype compared to the tissues from which they arise. This subject has received much attention and has been the subject of numerous reviews [91-94]. Overall the epigenetic switch is summarized as changes in the level and placement of both DNA methylation and histone modifications. Many cancers acquire or increase the expression of epigenetic enzymes, yet the products of their reactions (i.e. methylated cytosine and modified histones) do not correlate and suggest there are other factors affecting their activity [95]. For over 20 years it has been known that tumor cells are globally hypomethyalted at CpGs compared to the tissues from which they arise, while at the same time cytosine methylation is increased in specific parts of the genome [96, 97]. In normal cells, CpGs within repetitive DNA elements and coding regions of genes are methylated. In tumor cells LINE-1 repeats, satellite DNA, and moderately repeated DNA sequences become unmethylated, while genes containing CpG cluster become hypermethylated, rendering them transcriptionally silent (For review see [35]). Prime candidates for this type of repression are tumor suppressor genes such as p16 and 14-3-3 sigma [98-100]. Methylated CpGs also form mutation hot spots within coding regions of tumor suppressor genes like p53 [101, 102]. The deamination of methylated cytosine forms thymine, creating a lesion that is difficult to correct because our DNA repair mechanisms cannot easily discriminate which base is correct in the resulting G:T mismatch. Proportional changes in histone modifications are also observed in cancer. Work from Manel Esteller's group has shown that loss of acetylation of lysine 16 and trimethylation of lysine 20 of histone H4 are common events in cancer cells [103]. Others have reported global decreases in H3K4me3, H3K9me3 and H3K27me3 in cancer [104, 105]. In many caners loss of these histone modifications is a predictive marker of disease outcome [104, 106]. Histone hypoacetylation can also silence tumor suppressor genes, while hyperacetylation can potentially activate oncogenes [107, 108]. Epigenetic alterations in cancer could also be affecting the stability of the genome, given the link between the organization of the genome and its repair and replication [27, 28]. While much descriptive work has shown the nature of such changes, the cause(s) of the epigenetic switch in cancer remains unknown. At the core of the epigenetic switch is likely the inability of enzymes like DNMTs, HMTs, HDACs and KDMs to maintain the epigenome of the tissue of origin. One means by which these enzymes may be affected in cancer is through loss of their cofactors.
Epigenetic enzymes are reliant upon metabolic cofactors
Creating and maintaining the epigenome requires the enzymes mentioned above and their metabolic cofactors. Transmethylation by DNMTs and HMTs requires Sadenosylmethionine (SAM) as a methyl group donor, a cofactor whose level and availability is linked to metabolism and redox. Removing histone modifications requires cofactors linked to glycolysis and oxidative phosphorylation. Histone deacetylation by class III HDACs use nicotinamide adenine dinucleotide (NAD+) to accept acetyl groups from lysines [109]. Likewise, KDMs remove methyl groups by using α-ketoglutarate as an electron donor [110]. Redox status may also influence the function of epigenetic enzymes and the effect proteins that bind their products. From this brief description, it is apparent that the epigenome relies heavily upon metabolic and dietary cofactors. Below we will discuss how the metabolic defects of cancer metabolism influences each of these cofactors to flip the epigenetic switch in cancer.
Once carbon metabolism and epigenetics
In the past few years a push towards understanding the connection between diet and gene expression has revealed that one carbon metabolism can influence epigenetics. These studies have focused on two metabolites, folate and SAM. Both cofactors are central to methylation reactions in cells. Studies by Randy Jirtle's group have shown that dietary folate can impact the methylation of specific genes, while a growing number of studies have begun to investigate the role of SAM in controlling gene expression during liver injury and carcinogenesis [111-113]. SAM's level is governed by folate and the needs of cells at any given time. In mammalian cells, dietary folate serves as a cofactor to assimilate carbon groups from glycine into one carbon metabolism. Prior to folate's use in cells it is converted to tetrahydrofolate. Inhibition of folate metabolism by methotrexate blocks the conversion of deoxyuridylic acid to thymidylic acid [114, 115]. Other than nucleotide biosynthesis, folate is used to produce methionine from homocysteine, and eventually to SAM [116]. SAM is used as the methyl donor in biochemical reactions because the methyl group bound to sulfur can be removed with relative biological ease, an essential requirement for such a donor. SAM is an essential cofactor that is required by several biochemical processes (for review see [117]) (Fig. 3). If SAM levels become too low, methylation reactions participating in epigenetic processes may no longer function properly. The overlying concept is that if folate status is interrupted, cells will no longer be able to maintain epigenetic control and have altered gene expression. Mutations in genes that metabolize folate prior to the synthesis of SAM disrupt genomic DNA methylation [118]. Limiting folate intake decreases the SAM/SAH ratio. Decreasing SAM/SAH ratio also inhibits DNA methylation by affecting the activity of DNMTs [119]. DNA hypomethylation induced by folate deficiency is another risk factor for increased cancer susceptibility [116]. One carbon metabolism appears to have the same impact on histone modifications. Feeding rats a methyl deficient diet induces a global decrease of H3K9me3, H3K9ac, H4K16ac, and H4K20me3 histones and promotes carcinogenesis [105].
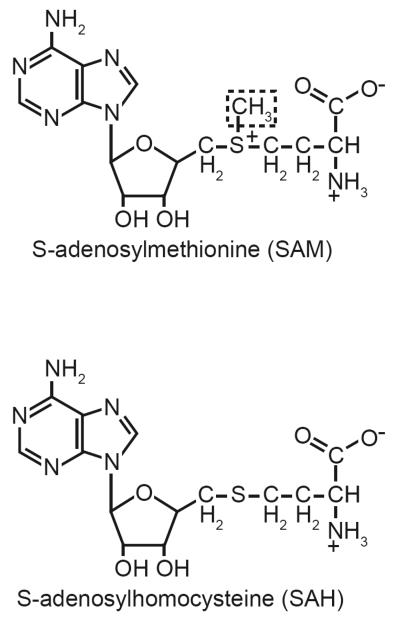
Fig. 3. The structures of the epigenetic metabolites S-adenosylmethionine and S-adenosylhomocysteine.
During transmethylation reactions the methyl group of S-adenosylmethionine (dashed box) serves as a nucleophile to attack C-5 of cytosine, and the ε-N of lysine. Once the reaction is complete, S-adenosylhomocysteine is released and utilized in other biochemical reactions.
S-adenosylmethionine synthesis
Mammalian cells produce SAM by the addition of ATP to methionine by SAM synthetases. Humans have three forms of SAM sythetases: MATI, MATII and MATIII. Both MATI and MATIII are encoded by the mat1α gene and are primarily expressed in the liver [120]. While the primary amino acid sequence is identical between MATI and MATIII, the two enzymes exist as tetramers and dimers respectively [121]. All other tissues use MATII, which is a heterodimer of MAT2α and MAT2β subunits [122]. Regardless of the tissue, these enzymes are critical in maintaining cellular SAM levels and the activity of the methionine cycle. Here we will discuss the function of SAM and its role in carcinogenesis. We will then introduce how redox status may influence SAM and SAM levels in cancer.
SAM inhibits tumor formation
SAM levels in cells directly influence carcinogenesis by affecting methylation reactions. Tumor promotion studies using rat liver models have shown that SAM content and the SAM/SAH ratio are decreased in preneoplastic lesions [123]. Consequently, treating initiated animals with SAM decreases the size and frequency of these preneoplastic lesions after initiation [124]. SAM treatment also blocks the progression of these preneoplastic lesions into hepatocellular carcinoma [112, 124]. This has led to the conclusion that SAM may have chemopreventative properties [112]. The effects mentioned above can be attributed to SAM's influence on DNA methylation. Studies by Francesco Feo's group in the early 1990's showed that SAM inhibits liver preneoplastic lesion growth by influencing the expression of proto-oncogenes. Administering SAM to animals after initiation decreased the expression of c-myc, c-Haras and c-K-ras in proliferating liver and nodules [123, 125]. They determined that the decreased expression of these three proto-oncogenes was caused by increased DNA methylation at their promoters [123]. Similar observations have been observed when human model tumor cell lines are treated with SAM. For example, treating prostate cancer cell lines with SAM decreases their expression of two known tumor promoting genes, urokinase-type plasminogen activator and matrix metalloproteinase-2. The silencing of these two genes is casually linked to hypermethylation of CpGs within their promoters [126]. Furthermore, pretreatment of prostate cancer cells with SAM also decreases tumor growth rate in mouse xenografts [126]. Given these studies, it seems of great importance to understand what fundamental changes in SAM biochemistry occur during carcinogenesis.
Glutathione production is linked to SAM
Mitochondrial defects lead to the aberrant production of reactive oxygen species like superoxide and hydrogen peroxide. To counter this, cancer cells increase their production of small molecular weight antioxidants like glutathione [127]. Increasing the production of glutathione requires cells to tap their sulfur pools. Cells meet this need by increasing the flux of homocysteine into the transsulfuration pathway and away from the methionine cycle [128]. Homocysteine can enter the transsulfuration pathway or be recycled back into methionine by either methionine synthase or betaine homocysteine methyltransferase. This choice is dictated in part by the current needs of the cell at that time. During a pro-oxidant state, homocysteine is diverted away from the methionine cycle and into the transsulfuration pathway. A pro-oxidant sate accomplishes this by increasing the activity of cystathionine β-synthase [129]. The transsulfuration pathways can account for up to 50 percent of the glutathione produced in some tissues [129]. The synthesis of glutathione directly affects epigenetic processes. When glutathione is depleted by chemical means methyl donors become deficient, leading to genome wide DNA hypomethylation [128, 130, 131]. This change occurs because the level of SAM required by DNMTs and HMTs can no longer be met. A similar case would arise in tumor cells. The pro-oxidant state of tumor promotion would rob the methionine cycle of tumor cells to feed their need to produce glutathione. Countering mitochondrial oxidants in this manner would lead to a depletion of SAM in tumor cells. The result of which is the global loss of DNA and histone methylation observed in cancers.
Many cancers also increase production of unusual metabolites such as sarcosine that appear to be causally involved in malignant behavior [132]. The first step in the synthesis of this metabolite is catalyzed by glycine N-methyltransferase (GNMT), which transfers a methyl group from SAM to glycine to produce sarcosine and SAH. Unlike DNA and histone methyltransferases, GNMT is only weekly inhibited by SAH, which allows it to freely control the SAM/SAH ratio. It has also been proposed GNMT controls transmethylation reactions by siphoning carbon units from the methionine cycle and back into the folate cycle [120]. In normal cells it is likely this mechanism works efficiently as a way to regulate global transmethylation reactions, since the Km of GNMT for SAM is high and would only redirects carbon groups when folate levels are high [133]. This observation also explains the large SAM/SAH ratio of normal cells. In cancer cells GNMT activity could also create an environment that inhibits transmethylation. Tumor cells often over express GNMT, and could be one factor that creating their decreased SAM/SAH ratios [132, 134]. If this is the case, then the increased GNMT activity of tumor cells could potentially influence epigenetic processes in two ways. First, GNMT would decrease the SAM/SAH ratio, and thus inhibit the activities of DNA and histone methyltransferases, which are highly susceptible to inhibition by SAH. Secondly, GNMT would decrease transmethylation reactions by directly removing SAM from the methionine cycle. There is empirical evidence to support this connection between GNMT and epigenetic processes. Activating GNMT activity with glucocorticoids or retinoids results in global loss of DNA methylation [135]. Combined with our knowledge about GNMT levels in cancer it appears that disregulation of this enzyme could be causally involved in the epigenetic switch of cancer.
Other Metabolic intermediates influence epigenetics
The metabolic defect of cancer alters the levels and fluxes of metabolites through glycolysis and the electron transport chain. These fundamental metabolic changes could both influence the function of epigenetic processes in cancer. It was exactly this relationship between metabolism and redox that led Peter Cerutti to hypothesize that altered nicotinamide adenine dinucleotide (NAD+) was affecting epigenetic regulation of gene expression of [5]. Cerutti attributed these changes to the poly ADP-ribosylation of “chromosomal proteins”, which were one of the best-characterized histone modifications at that time. Here, with the benefit of our current understanding, we extend upon Cerutti's original ideas and speculate that the metabolic changes effect histone acetylation and methylation as well.
Histone acetylation and NAD+
Sirtuins utilize NAD+ to deacetylate histones and other acetylated proteins through the hydrolysis of NAD+, the results of which are O-acetyl-ADP-ribose, nicotinamide, and lysine [75]. An intimate association among the NAD+/NADH, sirtuins, and histone acetylation has also been well established by studies investigating lifespan and caloric restriction [75]. Caloric and/or glucose restriction effectively increases the NAD+/NADH ratio, and in turn dictates the HDAC activity of sirtuins [109]. Currently seven sirtuin family members have been identified in humans: SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6 and SIRT7 [109, 136]. Currently the role of sirtuins in cancer is only emerging. We speculate that the increased glycolytic activity of the Warburg effect may partially influence sirtuin activity by altering the NAD+/NADH ratio in tumor cells. In the early 1970's it was reported that immortalization decreases the NAD+/NADH ratio in fibroblasts, however current studies on this topic are lacking [137]. If NAD+ levels decrease during carcinogenesis a concomitant change in sirtuin activity and histone acetylation must also be occurring. During development, changing NAD+/NADH ratio of muscle cells has already been established as a means to alter sirtuin activity and affect chromatin structure [138-140]. With cancer, the Warburg effect would drive down the NAD+/NADH ratio, decrease sirtuin activity, and lead to aberrant gene expression through histone hypoacetylation. Connecting the Warburg effect with gene expression via sirtuins is potentially astounding. Because increased glucose consumption is a common trait in cancer, it has been seen as an avenue for creating novel therapies in cancer. The addition of an epigenetic component to the Warburg effect lends further credence to the development of such therapies for cancer.
Kreb's cycle intermediates also have an intimate link with enzymes that regulate epigenetic processes. As we discussed above, some KDMs require oxygen, Fe2+, α-ketoglutarate and ascorbate to demethylate histones [85, 90]. Increased glutamine consumption by tumor cells would affect their concentration of α-ketoglutarate, KDM activity, and histone methylation. The impact of increased glutamine consumption on KDMs would be further affected by dysfunctional electron transport. Defects in electron transport chain complexes are common events and coincide with ROS production, however they can also change the level of Krebs cycle intermediates [141]. For example, Complex II defects increase cellular levels of succinate, and stabilize HIF-1α through substrate inhibition of α-ketoglutarate requiring prolyl hydroxylases [142, 143]. This serves as an elegant example of how metabolism in the mitochondria can directly influence transcriptional activity in the nucleus. We speculate metabolic defects also influence nuclear transcription through KDMs that manipulate the epigenome. Aberrant glutamine consumption by tumor cells is compounded by their electron transport chain defects and creates a sizable pool of α-ketoglutarate to accelerate histone demethylation. This is consistent with the observed decreases of H3K9me3, H3K4me3 and H3K27me3 in cancer, all of which are targeted by KDMs [61, 103-106, 144, 145]. Likewise, mitochondrial defects, such as SDH mutations, could also increase succinate levels and potentially inhibit KDM activity. Regardless if metabolism is activating or inhibiting KDMs, from our discussion above there appears to be strong evidence to support a causal link between Krebs cycle imbalance and altered histone methylation in cancer.
Redox regulation of epigenetics
Cancer cells have an atypical redox status that is dictated by ratio of glutathione to glutathione disulfide (GSH/GSSG). Normal cells have an almost infinite GSH/GSSG ratio because their concentration of GSSG is nearly zero. This high GSH/GSSG ratio makes a good redox buffer that favors healthy biological activity in the reduced state [146]. Tumor cells on the other hand have appreciable amounts of GSSG, which effectively decreases their GSH/GSSG ratio. The result is a change in the redox buffering capacity of tumor cells that alters biological activity by affecting enzyme function. Not all enzymes would be subjected to redox regulation in this manner. Candidates for redox regulation are enzymes or factors that utilize oxidizable amino acids such cysteine in their enzymatic mechanisms or functional motifs. The atypical redox state of tumor cells influences epigenetic processes by affecting the production of SAM, epigenetic enzymes, and effector proteins that bind modified histones.
The synthesis of SAM by SAM synthetases is a redox regulated process. Controlling the tertiary structure of SAM synthetase is its oxidation state and the GSH/GSSG ratio [147]. Blocking glutathione synthesis with BSO reduces the GSH/GGSH ratio and MAT activity. However, treatment with SAM reverses BSO's inhibition of MATs by restoring the redox status of cells. [113, 148]. These observations reveal an interesting link between SAM levels, MAT activity, and redox state. Redox buffering also influences MAT activity. High GSH/GSSG ratios have a positive influence on MAT activity by keeping the enzyme in a reduced state, however when the GSH/GSSG ratio falls below 10:1, its activity becomes decreased [149]. These enzymes are amendable to redox regulation because of a conserved critical cysteine residue located within their active sites. Work by Pajares and others have identified cysteine 150 (C150) of rat MATI and III as a critical cysteine residue whose oxidation is believed to be a contributing factor in the development of liver cirrhosis [150, 151]. While these studies have focused primarily on liver disease, and not cancer, we can speculate that the influence of redox biology on this process would transition between pathologies. We can draw this conclusion because cysteine 150 is conserved between rat MATI and MATII, meaning redox exerts an influence over SAM levels in tissues other than liver. The GSH/GSSG ratios of cancer cells vary, but are generally lower than 10:1. Based on how redox buffering influences SAM synthetase activity we would expect cancer cells to exhibit lower SAM synthetase activity even if the expression of this enzyme is unaffected. This makes sense because of the metabolic network that exists between SAM and glutathione. Oxidizing conditions would slow down the methionine cycle to feed sulfur into the production of glutathione via the transsulfuration pathway. The chronic redox changes in cancer cells would create a SAM deficit in this manner, leaving insufficient fuel for HMTs and DNMTs. The result would be global epigenetic alterations similar to what is observed when glutathione is depleted.
Enzymes that initiate and perpetuate epigenetic events are also subject to redox regulation by the disruption of their enzymatic mechanisms. When Thomas Jenuwein's group characterized the first mammalian homologue of SUV39H1, they identified a cysteine rich region near its SET domain whose presence was necessary for the enzyme's activity [64]. These cysteine rich “Post-SET domains” are found in several HMTs and often have a role in substrate recognition and enzyme activity [81]. DNA methyltransferases also have highly conserved cysteine residues in their active sites. This active site cysteine is generally deprotonated and serves as a catalytic nucleophile by forming an intermediary covalent bond with cytosine that primes the 5 position of its pyrimidine ring to accept a methyl group form SAM (Fig. 4) [152]. Mutation of this cysteine in DNMT1, DNMT3a and DNMT3b results in a catalytically dead methyltransferase [153, 154]. Sulfur nucleophiles like the one in DNMTs are amendable to oxidation, resulting in loss of enzyme activity. The potential for redox to alter the activity of DNMTs via cysteine oxidation is best exemplified by 5-aza-2′-deoxycytidine, a potent inhibitor of DNA methylation [155]. When DNMTs attempt to methylate 5-aza-2′-deoxycytidine that has been incorporated into double stranded DNA, an irreversible covalent bond is formed between enzyme and inhibitor via this catalytic cysteine, resulting in loss of DNMT function and a global decrease in methylated DNA [156]. Oxidation of the catalytic cysteine would stop it from serving as a nucleophile, essentially removing the enzyme from the active pool of DNMTs like 5-aza-2′-deoxycytidine. These examples allow us to speculate that the prooxidant state of cancer cells would render DNMTs and HMTs less active. Both enzyme types often exhibit increased expression in cancer, yet the product of their reactions (i.e. methylated cytosine and lysine) are paradoxically decreased [95]. A mechanism that decreases enzyme activity, but not expression, could reconcile these observations. Oxidation of these cysteine rich domains could be one means by which the pro-oxidant state of cancer impinges on the activity of these enzymes.
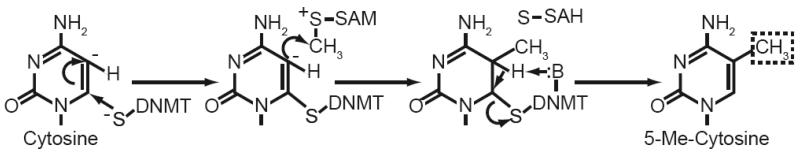
Fig. 4. Enzymatic mechanism of DNA methyltransferases (DNMTs).
Methylation of cytosine begins with the nucleophillic attack of position 6 by a thiolate nucleophile. The resulting electron rich region at position 5 is then at attacked by the methyl group of Sadenosylmethionine (SAM). The reaction then proceeds with the removal of the hydrogen at position 5 by a basic amino acid in the active site of DNMT. In the final step, the double bond is reformed in the pyrimidine ring, resulting in elimination of bond between position 6 and the DNMT.
Histone modifying enzymes and their effector proteins contain several conserved domains which have cysteine rich regions within them. Plant homeo domains (PHDs) are an ∼50-80 amino acid domain that contain a conserved Cys4-HisCys3 zinc finger motif [157]. PHDs are a common motif in several nuclear proteins, many of which have a profound role in regulating chromatin structure and function. Nucleosome binding of these domains was initially characterized in the HAT p300 and the chromatin remodeling complex subunits ACF1 and NUR301 [158-160]. PHD domains are also present in some DNMTs and may influence the location and specificity of DNA methylation [161]. In the past two years several groups have reported that these domains also bind methylated histone tails, specifically H3K4me2, H3K4me3, and H3R2me2 to regulate transcription and possibly genetic recombination [162-165]. Loss of the PHD domain within RAG2 by deletion or mutation of its cysteine abrogates its function and manifests itself in vivo as severe combined immunodeficiency [162]. It is interesting to note that H3K4me2 and H3K4me3 are prevalent histone modifications within the human genome, and many genes are regulated by p300/CBP type HATS. Given this information it seems likely that some signaling mechanism controls local function of proteins to stipulate when their activities are required. We suggest that cysteine rich domains are common at the contact sites between chromatin-associated proteins and the factors that recruit them (i.e. modified histone or transcription factor). One example is the PHD domain bearing protein p300 and its interaction with CREB/ATF transcription factors. From the example of PHDs we surmise that redox status could in part regulate the interactions between different histone modifying and binding proteins. Such a regulatory role for redox status is not without precedence. The REF-1 redox switch controls the function of several transcription factors (for reviews on this topic see [166, 167]). For transcription factors, redox switches modulate their binding to cognate sites throughout the genome. Is it possible that the contacts between modified histones and their effector proteins are regulated in a similar manner? The presence of a particular histone modification isn't synonymous with its effector protein binding. For example, phosporylation of 53BP1 is required prior to binding H4K20me2, a histone modification present on 80% of all nucleosomes [83, 168]. Because redox switches and ROS have the ability to activate transcription factors, it seems very likely that affecting histone modifications is another avenue by which they can influence gene expression [3, 166, 169, 170]. Such an investigation in cancer, or chromatin biology remains unexplored and open for speculation.
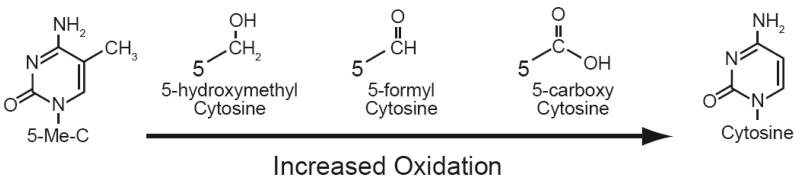
Oxidants also influence epigenetic processes by oxidizing nucleotide bases and disrupting higher ordered chromatin structure. The oxidation of nucleotide bases by free radicals creates several unique bases, most common of which is 8-oxoguanine (8-oxoG). This oxidized base can directly inhibit DNMTs and possibly induce DNA demethylation at this site [171, 172]. Such a case arises when 8-oxoG is in either strand of a hemimethylated CpG doublet, or the base adjacent to a hemimethylated CpG [171, 172] (Fig. 5 A, B). The presence of 8-oxoG within CpG doubles occurs via the direct oxidation of guanine in double stranded DNA, or through incorporation of 8-oxoGTP from the nucleotide pool during S phase [173]. In normal cells 8-oxoG is evicted from double stranded DNA by 8-oxoguanine glycosylase 1 (OGG1) and removed from the nucleotide pool by the 8-oxoGTPase hMTH1 (For review of these topics see [173, 174]). However, deleterious mutations have been identified for both OGG1 and hMTH1 in many cancers, thus allowing 8-oxoG to persist as a mutagen, and potentially influence the epigenotype and progression of these diseases [175, 176]. Oxidation of 5-meC is another type of base damage that can influence DNA methylation [177]. 5-meC accounts for approximately 3-5% of all cytosine within a cell but is still a target for oxidation by reactive oxygen species [178]. Progressive oxidation of 5-meC can lead to its demethylation to form cytosine (Fig. 6). While this reaction is chemically plausible, its progressive oxidation is complex and has been speculated to lead to the demethylation of CpGs in vivo [179, 180]. The long-term effects of oxidized 5-meC during carcinogenesis are likely attributable to 5-formylcytosine (5-fC), or 5-hydroxymethylcytosine (5-hmC) both of which are detectable in vivo [181-183]. Like 8-oxoG, 5-hmC is removed from DNA by its own specific glycosylase, HMC glycosylase [184]. HMC glycosylases are unique enzymes because they simultaneously correct genetic and epigenetic defects. Work from Lawrence Sower's laboratory has revealed that oxidation of 5meC could potentially hinder the ability of DNMT1 to methylate the nascent strand of DNA following replication [185]. Such changes could lead to demethylation at a specific region or locus and dramatically affect the epigenome. The Sowers group has also shown that oxidation of cytosine or guanine on either strand of a CpG affects the epigenome by inhibiting the activity binding of MBPs such as MBP1, MBP2 and MeCP2 and the chromatin remodeling complexes associated with them [183] (Fig. 5C). Thus, the oxidation of individual methylated CpGs could exert strong effects on local chromatin structure and possibly gene expression. But is this a viable mechanism to alter gene expression? Recent findings suggest that ROS production can lead to demethylation of the E-cadherin promoter [186]. While such studies begin to demonstrate a link between ROS and epigenetic processes, they lack a mechanistic component. Additional studies are required to dissect the fundamental mechanisms behind these observations. Aberrant production of oxidants such as hydrogen peroxide also triggers the degradation of higher ordered chromatin structure through the mobilization of HATS such as CBP/p300 [187, 188]. Direct oxidation also influences the interaction between nucleosomes. The addition of reducing agents increases the susceptibility of chromatin to nuclease digestion, however the mechanism by which this occurs remains undefined [189-191]. Base oxidation created by ROS is appreciated as a means to induce mutations during carcinogenesis. From our discussion here it likely affects epigenetic processes as well, and establishes another means by which free radicals can influence carcinogenesis.
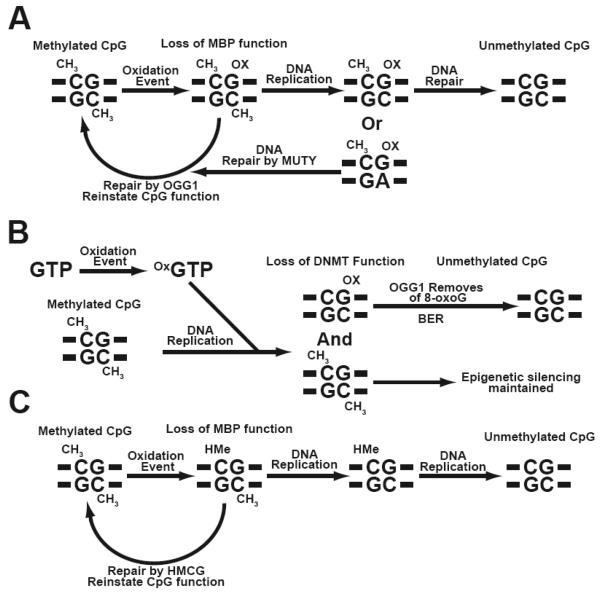
Fig. 5. Oxidation of nucleotide bases within methylated CpGs alters epigenetic processes.
(A) Oxidation of guanine within a methylated CpG doublet abrogates MBP function, resulting in an epigenetic change. If these oxidized bases are not removed by 8-oxoguanine glycosylase 1 (OGG1) the epigenetic defect can be passed on during DNA replication and result in an unmethylated CpG. (B) Incorporation of oxidized GTP during DNA replication results in a methylated strand, and a hemimethylated CpG that is resistant to DNMTs. Removal of 8-oxoG by OGG1 and repair by base excision repair machinery (BER), creates an unmethylated CpG. (C) Oxidation of 5-methylcytosine creates 5-hydroxymethylcytosine (HMe). If this base is not removed by HMC glycosylases (HMCG) MBP function is lost at the site of oxidation and can create a hemimethylated strand following DNA replication. This epigenetic change can then persist as an unmethylated CpG in subsequent cell divisions.
Fig. 6. The oxidative demethylation of 5-Me-cytosine to cytosine.
Progressive oxidation of the carbon within the methyl group of 5-methyl cytosine (5-Me-C) results in its demethylation, and formation of cytosine. Each of the intermediates produced during the oxidation process are stable within DNA and affect epigenetic processes in a unique manner.
Summary
The nuclei of eukaryotes contain two sets of information, genetic and epigenetic, that regulate gene expression. Until recently, the focus of cancer biology has been genetic changes as a means to drive carcinogenesis. Oberley and Buettner hypothesized 30 years ago that ROS could be causal in carcinogenesis [1]. Today, there is strong empirical evidence to support the ability of ROS, redox state, and metabolic changes to create genetic mutations and the mutator phenotype of cancer [1-4, 192]. With our increased understanding of epigenetics we now know that it can accomplish many of the same functions as genetic changes in cancer: loss of gene function by silencing, activation of oncogene expression, and inducing chromosomal aberrations [91-94]. In multi-cellular organisms, epigenetics controls the function of the genome to create various cell types without genetic diversity, while in cancer, genetic and epigenetic alterations most likely collaborate to manifest the malignant phenotype. Is it possible that genetic and epigenetic defects in cancer share a common cause? Peter Cerutti was the first to propose a relationship between epigenetics and the prooxidant state of cancer, and we have described a similar connection in development [5, 6]. Our current model for the how each of these metabolic defects influences epigenetic control of gene expression is shown in Fig. 7. In this review we have sought to expand upon current theories regarding the causal role for metabolic defects during carcinogenesis. Given the connections we have outlined above, such theories should be amended to include our new understanding of epigenetic processes in cancer.
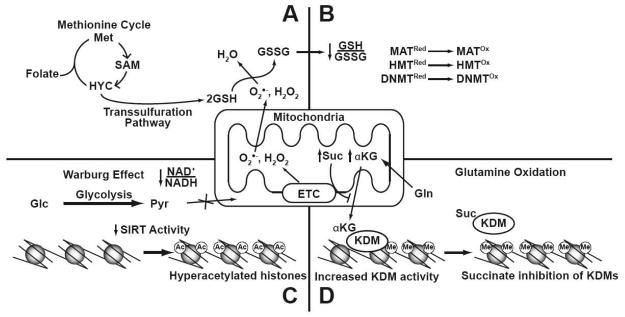
Fig. 7. The current model for the relationship between cancer metabolic defects and epigenetic processes.
(A) Tumor cells increase their production of GSH to counter mitochondrially derived oxidants such as O2•− and H2O2. To sustain GSH production, cancer cells divert metabolites away from the methionine cycle into the transsulfuration pathway, resulting in decreased SAM production. (B) Aberrant production of oxidants creates an atypical redox state by decreasing the GSH/GSSG ratio which affects the activities of SAM synthetases, DNMTs and HMTs. (C) The increased Glc (glucose) consumption of the Warburg effect decreases the NAD+/NADH ratio and produces Pyr (pyruvate). Decreasing this ratio creates an environment that inhibits the activity of sirtuins, and liberates genes from their negative regulation. (D) Oxidation of glutamine (Gln), and dysfunctional electron transport, alters the flow of α-KG (α-ketoglutarate) and Suc (succinate) metabolites within the Krebs cycle. These metabolites can then influence transcription in the nucleus by affecting the activity of KDMs (lysine demethylases).
Acknowledgements
The authors are deeply indebted to the late Larry Oberley for his unending enthusiasm for and promotion of many of the concepts herein. The work was supported by NIH grants CA73612 and CA115438 to FED. MJH received salary support from T32078586.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Oberley LW, Buettner GR. Role of superoxide dismutase in cancer: a review. Cancer Res. 1979;39:1141–1149. [PubMed] [Google Scholar]
- 2.Oberley LW, Oberley TD, Buettner GR. Cell division in normal and transformed cells: the possible role of superoxide and hydrogen peroxide. Med Hypotheses. 1981;7:21–42. doi: 10.1016/0306-9877(81)90018-9. [DOI] [PubMed] [Google Scholar]
- 3.Gius D, Spitz DR. Redox signaling in cancer biology. Antioxidants & redox signaling. 2006;8:1249–1252. doi: 10.1089/ars.2006.8.1249. [DOI] [PubMed] [Google Scholar]
- 4.Spitz DR, Sim JE, Ridnour LA, Galoforo SS, Lee YJ. Glucose deprivation-induced oxidative stress in human tumor cells. A fundamental defect in metabolism? Annals of the New York Academy of Sciences. 2000;899:349–362. doi: 10.1111/j.1749-6632.2000.tb06199.x. [DOI] [PubMed] [Google Scholar]
- 5.Cerutti PA. Prooxidant states and tumor promotion. Science. 1985;227:375–381. doi: 10.1126/science.2981433. [DOI] [PubMed] [Google Scholar]
- 6.Hitchler MJ, Domann FE. An epigenetic perspective on the free radical theory of development. Free radical biology & medicine. 2007;43:1023–1036. doi: 10.1016/j.freeradbiomed.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warburg OH, Dickens F. The metabolism of tumours. R. R. Smith; New York: 1931. Berlin. Kaiser Wilhelm-institut für biologie. [from old catalog] [Google Scholar]
- 8.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 9.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 10.Kim JW, Dang CV. Cancer's molecular sweet tooth and the Warburg effect. Cancer research. 2006;66:8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- 11.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 12.Bonnet S, Archer SL, Allalunis-Turner J, Haromy A, Beaulieu C, Thompson R, Lee CT, Lopaschuk GD, Puttagunta L, Harry G, Hashimoto K, Porter CJ, Andrade MA, Thebaud B, Michelakis ED. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell. 2007;11:37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 13.Patel MS, Korotchkina LG. Regulation of mammalian pyruvate dehydrogenase complex by phosphorylation: complexity of multiple phosphorylation sites and kinases. Exp Mol Med. 2001;33:191–197. doi: 10.1038/emm.2001.32. [DOI] [PubMed] [Google Scholar]
- 14.Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. The Journal of biological chemistry. 1979;254:2669–2676. [PubMed] [Google Scholar]
- 15.Lazo PA. Amino acids and glucose utilization by different metabolic pathways in ascites-tumour cells. European journal of biochemistry / FEBS. 1981;117:19–25. doi: 10.1111/j.1432-1033.1981.tb06297.x. [DOI] [PubMed] [Google Scholar]
- 16.Moreadith RW, Lehninger AL. Purification, kinetic behavior, and regulation of NAD(P)+ malic enzyme of tumor mitochondria. The Journal of biological chemistry. 1984;259:6222–6227. [PubMed] [Google Scholar]
- 17.Baggetto LG. Deviant energetic metabolism of glycolytic cancer cells. Biochimie. 1992;74:959–974. doi: 10.1016/0300-9084(92)90016-8. [DOI] [PubMed] [Google Scholar]
- 18.Gomez-Fabre PM, Aledo JC, Del Castillo-Olivares A, Alonso FJ, Nunez De Castro I, Campos JA, Marquez J. Molecular cloning, sequencing and expression studies of the human breast cancer cell glutaminase. The Biochemical journal. 2000;345(Pt 2):365–375. [PMC free article] [PubMed] [Google Scholar]
- 19.Metzler DE, Metzler CM. Biochemistry : the chemical reactions of living cells. Harcourt/Academic Press; San Diego, Calif.: 2001. [Google Scholar]
- 20.Jeffree GM. Hydrogen peroxide and cancer. Nature. 1958;182:892. doi: 10.1038/182892a0. [DOI] [PubMed] [Google Scholar]
- 21.Spitz DR, Azzam EI, Li JJ, Gius D. Metabolic oxidation/reduction reactions and cellular responses to ionizing radiation: a unifying concept in stress response biology. Cancer metastasis reviews. 2004;23:311–322. doi: 10.1023/B:CANC.0000031769.14728.bc. [DOI] [PubMed] [Google Scholar]
- 22.Ridnour LA, Oberley TD, Oberley LW. Tumor suppressive effects of MnSOD overexpression may involve imbalance in peroxide generation versus peroxide removal. Antioxidants & redox signaling. 2004;6:501–512. doi: 10.1089/152308604773934260. [DOI] [PubMed] [Google Scholar]
- 23.Oberley LW. Mechanism of the tumor suppressive effect of MnSOD overexpression. Biomed Pharmacother. 2005;59:143–148. doi: 10.1016/j.biopha.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Das UN. Pyruvate is an endogenous anti-inflammatory and anti-oxidant molecule. Med Sci Monit. 2006;12:RA79–84. [PubMed] [Google Scholar]
- 25.Waddington CH. The epigenetics of birds. University Press; Cambridge [Eng.]: 1952. [Google Scholar]
- 26.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 27.Downs JA. Chromatin structure and DNA double-strand break responses in cancer progression and therapy. Oncogene. 2007;26:7765–7772. doi: 10.1038/sj.onc.1210874. [DOI] [PubMed] [Google Scholar]
- 28.Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nature reviews. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Browne MJ, Turnbull JF, McKay EL, Adams RL, Burdon RH. The sequence specificity of a mammalian DNA methylase. Nucleic Acids Res. 1977;4:1039–1045. doi: 10.1093/nar/4.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell GJ, Walker PM, Elton RA, Subak-Sharpe JH. Doublet frequency analysis of fractionated vertebrate nuclear DNA. Journal of molecular biology. 1976;108:1–23. doi: 10.1016/s0022-2836(76)80090-3. [DOI] [PubMed] [Google Scholar]
- 31.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. Journal of molecular biology. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 32.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 33.Caiafa P, Zampieri M. DNA methylation and chromatin structure: the puzzling CpG islands. J Cell Biochem. 2005;94:257–265. doi: 10.1002/jcb.20325. [DOI] [PubMed] [Google Scholar]
- 34.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science (New York, N.Y. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 35.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 36.Ehrlich M. The ICF syndrome, a DNA methyltransferase 3B deficiency and immunodeficiency disease. Clinical immunology (Orlando, Fla. 2003;109:17–28. doi: 10.1016/s1521-6616(03)00201-8. [DOI] [PubMed] [Google Scholar]
- 37.DiNardo DN, Butcher DT, Robinson DP, Archer TK, Rodenhiser DI. Functional analysis of CpG methylation in the BRCA1 promoter region. Oncogene. 2001;20:5331–5340. doi: 10.1038/sj.onc.1204697. [DOI] [PubMed] [Google Scholar]
- 38.Mancini DN, Rodenhiser DI, Ainsworth PJ, O'Malley FP, Singh SM, Xing W, Archer TK. CpG methylation within the 5′ regulatory region of the BRCA1 gene is tumor specific and includes a putative CREB binding site. Oncogene. 1998;16:1161–1169. doi: 10.1038/sj.onc.1201630. [DOI] [PubMed] [Google Scholar]
- 39.Comb M, Goodman HM. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18:3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bacolla A, Pradhan S, Larson JE, Roberts RJ, Wells RD. Recombinant human DNA (cytosine-5) methyltransferase. III. Allosteric control, reaction order, and influence of plasmid topology and triplet repeat length on methylation of the fragile X CGG.CCG sequence. J Biol Chem. 2001;276:18605–18613. doi: 10.1074/jbc.M100404200. [DOI] [PubMed] [Google Scholar]
- 41.Beaulieu N, Morin S, Chute IC, Robert MF, Nguyen H, MacLeod AR. An essential role for DNA methyltransferase DNMT3B in cancer cell survival. J Biol Chem. 2002;277:28176–28181. doi: 10.1074/jbc.M204734200. [DOI] [PubMed] [Google Scholar]
- 42.Bolden AH, Nalin CM, Ward CA, Poonian MS, Weissbach A. Primary DNA sequence determines sites of maintenance and de novo methylation by mammalian DNA methyltransferases. Molecular and cellular biology. 1986;6:1135–1140. doi: 10.1128/mcb.6.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robertson KD, Keyomarsi K, Gonzales FA, Velicescu M, Jones PA. Differential mRNA expression of the human DNA methyltransferases (DNMTs) 1, 3a and 3b during the G(0)/G(1) to S phase transition in normal and tumor cells. Nucleic Acids Res. 2000;28:2108–2113. doi: 10.1093/nar/28.10.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kautiainen TL, Jones PA. DNA methylation in mammalian nuclei. Biochemistry. 1985;24:5575–5581. doi: 10.1021/bi00341a043. [DOI] [PubMed] [Google Scholar]
- 45.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 46.Weisenberger DJ, Velicescu M, Preciado-Lopez MA, Gonzales FA, Tsai YC, Liang G, Jones PA. Identification and characterization of alternatively spliced variants of DNA methyltransferase 3a in mammalian cells. Gene. 2002;298:91–99. doi: 10.1016/s0378-1119(02)00976-9. [DOI] [PubMed] [Google Scholar]
- 47.Liu K, Wang YF, Cantemir C, Muller MT. Endogenous assays of DNA methyltransferases: Evidence for differential activities of DNMT1, DNMT2, and DNMT3 in mammalian cells in vivo. Mol Cell Biol. 2003;23:2709–2719. doi: 10.1128/MCB.23.8.2709-2719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 49.Xie S, Wang Z, Okano M, Nogami M, Li Y, He WW, Okumura K, Li E. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236:87–95. doi: 10.1016/s0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- 50.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 51.Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Current opinion in cell biology. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- 52.Prigent C, Dimitrov S. Phosphorylation of serine 10 in histone H3, what for? Journal of cell science. 2003;116:3677–3685. doi: 10.1242/jcs.00735. [DOI] [PubMed] [Google Scholar]
- 53.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes & development. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 54.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 55.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 56.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Molecular and cellular biology. 2006;26:9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 58.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 59.Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes & development. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benetti R, Gonzalo S, Jaco I, Schotta G, Klatt P, Jenuwein T, Blasco MA. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. The Journal of cell biology. 2007;178:925–936. doi: 10.1083/jcb.200703081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 62.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 63.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 64.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 65.Kalakonda N, Fischle W, Boccuni P, Gurvich N, Hoya-Arias R, Zhao X, Miyata Y, Macgrogan D, Zhang J, Sims JK, Rice JC, Nimer SD. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene. 2008;27:4293–4304. doi: 10.1038/onc.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trojer P, Li G, Sims RJ, 3rd, Vaquero A, Kalakonda N, Boccuni P, Lee D, Erdjument-Bromage H, Tempst P, Nimer SD, Wang YH, Reinberg D. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 67.Min J, Allali-Hassani A, Nady N, Qi C, Ouyang H, Liu Y, MacKenzie F, Vedadi M, Arrowsmith CH. L3MBTL1 recognition of mono- and dimethylated histones. Nat Struct Mol Biol. 2007;14:1229–1230. doi: 10.1038/nsmb1340. [DOI] [PubMed] [Google Scholar]
- 68.Tan S. One HAT size fits all? Nature structural biology. 2001;8:8–10. doi: 10.1038/83098. [DOI] [PubMed] [Google Scholar]
- 69.Racey LA, Byvoet P. Histone acetyltransferase in chromatin. Evidence for in vitro enzymatic transfer of acetate from acetyl-coenzyme A to histones. Exp Cell Res. 1971;64:366–370. doi: 10.1016/0014-4827(71)90089-9. [DOI] [PubMed] [Google Scholar]
- 70.Noland BJ, Hardin JM, Shepherd GR. Histone acetyltransferase activity in synchronized mammalian cells. Biochim Biophys Acta. 1971;246:263–268. doi: 10.1016/0005-2787(71)90136-5. [DOI] [PubMed] [Google Scholar]
- 71.Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26:5521–5527. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]
- 72.Libby PR. Activity of histone deacetylase in rat liver and Novikoff hepatoma. Biochim Biophys Acta. 1970;213:234–236. doi: 10.1016/0005-2787(70)90027-4. [DOI] [PubMed] [Google Scholar]
- 73.Hay CW, Candido EP. Histone deacetylase. Association with a nuclease resistant, high molecular weight fraction of HeLa cell chromatin. J Biol Chem. 1983;258:3726–3734. [PubMed] [Google Scholar]
- 74.Zhang K, Dent SY. Histone modifying enzymes and cancer: going beyond histones. Journal of cellular biochemistry. 2005;96:1137–1148. doi: 10.1002/jcb.20615. [DOI] [PubMed] [Google Scholar]
- 75.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 76.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 77.Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 78.An W. Histone acetylation and methylation: combinatorial players for transcriptional regulation. Subcell Biochem. 2007;41:351–369. [PubMed] [Google Scholar]
- 79.Qian C, Zhou MM. SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell Mol Life Sci. 2006;63:2755–2763. doi: 10.1007/s00018-006-6274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilson JR, Jing C, Walker PA, Martin SR, Howell SA, Blackburn GM, Gamblin SJ, Xiao B. Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell. 2002;111:105–115. doi: 10.1016/s0092-8674(02)00964-9. [DOI] [PubMed] [Google Scholar]
- 81.Zhang X, Yang Z, Khan SI, Horton JR, Tamaru H, Selker EU, Cheng X. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, Lis JT, Allis CD, Reinberg D. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 83.Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Molecular and cellular biology. 2008;28:468–486. doi: 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 85.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nature reviews. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 86.Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 87.Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y, Shi Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 88.You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc Natl Acad Sci U S A. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 90.Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 91.Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene. 2002;21:5462–5482. doi: 10.1038/sj.onc.1205606. [DOI] [PubMed] [Google Scholar]
- 92.Laird PW. Cancer epigenetics. Human molecular genetics. 2005;14(Spec No 1):R65–76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- 93.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nature reviews. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 94.Hake SB, Xiao A, Allis CD. Linking the epigenetic ‘language’ of covalent histone modifications to cancer. British journal of cancer. 2004;90:761–769. doi: 10.1038/sj.bjc.6601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ehrlich M, Woods CB, Yu MC, Dubeau L, Yang F, Campan M, Weisenberger DJ, Long T, Youn B, Fiala ES, Laird PW. Quantitative analysis of associations between DNA hypermethylation, hypomethylation, and DNMT RNA levels in ovarian tumors. Oncogene. 2006;25:2636–2645. doi: 10.1038/sj.onc.1209145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goelz SE, Vogelstein B, Hamilton SR, Feinberg AP. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science. 1985;228:187–190. doi: 10.1126/science.2579435. [DOI] [PubMed] [Google Scholar]
- 97.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 98.Cody DT, 2nd, Huang Y, Darby CJ, Johnson GK, Domann FE. Differential DNA methylation of the p16 INK4A/CDKN2A promoter in human oral cancer cells and normal human oral keratinocytes. Oral oncology. 1999;35:516–522. doi: 10.1016/s1368-8375(99)00026-3. [DOI] [PubMed] [Google Scholar]
- 99.Oshiro MM, Futscher BW, Lisberg A, Wozniak RJ, Klimecki WT, Domann FE, Cress AE. Epigenetic regulation of the cell type-specific gene 14-3-3sigma. Neoplasia. 2005;7:799–808. doi: 10.1593/neo.05274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lal G, Padmanabha L, Provenzano M, Fitzgerald M, Weydert J, Domann FE. Regulation of 14-3-3sigma expression in human thyroid carcinoma is epigenetically regulated by aberrant cytosine methylation. Cancer letters. 2008;267:165–174. doi: 10.1016/j.canlet.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Magewu AN, Jones PA. Ubiquitous and tenacious methylation of the CpG site in codon 248 of the p53 gene may explain its frequent appearance as a mutational hot spot in human cancer. Molecular and cellular biology. 1994;14:4225–4232. doi: 10.1128/mcb.14.6.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tornaletti S, Pfeifer GP. Complete and tissue-independent methylation of CpG sites in the p53 gene: implications for mutations in human cancers. Oncogene. 1995;10:1493–1499. [PubMed] [Google Scholar]
- 103.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, Iyer NG, Perez-Rosado A, Calvo E, Lopez JA, Cano A, Calasanz MJ, Colomer D, Piris MA, Ahn N, Imhof A, Caldas C, Jenuwein T, Esteller M. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nature genetics. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 104.Wei Y, Xia W, Zhang Z, Liu J, Wang H, Adsay NV, Albarracin C, Yu D, Abbruzzese JL, Mills GB, Bast RC, Jr., Hortobagyi GN, Hung MC. Loss of trimethylation at lysine 27 of histone H3 is a predictor of poor outcome in breast, ovarian, and pancreatic cancers. Mol Carcinog. 2008;47:701–706. doi: 10.1002/mc.20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pogribny IP, Tryndyak VP, Muskhelishvili L, Rusyn I, Ross SA. Methyl deficiency, alterations in global histone modifications, and carcinogenesis. J Nutr. 2007;137:216S–222S. doi: 10.1093/jn/137.1.216S. [DOI] [PubMed] [Google Scholar]
- 106.Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 107.Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:10014–10019. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1241–1246. doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006;444:868–874. doi: 10.1038/nature05486. [DOI] [PubMed] [Google Scholar]
- 110.Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Current opinion in cell biology. 2008;20:316–325. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Molecular and cellular biology. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pascale RM, Marras V, Simile MM, Daino L, Pinna G, Bennati S, Carta M, Seddaiu MA, Massarelli G, Feo F. Chemoprevention of rat liver carcinogenesis by S-adenosyl-L-methionine: a long-term study. Cancer research. 1992;52:4979–4986. [PubMed] [Google Scholar]
- 113.Corrales F, Gimenez A, Alvarez L, Caballeria J, Pajares MA, Andreu H, Pares A, Mato JM, Rodes J. S-adenosylmethionine treatment prevents carbon tetrachloride-induced S-adenosylmethionine synthetase inactivation and attenuates liver injury. Hepatology (Baltimore, Md. 1992;16:1022–1027. doi: 10.1002/hep.1840160427. [DOI] [PubMed] [Google Scholar]
- 114.Roberts D, Hall TC. The reduction of folate and of dihydrofolate by homogenates of leukocytes from patients with leukemia or with myeloid metaplasia. Cancer research. 1967;27:994–999. [PubMed] [Google Scholar]
- 115.Jenh CH, Rao LG, Johnson LF. Regulation of thymidylate synthase enzyme synthesis in 5-fluorodeoxyuridine-resistant mouse fibroblasts during the transition from the resting to growing state. J Cell Physiol. 1985;122:149–154. doi: 10.1002/jcp.1041220122. [DOI] [PubMed] [Google Scholar]
- 116.Davis CD, Uthus EO. Dietary folate and selenium affect dimethylhydrazine-induced aberrant crypt formation, global DNA methylation and one-carbon metabolism in rats. J Nutr. 2003;133:2907–2914. doi: 10.1093/jn/133.9.2907. [DOI] [PubMed] [Google Scholar]
- 117.Grillo MA, Colombatto S. S-adenosylmethionine and its products. Amino Acids. 2008;34:187–193. doi: 10.1007/s00726-007-0500-9. [DOI] [PubMed] [Google Scholar]
- 118.Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, Olivieri O, Jacques PF, Rosenberg IH, Corrocher R, Selhub J. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:5606–5611. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Balaghi M, Wagner C. DNA methylation in folate deficiency: use of CpG methylase. Biochem Biophys Res Commun. 1993;193:1184–1190. doi: 10.1006/bbrc.1993.1750. [DOI] [PubMed] [Google Scholar]
- 120.Stipanuk MH. Sulfur amino acid metabolism: pathways for production and removal of homocysteine and cysteine. Annual review of nutrition. 2004;24:539–577. doi: 10.1146/annurev.nutr.24.012003.132418. [DOI] [PubMed] [Google Scholar]
- 121.Cai J, Sun WM, Hwang JJ, Stain SC, Lu SC. Changes in S-adenosylmethionine synthetase in human liver cancer: molecular characterization and significance. Hepatology (Baltimore, Md. 1996;24:1090–1097. doi: 10.1002/hep.510240519. [DOI] [PubMed] [Google Scholar]
- 122.Halim AB, LeGros L, Geller A, Kotb M. Expression and functional interaction of the catalytic and regulatory subunits of human methionine adenosyltransferase in mammalian cells. The Journal of biological chemistry. 1999;274:29720–29725. doi: 10.1074/jbc.274.42.29720. [DOI] [PubMed] [Google Scholar]
- 123.Garcea R, Daino L, Pascale R, Simile MM, Puddu M, Ruggiu ME, Seddaiu MA, Satta G, Sequenza MJ, Feo F. Protooncogene methylation and expression in regenerating liver and preneoplastic liver nodules induced in the rat by diethylnitrosamine: effect of variations of S-adenosylmethionine:S-adenosylhomocysteine ratio. Carcinogenesis. 1989;10:1183–1192. doi: 10.1093/carcin/10.7.1183. [DOI] [PubMed] [Google Scholar]
- 124.Pascale RM, Simile MM, Satta G, Seddaiu MA, Daino L, Pinna G, Vinci MA, Gaspa L, Feo F. Comparative effects of L-methionine, S-adenosyl-L-methionine and 5′-methylthioadenosine on the growth of preneoplastic lesions and DNA methylation in rat liver during the early stages of hepatocarcinogenesis. Anticancer research. 1991;11:1617–1624. [PubMed] [Google Scholar]
- 125.Simile MM, Pascale R, De Miglio MR, Nufris A, Daino L, Seddaiu MA, Gaspa L, Feo F. Correlation between S-adenosyl-L-methionine content and production of c-myc, c-Ha-ras, and c-Ki-ras mRNA transcripts in the early stages of rat liver carcinogenesis. Cancer letters. 1994;79:9–16. doi: 10.1016/0304-3835(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 126.Shukeir N, Pakneshan P, Chen G, Szyf M, Rabbani SA. Alteration of the methylation status of tumor-promoting genes decreases prostate cancer cell invasiveness and tumorigenesis in vitro and in vivo. Cancer research. 2006;66:9202–9210. doi: 10.1158/0008-5472.CAN-06-1954. [DOI] [PubMed] [Google Scholar]
- 127.Huang ZZ, Chen C, Zeng Z, Yang H, Oh J, Chen L, Lu SC. Mechanism and significance of increased glutathione level in human hepatocellular carcinoma and liver regeneration. Faseb J. 2001;15:19–21. doi: 10.1096/fj.00-0445fje. [DOI] [PubMed] [Google Scholar]
- 128.Lertratanangkoon K, Savaraj N, Scimeca JM, Thomas ML. Glutathione depletion-induced thymidylate insufficiency for DNA repair synthesis. Biochem Biophys Res Commun. 1997;234:470–475. doi: 10.1006/bbrc.1997.6623. [DOI] [PubMed] [Google Scholar]
- 129.Mosharov E, Cranford MR, Banerjee R. The quantitatively important relationship between homocysteine metabolism and glutathione synthesis by the transsulfuration pathway and its regulation by redox changes. Biochemistry. 2000;39:13005–13011. doi: 10.1021/bi001088w. [DOI] [PubMed] [Google Scholar]
- 130.Lertratanangkoon K, Orkiszewski RS, Scimeca JM. Methyl-donor deficiency due to chemically induced glutathione depletion. Cancer research. 1996;56:995–1005. [PubMed] [Google Scholar]
- 131.Lertratanangkoon K, Wu CJ, Savaraj N, Thomas ML. Alterations of DNA methylation by glutathione depletion. Cancer letters. 1997;120:149–156. doi: 10.1016/s0304-3835(97)00300-5. [DOI] [PubMed] [Google Scholar]
- 132.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 133.Takata Y, Huang Y, Komoto J, Yamada T, Konishi K, Ogawa H, Gomi T, Fujioka M, Takusagawa F. Catalytic mechanism of glycine N-methyltransferase. Biochemistry. 2003;42:8394–8402. doi: 10.1021/bi034245a. [DOI] [PubMed] [Google Scholar]
- 134.Guerinot F, Bohuon C. Glycine-N-methyltransferase levels in human breast cancer tissue. Eur J Cancer. 1977;13:1257–1259. doi: 10.1016/0014-2964(77)90033-0. [DOI] [PubMed] [Google Scholar]
- 135.Rowling MJ, McMullen MH, Schalinske KL. Vitamin A and its derivatives induce hepatic glycine N-methyltransferase and hypomethylation of DNA in rats. J Nutr. 2002;132:365–369. doi: 10.1093/jn/132.3.365. [DOI] [PubMed] [Google Scholar]
- 136.Saunders LR, Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene. 2007;26:5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- 137.Schwartz JP, Passonneau JV, Johnson GS, Pastan I. The effect of growth conditions on NAD+ and NADH concentrations and the NAD+:NADH ratio in normal and transformed fibroblasts. The Journal of biological chemistry. 1974;249:4138–4143. [PubMed] [Google Scholar]
- 138.Backesjo CM, Li Y, Lindgren U, Haldosen LA. Activation of Sirt1 decreases adipocyte formation during osteoblast differentiation of mesenchymal stem cells. J Bone Miner Res. 2006;21:993–1002. doi: 10.1359/jbmr.060415. [DOI] [PubMed] [Google Scholar]
- 139.Li H, Rajendran GK, Liu N, Ware C, Rubin BP, Gu Y. SirT1 modulates the estrogen-insulin-like growth factor-1 signaling for postnatal development of mammary gland in mice. Breast Cancer Res. 2007;9:R1. doi: 10.1186/bcr1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 141.Schiavi F, Savvoukidis T, Trabalzini F, Grego F, Piazza M, Amista P, Dematte S, Del Piano A, Cecchini ME, Erlic Z, De Lazzari P, Mantero F, Opocher G. Paraganglioma syndrome: SDHB, SDHC, and SDHD mutations in head and neck paragangliomas. Annals of the New York Academy of Sciences. 2006;1073:190–197. doi: 10.1196/annals.1353.020. [DOI] [PubMed] [Google Scholar]
- 142.Pollard PJ, Briere JJ, Alam NA, Barwell J, Barclay E, Wortham NC, Hunt T, Mitchell M, Olpin S, Moat SJ, Hargreaves IP, Heales SJ, Chung YL, Griffiths JR, Dalgleish A, McGrath JA, Gleeson MJ, Hodgson SV, Poulsom R, Rustin P, Tomlinson IP. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Human molecular genetics. 2005;14:2231–2239. doi: 10.1093/hmg/ddi227. [DOI] [PubMed] [Google Scholar]
- 143.Lu H, Dalgard CL, Mohyeldin A, McFate T, Tait AS, Verma A. Reversible inactivation of HIF-1 prolyl hydroxylases allows cell metabolism to control basal HIF-1. The Journal of biological chemistry. 2005;280:41928–41939. doi: 10.1074/jbc.M508718200. [DOI] [PubMed] [Google Scholar]
- 144.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 145.Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- 146.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free radical biology & medicine. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 147.Pajares MA, Corrales F, Duran C, Mato JM, Alvarez L. How is rat liver S-adenosylmethionine synthetase regulated? FEBS Lett. 1992;309:1–4. doi: 10.1016/0014-5793(92)80726-w. [DOI] [PubMed] [Google Scholar]
- 148.Corrales F, Ochoa P, Rivas C, Martin-Lomas M, Mato JM, Pajares MA. Inhibition of glutathione synthesis in the liver leads to S-adenosyl-L-methionine synthetase reduction. Hepatology (Baltimore, Md. 1991;14:528–533. [PubMed] [Google Scholar]
- 149.Pajares MA, Duran C, Corrales F, Pliego MM, Mato JM. Modulation of rat liver S-adenosylmethionine synthetase activity by glutathione. The Journal of biological chemistry. 1992;267:17598–17605. [PubMed] [Google Scholar]
- 150.Pajares MA, Corrales FJ, Ochoa P, Mato JM. The role of cysteine-150 in the structure and activity of rat liver S-adenosyl-L-methionine synthetase. The Biochemical journal. 1991;274(Pt 1):225–229. doi: 10.1042/bj2740225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mato JM, Alvarez L, Ortiz P, Mingorance J, Duran C, Pajares MA. S-adenosyl-L-methionine synthetase and methionine metabolism deficiencies in cirrhosis. Adv Exp Med Biol. 1994;368:113–117. doi: 10.1007/978-1-4615-1989-8_11. [DOI] [PubMed] [Google Scholar]
- 152.Chen L, MacMillan AM, Chang W, Ezaz-Nikpay K, Lane WS, Verdine GL. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991;30:11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- 153.Bestor TH. Cloning of a mammalian DNA methyltransferase. Gene. 1988;74:9–12. doi: 10.1016/0378-1119(88)90238-7. [DOI] [PubMed] [Google Scholar]
- 154.Hsieh CL. In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Molecular and cellular biology. 1999;19:8211–8218. doi: 10.1128/mcb.19.12.8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jones PA, Taylor SM, Wilson VL. Inhibition of DNA methylation by 5-azacytidine. Recent results in cancer research. Fortschritte der Krebsforschung. 1983;84:202–211. doi: 10.1007/978-3-642-81947-6_15. [DOI] [PubMed] [Google Scholar]
- 156.Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pascual J, Martinez-Yamout M, Dyson HJ, Wright PE. Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor. Journal of molecular biology. 2000;304:723–729. doi: 10.1006/jmbi.2000.4308. [DOI] [PubMed] [Google Scholar]
- 158.Ragvin A, Valvatne H, Erdal S, Arskog V, Tufteland KR, Breen K, AM OY, Eberharter A, Gibson TJ, Becker PB, Aasland R. Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. Journal of molecular biology. 2004;337:773–788. doi: 10.1016/j.jmb.2004.01.051. [DOI] [PubMed] [Google Scholar]
- 159.Eberharter A, Vetter I, Ferreira R, Becker PB. ACF1 improves the effectiveness of nucleosome mobilization by ISWI through PHD-histone contacts. The EMBO journal. 2004;23:4029–4039. doi: 10.1038/sj.emboj.7600382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Xiao H, Sandaltzopoulos R, Wang HM, Hamiche A, Ranallo R, Lee KM, Fu D, Wu C. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell. 2001;8:531–543. doi: 10.1016/s1097-2765(01)00345-8. [DOI] [PubMed] [Google Scholar]
- 161.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ramon-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, Gozani O, Yang W. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Martin DG, Baetz K, Shi X, Walter KL, MacDonald VE, Wlodarski MJ, Gozani O, Hieter P, Howe L. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Molecular and cellular biology. 2006;26:7871–7879. doi: 10.1128/MCB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sun Y, Oberley LW. Redox regulation of transcriptional activators. Free radical biology & medicine. 1996;21:335–348. doi: 10.1016/0891-5849(96)00109-8. [DOI] [PubMed] [Google Scholar]
- 167.Tell G, Damante G, Caldwell D, Kelley MR. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxidants & redox signaling. 2005;7:367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- 168.Anderson L, Henderson C, Adachi Y. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Molecular and cellular biology. 2001;21:1719–1729. doi: 10.1128/MCB.21.5.1719-1729.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Goswami R, Tandon N, Singh B, Kochupillai N. Adrenal tumour, congestive heart failure and hemiparesis in an 18-year-old male. A clinical-pathological conference. Int J Cardiol. 1995;49:233–238. doi: 10.1016/0167-5273(95)02302-d. [DOI] [PubMed] [Google Scholar]
- 170.Starkov AA. The role of mitochondria in reactive oxygen species metabolism and signaling. Ann N Y Acad Sci. 2008;1147:37–52. doi: 10.1196/annals.1427.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Turk PW, Laayoun A, Smith SS, Weitzman SA. DNA adduct 8-hydroxyl-2′-deoxyguanosine (8-hydroxyguanine) affects function of human DNA methyltransferase. Carcinogenesis. 1995;16:1253–1255. doi: 10.1093/carcin/16.5.1253. [DOI] [PubMed] [Google Scholar]
- 172.Weitzman SA, Turk PW, Milkowski DH, Kozlowski K. Free radical adducts induce alterations in DNA cytosine methylation. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:1261–1264. doi: 10.1073/pnas.91.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nakabeppu Y. Regulation of intracellular localization of human MTH1, OGG1, and MYH proteins for repair of oxidative DNA damage. Prog Nucleic Acid Res Mol Biol. 2001;68:75–94. doi: 10.1016/s0079-6603(01)68091-7. [DOI] [PubMed] [Google Scholar]
- 174.Fortini P, Pascucci B, Parlanti E, D'Errico M, Simonelli V, Dogliotti E. 8-Oxoguanine DNA damage: at the crossroad of alternative repair pathways. Mutat Res. 2003;531:127–139. doi: 10.1016/j.mrfmmm.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 175.Tsuzuki T, Nakatsu Y, Nakabeppu Y. Significance of error-avoiding mechanisms for oxidative DNA damage in carcinogenesis. Cancer Sci. 2007;98:465–470. doi: 10.1111/j.1349-7006.2007.00409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Nohmi T, Kim SR, Yamada M. Modulation of oxidative mutagenesis and carcinogenesis by polymorphic forms of human DNA repair enzymes. Mutat Res. 2005;591:60–73. doi: 10.1016/j.mrfmmm.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 177.Castro GD, Stamato CJ, Castro JA. 5-methylcytosine attack by free radicals arising from bromotrichloromethane in a model system: structures of reaction products. Free radical biology & medicine. 1994;17:419–428. doi: 10.1016/0891-5849(94)90168-6. [DOI] [PubMed] [Google Scholar]
- 178.Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pratt JR, Parker MD, Affleck LJ, Corps C, Hostert L, Michalak E, Lodge JP. Ischemic epigenetics and the transplanted kidney. Transplant Proc. 2006;38:3344–3346. doi: 10.1016/j.transproceed.2006.10.112. [DOI] [PubMed] [Google Scholar]
- 180.Parker MD, Chambers PA, Lodge JP, Pratt JR. Ischemia- reperfusion injury and its influence on the epigenetic modification of the donor kidney genome. Transplantation. 2008;86:1818–1823. doi: 10.1097/TP.0b013e31818fe8f9. [DOI] [PubMed] [Google Scholar]
- 181.Penn NW, Suwalski R, O'Riley C, Bojanowski K, Yura R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. The Biochemical journal. 1972;126:781–790. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Castro GD, Diaz Gomez MI, Castro JA. 5-Methylcytosine attack by hydroxyl free radicals and during carbon tetrachloride promoted liver microsomal lipid peroxidation: structure of reaction products. Chem Biol Interact. 1996;99:289–299. doi: 10.1016/0009-2797(95)03680-6. [DOI] [PubMed] [Google Scholar]
- 183.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cannon SV, Cummings A, Teebor GW. 5-Hydroxymethylcytosine DNA glycosylase activity in mammalian tissue. Biochem Biophys Res Commun. 1988;151:1173–1179. doi: 10.1016/s0006-291x(88)80489-3. [DOI] [PubMed] [Google Scholar]
- 185.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer research. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 186.Lim SO, Gu JM, Kim MS, Kim HS, Park YN, Park CK, Cho JW, Park YM, Jung G. Epigenetic changes induced by reactive oxygen species in hepatocellular carcinoma: methylation of the E-cadherin promoter. Gastroenterology. 2008;135:2128–2140. doi: 10.1053/j.gastro.2008.07.027. 2140 e2121-2128. [DOI] [PubMed] [Google Scholar]
- 187.Bai H, Konat GW. Hydrogen peroxide mediates higher order chromatin degradation. Neurochem Int. 2003;42:123–129. doi: 10.1016/s0197-0186(02)00072-4. [DOI] [PubMed] [Google Scholar]
- 188.Rahman I, Marwick J, Kirkham P. Redox modulation of chromatin remodeling: impact on histone acetylation and deacetylation, NF-kappaB and pro-inflammatory gene expression. Biochem Pharmacol. 2004;68:1255–1267. doi: 10.1016/j.bcp.2004.05.042. [DOI] [PubMed] [Google Scholar]
- 189.Tas S, Tam CF, Walford RL. Disulfide bonds and the structure of the chromatin complex in relation to aging. Mech Ageing Dev. 1980;12:65–80. doi: 10.1016/0047-6374(80)90030-5. [DOI] [PubMed] [Google Scholar]
- 190.Tas S, Walford RL. Influence of disulfide-reducing agents on fractionation of the chromatin complex by endogenous nucleases and deoxyribonuclease I in aging mice. J Gerontol. 1982;37:673–679. doi: 10.1093/geronj/37.6.673. [DOI] [PubMed] [Google Scholar]
- 191.Tas S, Walford RL. Increased disulfide-mediated condensation of the nuclear DNA-protein complex in lymphocytes during postnatal development and aging. Mech Ageing Dev. 1982;19:73–84. doi: 10.1016/0047-6374(82)90052-5. [DOI] [PubMed] [Google Scholar]
- 192.Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer research. 1991;51:3075–3079. [PubMed] [Google Scholar]