Abstract
In vitro production of the obligate intracellular bacterium, Wolbachia pipientis, is essential to its manipulation as a genetic tool to spread transgenes within vector populations. We have adapted the Wolbachia-infected Aa23 Aedes albopictus mosquito cell line to Eagle’s minimal medium, supplemented with nonessential amino acids, glutamine, and 20% fetal bovine serum. When plated at low densities, Aa23E cells grew as patchy monolayers, comprised of non-contiguous clusters of cells that gave rise to solid clumps of tightly adherent cells. Multicellular clumps eventually detached from the substrate and floated freely in the medium. Removal of Wolbachia by treatment with tetracycline did not alter the cytological properties of the host cells, which had a population doubling time of 4–5 d. The presence of Wolbachia was monitored by Giemsa staining of cytological preparations, polymerase chain reaction (PCR) amplification of Wolbachia 16S ribosomal DNA, and by simultaneous PCR amplification of ribosomal protein genes from Wolbachia and mosquito host cell genomes. Wolbachia morphology was pleomorphic, and Wolbachia DNA persisted in the culture medium for several weeks after degradation of PCR-amplifiable host cell DNA.
Keywords: Wolbachia, Aedes albopictus, Mosquito cell line, Intracellular bacteria, Giemsa stain, Polymerase chain reaction (PCR)
Introduction
Wolbachia were first described as Rickettsia-like intracellular bacteria in cytological preparations from mosquito reproductive tissues (Hertig 1936). More recently, diverse strains of Wolbachia, isolated from a wide distribution of arthropods and from filarial worms, have been described (Lo et al. 2002). In arthropods, the association between the host and Wolbachia is considered commensal or somewhat parasitic, while in filarial worms, the relationship is symbiotic. Although taxonomic and molecular analyses have not established criteria for species designations of Wolbachia, in vitro studies suggest that strains of Wolbachia that occur in insects can be readily transferred between insect hosts (Sinkins and Gould 2006) and to insect cell lines representing diverse orders (Dobson et al. 2002).
In mosquitoes, Wolbachia cause a reproductive distortion known as CI, or cytoplasmic incompatibility (Yen and Barr 1971). In its simplest form, CI decreases or eliminates viable progeny when sperm from an infected male fertilize eggs from an uninfected female. Infected females produce viable, infected progeny regardless of the infection status of the male, thus facilitating spread of Wolbachia through a naïve host population. In this context, Wolbachia provides a potentially powerful genetic drive mechanism for spread of transgenes designed to reduce disease transmission by insect vectors (Sinkins and Gould 2006). Development of reproducible approaches for systematic manipulation of Wolbachia in vitro is essential to effective manipulation of Wolbachia as an agent for insect population control.
The cell line Aa23 (O’Neill et al. 1997) is the first example of a mosquito cell line persistently infected with Wolbachia. The line represents an uncloned population of cells derived from Aedes albopictus (Houston strain) embryonic tissue. These cells retain wAlbB, one of two Wolbachia strains that infected the host mosquitoes (O’Neill et al. 1997; Sinkins 2004). The Aa23 line has been reported to consist of at least two morphological cell types, both of which support infection. O’Neill et al. (1997) further noted that the degree of Wolbachia infection varied, with respect to both the level of infection among individual cells and the overall level of infection in a population.
Cell culture provides an important means of recovering and manipulating Wolbachia in the quantities required for development of genetic tools. Towards this end, we adapted Aa23 cells to supplemented Eagle’s medium, in which we maintain the well-characterized C7-10 A. albopictus cell line (Fallon 1997; Fallon and Kurtti 2005). Because it lacks undefined components such as yeastolate and lactalbumin hydrolysate, Eagle’s medium offers advantages for metabolic labeling and selection of somatic cell variants. Here we describe the growth characteristics and doubling time of Aa23 cells in Eagle’s medium, verify that this medium supports retention of Wolbachia over several passages, and provide polymerase chain reaction (PCR)-based evidence to suggest long-term persistence of Wolbachia in the culture medium after degradation of host cell DNA.
Materials and Methods
Cells and culture conditions
Aa23 cells derived from a culture initiated by O’Neill et al. (1997) were a generous gift from Dr. Stephen Dobson, Department of Entomology, University of Kentucky. The cells were adapted to and subsequently maintained in Eagle’s medium, supplemented with non-essential amino acids, glutamine, vitamins, penicillin, and streptomycin, and 20 % heat-inactivated fetal bovine serum, essentially as described previously (Shih et al. 1998). This formulation of the medium with 20% serum is called E-20. Cells were subcultured at 2-wk intervals by resuspending attached cells and partially disrupting clusters by repeated pipetting; 0.5 ml of the resuspended material was distributed into 4.5 ml of fresh medium. Cells were maintained at 28° C under a 5% CO2 atmosphere. In the text below, adapted cells are called Aa23E, in recognition of possible selection of a subpopulation of the Aa23 cells during adaptation to E-20 medium.
To evaluate effects of tetracycline, uninfected A. albopictus C7-10 cells (Fallon and Kurtti 2005) were plated at 1×105 cells per milliliter in 35 mm dishes, containing 2 ml of E-5 medium (containing 5% fetal bovine serum) per dish and tetracycline at final concentrations from 0 to 100 μg/ml. Cells were allowed to grow until cells in control plates without tetracycline became confluent. Cells were resuspended and counted in a Coulter electronic cell counter.
Doubling times for Aa23E and Aa23E(T) cells
Prior to the experiment, Aa23E(T) cells had been maintained in medium containing tetracycline (10 μg/ml) for six passages. Cells were plated in 35 mm culture dishes in 2 ml of E-20 medium; for Aa23E(T) cells, tetracycline was included in the medium. At each time point, 10% NP-40 (0.1 ml) was added directly to the culture medium and mixed. After 5 min, cells were disrupted thoroughly with a pasteur pipet, and the entire sample was added to 8 ml of phosphate-buffered saline (Dulbecco and Vogt 1954). Nuclei were counted using a Coulter electronic cell counter.
Wolbachia staining
Cells were collected onto glass slides in a cytospin centrifuge (Shandon, Pittsburgh, PA), dried, and fixed for 10 min in 100% acetone. After drying, cells were stained by adding a few drops of freshly prepared staining solution onto the slide, followed by incubation for 10 min at 37° C in a humidified chamber. The staining solution was made by adding 4 ml of Harleco Giemsa stain (EM Science, Gibbstown, NJ) to distilled water (31 ml) to which had been added 3 ml of acetone and 2 ml of 80-mM sodium phosphate, pH 6.8. Excess stain was removed with water, and slides were viewed under oil, using a ×100 objective.
DNA extraction and PCR analyses
Cells (and Wolbachia) were harvested from 1 ml of resuspended culture medium by centrifugation for 10 min at 14,000 rpm in a microcentrifuge. The supernatant was removed by aspiration, and the cell pellet was stored frozen at −20° C. Pellets were resuspended in 0.20 ml of distilled water, and an equal volume of 20 mM Tris–HCl, pH 8.0, containing 0.2 M NaCl, 2 mM ethylenediamine tetraacetic acid, 1% sodium dodecyl sulfate, and 10 μg DNAse-free RNAse A, was added. After 1 h at 37° C, proteinase K (20 μg) was added, and incubation continued at 56° C for 1 h. The samples were heated at 95° C for 10 min, extracted with phenol, and precipitated with ethanol. DNA was recovered by centrifugation, washed in 70% ethanol, dried, and resuspended in 0.1 ml of double-distilled water.
Mosquito PCR primers were based on the single-copy gene for ribosomal protein RpS6 (Hernandez et al. 2003), which has unusual variation among mosquito species (Fallon and Li 2007). Wolbachia primers were based on ribosomal proteins rpS12 (rpsL) and rpS7 (rpsG), which are members of the “str operon” in Escherichia coli (Saito et al. 1994), and whose gene order is conserved in the genome of Drosophila melanogaster wMel (Wu et al. 2004). Primer sequences are given in Table 1.
Table 1.
PCR primers
| Name | Sequence | Target DNA |
|---|---|---|
| 99F | 5′-TTG TAG CCT GCT ATG GTA TAA CT | W 16S rDNA |
| 994R | 5′-GAA TAG GTA TGA TTT TCA TGT | W 16S rDNA |
| S12F | 5′-GCA CTA AGG TGT ATA CTA CAA CTC C | W str operon |
| S7R | 5′-GCC TTA TTA GCT TCA GCC AT | W str operon |
| S6.97F | 5′-ACT TCT ACG ACA AGC GTA TGG | Aedes RpS6 |
| S6.560R | 5′-TGC AGC ACA ACA GGA GTG ATC | Aedes RPS6 |
Primers 99F and 994R were designed by O’Neill et al. 1992. Primers S12F, S7R, S6.97F and S6.560R were designed specifically for this study.
PCR was done in a total reaction volume of 20 μl, containing final concentrations of magnesium chloride at 2.5 mM, each of the four deoxyribonucleotide triphosphates at 0.20 mM, primers at 400 nM, template, and Taq polymerase (Promega, Madison, WI; Catalog # M1661; 2.5 units per reaction), and 4 μl of DNA template (containing up to 1.5 μg DNA in double-distilled water as described above). Controls (not shown) included samples with water in place of template, DNA from uninfected C7-10 cells, and a standardized DNA sample from Wolbachia-infected cells. For primers 99F/994R, reaction conditions involved an initial denaturation at 95° C for 2 min, followed by 35 cycles of 95° C denaturation, 52° C annealing, and 72° C extension, with a final extension at 72° C for 2 min. For the primer mixture containing S12F/S7R and S6.97F/S6.560R, the reaction conditions were 1 cycle at 95° C for 2 min, 35 cycles at 95° C for 1 min, 63° C for 1 min, 72° C for 1 min, followed by 1 cycle at 72° C for 3 min. Samples were electrophoresed on 2% agarose gels in buffer containing ethidium bromide and photographed with ultraviolet light illumination. Images were “inverted” electronically to show dark bands on a white background.
Results
Growth characteristics of Aa23 cells
Aa23 cells were originally maintained in a culture medium made from equal volumes of Mitsuhashi–Maramorosch and VP12 media, containing 15% heat-inactivated fetal bovine serum (Varma and Pudney 1969; O’Neill et al. 1997). More recently, Rasgon et al. (2006) maintained Aa23 cells in Schneider’s Drosophila medium (Schneider 1964). Because these media contain undefined components such as yeastolate and lactalbumin hydrolysate, which can compromise isotope incorporation and use of metabolic inhibitors, we adapted Aa23 cells to a supplemented version of Eagle’s (E) medium containing 20% heat-inactivated, fetal bovine serum (E-20). Apart from the serum, Eagle’s medium with additional vitamins, non-essential amino acids, and glutamine (Shih et al. 1998) is chemically defined.
In Eagles medium, Aa23E cells retained the morphological heterogeneity originally described by O’Neill et al. (1997). The overall appearance of the culture was affected by the degree of dilution at the time of subculture. Because the duration of exponential growth is affected by cell density (Gerenday and Fallon 1996), Aa23E cells were routinely subcultured at a 1:10 dilution into fresh E-20 medium at 2- to 3-wk intervals. Unlike A. albopictus C7-10 cells, which represent a clonal population (Fallon and Kurtti 2005), Aa23E cells tended to grow as tightly adhesive cell clusters, which did not disperse evenly by repeated passage through a pipet. The diluted Aa23E cell suspension formed patchy monolayers, in which individual cells did not make uniform, confluent contact with adjacent patches of cells (Fig. 1a). Cell growth was most robust in foci of aggregated cells (Fig. 1a, arrow), from which grew solid clusters of tightly adherent cells (Fig. 1b). As the clusters enlarged in size, they detached from the substrate and floated freely in the medium. Meanwhile, the proportion of attached cells decreased (compare Fig. 1b, c with Fig. 1d), with accompanying accumulation of amorphous sub-cellular debris, barely visible by light microscopy, in the culture medium. At least a proportion of the cells remained viable over several weeks, even when the majority of cells were contained within floating clumps. These cells regenerated patchy monolayers (Fig. 1a) when diluted into fresh medium after dispersal by various means, ranging from gentle pipetting to brief sonication.
Figure 1.
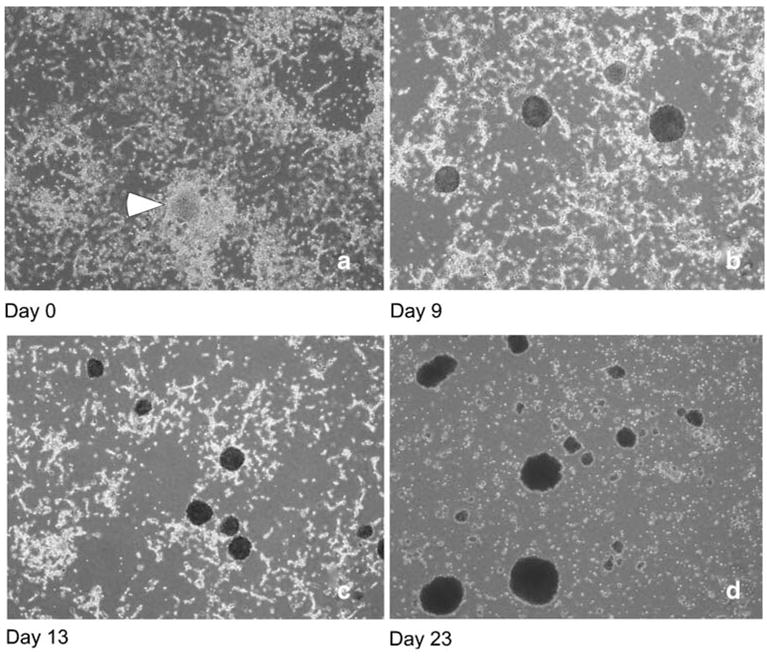
Changes in cytological characteristics of Aa23E cells during time in culture. Photographs were taken with a ×4 objective.
Typically, cultures were maintained without addition of uninfected cells or replenishment of the medium. However, when the medium was replenished, the proportion of attached cells increased, with eventual formation of additional clusters. When cells were subcultured at relatively high density, such as a 1:3 dilution, they formed more contiguous monolayers than those shown in Fig. 1 but with increasing dilution, the proportion of surviving cells represented by clusters also increased, preventing establishment of clonal populations originating from single cells. Attempts to “subclone” Aa23E cells, by allowing the largest cell clusters to settle by gravity and re-plating only those cells that remained in suspension, did not reduce their tendency to form adherent clusters.
Cytological detection of Wolbachia infection
When levels of Wolbachia in the Aa23E cells were evaluated in cytospin preparations stained with Giemsa, high levels of infection were particularly apparent in the older, deteriorating cultures (Fig. 1d). In highly infected cells (Fig. 2a), Wolbachia appeared as darkly stained, roughly round particles of varying size, at an abundance comparable to that observed using a monoclonal antibody (see Fig. 2 in O’Neill et al. 1997). Although it remains possible that apparent extracellular forms arise from particularly fragile cells during processing, these included distinct rod-like structures (Fig. 2b) reminiscent of the diverse pleomorphic forms described by Hertig (1936). Despite the ease with which highly infected cells could be identified in stained preparations by light microscopy, the relatively dense cytoplasm in uninfected cultures made it difficult to ascertain the absolute absence of Wolbachia.
Figure 2.
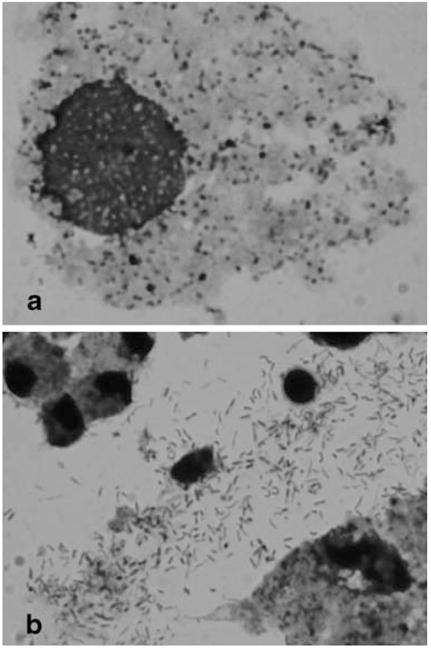
Appearance of Wolbachia in Giemsa-stained preparations. Samples were collected onto slides by centrifugation, dried, fixed in acetone, and stained with Giemsa stain as described in “Materials and Methods.” a Rounded forms of Wolbachia within a cell. b Extracellular, elongated forms. Photographs were taken with a ×100 objective.
Cultures can be cured with tetracycline
In earlier studies, O’Neill et al. (1997) used tetracycline (10 μg/ml) to eliminate Wolbachia from Aa23 cells. We used the uninfected A. albopictus C7-10 cell line (Fallon and Kurtti 2005) to verify that tetracycline itself, at the concentration established by O’Neill et al. (1997), had relatively little effect on cell growth in Eagle’s medium (Fig. 3a). In a standard test for toxicity, cells were plated at 1×105 cells per milliliter with increasing concentrations of tetracycline. After 4 d, when control cells without tetracycline reached confluency (~4×106 cells per plate), cells from all of the plates were counted. Note that cell numbers in control plates increased more than tenfold over a 4-d interval, consistent with the population doubling time of approximately 18 h established by Gerenday and Fallon (1996). In the presence of tetracycline at 5 and 10 μg/ml, cell numbers attained 95% and 94% of control values, respectively. At 25 μg/ml, tetracycline decreased cell accumulation by 44%.
Figure 3.
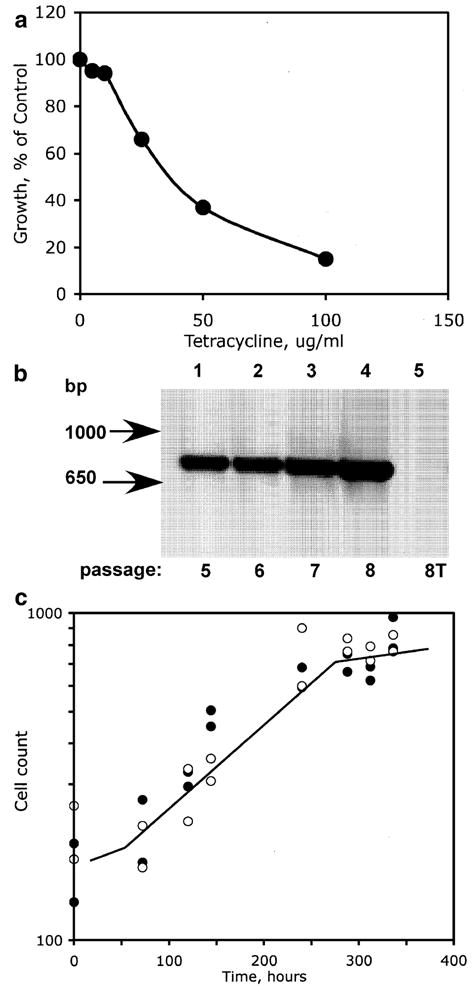
Removal of Wolbachia with tetracycline and effects on doubling time. (a) Toxicity curve for tetracycline. Growth of A. albopictus C7-10 cells after 4 d in E-5 medium containing various concentrations of tetracycline is expressed as a percentage of control values in the absence of tetracycline (1.9×106 cells per milliliter). Values are averaged from two counts from each of two duplicate plates; variation among counts was less than 5%. (b) DNA was extracted from Aa23E cells (passages 5, 6, 7, 8, established over a 7-wk interval) and assayed by PCR using primer pairs 99F/994R (Table 1). Tetracycline-cured cells (8T) had been maintained for 22 d in tetracycline. Base pair (bp) markers indicate band length. (c) Similar growth rates in Aa23E and Aa23E(T) cells. Solid symbols show the high and low values from a range of three to five replicate counts of nuclei from NP-40-lysed Aa23E cells; open symbols show similar data for Aa23E(T) cells. Counts ranged from 1.8×105 to 1×106 nuclei per milliliter.
To verify that growth in the presence of tetracycline eliminated Wolbachia from Aa23E cells, we used PCR to amplify 16S rDNA with the diagnostic primer pair 99F/994R (Table 1; O’Neill et al. 1992). Figure 3b shows levels of 16S rDNA amplification from Aa23E cells from passages 5, 6, 7, and 8 (lanes 1–4), maintained without tetracycline. Cells from passage 6 were subcultured in the presence of tetracycline (10 μg/ml) for two consecutive passages over a 22-d period. At passage 8, these Aa23E(T) cells were free of PCR-detectable Wolbachia DNA (Fig. 3b, lane 5, compare lanes 1–4). After removal of Wolbachia, Aa23E(T) cells showed the same morphological characteristics and a similar progression from patchy monolayers to floating clumps of cells as the cultures aged, as did the Aa23E cells shown in Fig. 1.
As was the case with Aa23E cells, we were unable to disperse tetracycline-cured Ae23E(T) cells into single cells by mechanical disruption. Nevertheless, we obtained a rough estimate of the population doubling time by lysing the cells with NP-40 and counting released nuclei with a Coulter electronic cell counter (Fig. 3c). By this criterion, growth rates of Aa23E and Aa23E(T) cells were comparable, with an exponential phase corresponding to a fourfold increase in cell number and a population doubling time on the order of 4–5 d. Computer-generated trendlines were nearly superimposable, with an R2 value of 0.907 for the Aa23E cells and 0.865 for the Aa23E(T) cells.
Long-term persistence of Wolbachia
PCR products shown in Fig. 3b, lanes 1–4, were amplified from DNA isolated from cultures that had been maintained undisturbed, without supplementation of the culture medium, from the date of the designated passage. These cultures gradually deteriorated, generating floating clusters of cells as shown in Fig. 1d. As shown above (Fig. 3b), Wolbachia DNA remained intact during long-term maintenance, and to a first approximation, levels of Wolbachia DNA were comparable in cultures held from passages 5 to 8. The intensity of the PCR product in relatively fresh cultures (passage 8) contrasted with our evaluation of Wolbachia density by Giemsa staining, wherein abundant Wolbachia were most easily detected in older cultures and more difficult to see in fresh cultures. The discrepancy between apparent Wolbachia abundance by Giemsa staining and PCR raises the possibility that the various forms of Wolbachia described by Hertig (1936) vary in their accessibility to stains, particularly during early stages of infection. Thus, for sensitive detection of Wolbachia in the early stages of infection, PCR is the more reliable approach.
When we amplified host cell and Wolbachia DNA using two sets of primers targeting single-copy, protein-coding genes (see “Discussion”), we found that the upper band, representing the Wolbachia product, and lower band, representing mosquito genomic DNA, increased in parallel with time (Fig 4a), with reasonable consistency in replicate plates (compare lanes 1–4; 5–8). Based on relative intensity of the PCR bands, overall densities of Wolbachia increased as the cells grew, and eventually stabilized. During the third month of cell maintenance without subculture and/or replenishment with fresh medium, host cell DNA degraded to the point that it was not amplifiable by PCR (Fig. 4b, compare lane 1 with lanes 2–5), while Wolbachia DNA, amplifiable by PCR, was detectable in the cultures for an additional 2.5 mo, in the apparent absence of metabolically active cells. We have repeated these PCR reactions on serially diluted samples to verify our interpretation of these data and have observed similar persistence of Wolbachia DNA in the absence of amplifiable mosquito DNA in cells grown in suspension culture (data not shown). Finally, we note that Rasgon et al. (2006) recently reported survival of wAlbB in cell-free medium. Loss of amplifiable host cell DNA suggests that once levels of Wolbachia attain a critical level, cellular control breaks down, consistent with the abundance of Wolbachia detectable by Giemsa staining in aged cultures. This differential stability of Wolbachia DNA suggests that extracellular forms remain intact, and possibly infectious, for extended periods.
Figure 4.
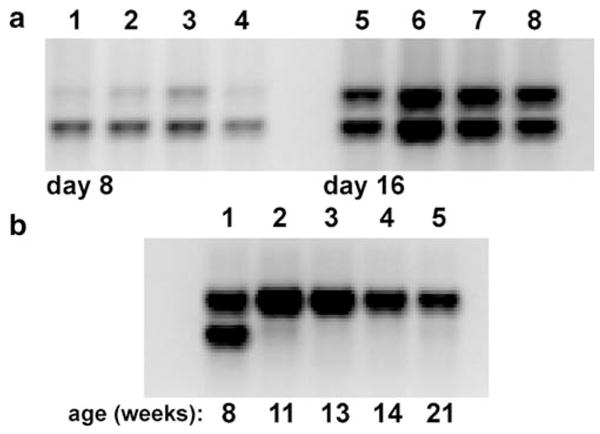
Changes in Wolbachia abundance. a Aa23E cells at passage 17 were seeded in sets of four replicate 35 mm plates at low density in E-20 medium and sampled at 8 and 16 d after plating. The upper band represents Wolbachia DNA amplified by primer pair S12F/S7R; the lower band represents host cell nuclear DNA amplified by S6.97F/S6.560R in the same reaction. b Relative stability of Wolbachia (upper band) and cellular (lower band) DNA as a function of weeks in culture, without passage.
Discussion
Cell lines persistently infected with obligate intracellular bacteria provide an important tool for investigating metabolic interactions that support host–microbe interactions. Among mosquito cell lines, the Aa23 line is unique in that it was intentionally developed from embryos of the naturally infected Houston strain of A. albopictus mosquitoes, with the goal of establishing a line persistently infected with Wolbachia pipientis (O’Neill et al. 1997). The mosquitoes from which Aa23 cells derive were coinfected with two strains of Wolbachia, wAlbA and wAlbB, which belong to classes A and B, respectively, as defined by Lo et al. (2002). However, Aa23 cells retained only wAlbB, which was the more abundant strain in the infected mosquitoes (Dutton and Sinkins 2004; Sinkins 2004). The wAlbB infection has remained in the cell line over nearly a decade, suggesting that it may be particularly well adapted to its host cell. Despite distribution of Aa23 cells to several laboratories, factors responsible for variability in overall levels of infection in individual cell populations and among individual cells within a population remain unknown.
To provide a basis for identifying metabolic interactions between Wolbachia and the host cells, we first adapted the Aa23 cell line to an Eagle’s-based medium, which aside from the serum is chemically defined. Although adaptation to new culture conditions may have selected for a subpopulation of cells, the Wolbachia infection remained stable in the adapted, Aa23E line. During more than 18 mo (32 passages to date) in E-20 medium, we have not lost the infection other than by deliberate treatment with tetracycline.
Because Aa23E cells grow primarily as tightly adherent clusters, their metabolic uniformity is difficult to assess. It is not clear, for example, how Wolbachia are distributed within a cluster or whether they preferentially grow in cells that, at least initially, are attached to the substrate. Moreover, because cells within a cluster are unevenly exposed to the culture medium, it seems likely that the cell population that forms each cluster generates a gradient of decreasing metabolic activity extending from the surface to the interior. Although the extent to which this growth pattern is required for stable maintenance of Wolbachia remains to be determined, elimination of Wolbachia by tetracycline treatment does not alter these characteristics of cell growth. Thus, the cytological properties of Aa23E cells are independent of Wolbachia infection per se.
We used standard PCR-based approaches to obtain a qualitative estimation of changes in Wolbachia abundance in experimental cell populations, representing on the order of 106 to 107 cells in a 60-mm plate. PCR primers 99F/994R, which amplify a region of Wolbachia rDNA, sometimes failed to give good agreement between replicate samples. Because variability was greatest when template levels were high, it seems likely that formation of secondary structure within the single stranded template DNA competes with primer annealing/extension.
We eliminated this difficulty by designing primers based on protein-coding genes, eventually including two primer pairs designed to amplify single-copy ribosomal protein genes from Wolbachia and from the host cell nuclear genome in the same reaction. The relative amounts of PCR products suggested that Wolbachia abundance gradually increases with increasing cell number, while the host cells remained viable. Giemsa staining suggests that forms of Wolbachia detectable by staining undergo rapid accumulation as the culture deteriorates, possibly resulting from liberation of precursors within dying cells that are scavenged by Wolbachia for growth and metabolism. Persistence of Wolbachia DNA for several weeks, even after the host cell DNA disappeared entirely, suggests survival of intact bacteria, but their infectivity relative to Wolbachia isolated directly from cells remains to be rigorously assessed. Along similar lines, however, Rasgon et al. (2006) reported that Wolbachia purified from Aa23 cells and stored extracellularly for up to 1 wk in culture medium remained infectious when inoculated at the relatively high level of 2,600 bacteria per tetracycline-cured [Aa23(T)] cell.
Efforts to understand initiation and maintenance of Wolbachia infections are particularly amenable to exploration in cell culture. For example, Sasaki et al. (1998) attempted to metabolically label Wolbachia-infected ovaries and testes in Drosophila simulans adults by injecting labeled precursor into insects that had been pre-treated with cycloheximide, which was expected to differentially inhibit eukaryotic (host cell) protein synthesis. Although results with intact Drosophila were inconclusive, cultured cells may provide a more tractable system for detecting qualitative differences between infected and uninfected cells that can be attributed to proteins synthesized by Wolbachia.
The potential importance of Wolbachia as an effective genetic drive system for insect vectors warrants further systematic investigation of its capacity for in vitro manipulation. Studies with mosquito embryos suggest that Wolbachia replication is dependent on host cell replication (Ruang-areerate et al. 2004), and if this is the case, we anticipate that detection of metabolic events reflecting the presence of Wolbachia may be facilitated by transfer of the infection to one of the more rapidly growing clonal populations of A. albopictus cell lines that we have described over the past several years (Fallon and Kurtti 2005).
Acknowledgments
This work was supported by NIH grant AI070913 and by the University of Minnesota Agricultural Experiment Station, St. Paul, MN. I thank Anna Gerenday for technical assistance and Drs. TJ Kurtti, UG Munderloh and A Gerenday for helpful discussions.
References
- Dobson SL, Marsland EJ, Veneti Z, Bourtzis K, O’Neill SL. Characterization of Wolbachia host cell range via the in vitro establishment of infections. Appl Environ Microbiol. 2002;68:656–660. doi: 10.1128/AEM.68.2.656-660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulbecco R, Vogt M. Plaque formation and isolation of pure lines with poliomyelitis virus. J Exp Med. 1954;99:167–182. doi: 10.1084/jem.99.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton TJ, Sinkins SP. Strain-specific quantitation of Wolbachia density in Aedes albopictus and effects on larval rearing conditions. Insect Mol Biol. 2004;13:317–322. doi: 10.1111/j.0962-1075.2004.00490.x. [DOI] [PubMed] [Google Scholar]
- Fallon AM. Transfection of cultured mosquito cells. In: Crampton JM, Beard CB, Louis C, editors. Molecular biology of insect disease vectors. Chapman and Hall; New York: 1997. pp. 430–443. [Google Scholar]
- Fallon AM, Kurtti TJ. Cultured cells as a tool for analysis of gene expression. In: Marquardt WC, editor. Biology of disease vectors. 2. Elsevier; New York: 2005. pp. 539–549. [Google Scholar]
- Fallon AM, Li L. The C-terminal extension that characterizes mosquito (Diptera: Culicidae) ribosomal protein S6 is widespread among the Culicomorpha. J Med Entomol. 2007;44:608–616. doi: 10.1603/0022-2585(2007)44[608:tcetcm]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Gerenday A, Fallon AM. Cell cycle parameters in Aedes albopictus mosquito cells. In Vitro Cell Dev Biol, Anim. 1996;32:307–312. doi: 10.1007/BF02723064. [DOI] [PubMed] [Google Scholar]
- Hernandez VP, Higgins LA, Schwientek MS, Fallon AM. The histone-like C-terminal extension in ribosomal protein S6 in Aedes and Anopheles mosquitoes is encoded within the distal portion of exon 3. Insect Biochem Mol Biol. 2003;33:901–910. doi: 10.1016/s0965-1748(03)00095-x. [DOI] [PubMed] [Google Scholar]
- Hertig M. The Rickettsia, Wolbachia pipientis (gen et sp. n.) and associated inclusions of the mosquito, Culex pipiens. Parasitology. 1936;28:453–486. [Google Scholar]
- Lo N, Casiraghi M, Salati E, Bazzocchi C, Bandi C. How many Wolbachia supergroups exist? Mol Biol Evol. 2002;19:341–346. doi: 10.1093/oxfordjournals.molbev.a004087. [DOI] [PubMed] [Google Scholar]
- O’Neill SL, Giordano R, Colbert AME, Karr TL, Robertson HM. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc Natl Acad Sci U S A. 1992;89:2699–2702. doi: 10.1073/pnas.89.7.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill SL, Pettigrew MM, Sinkins SP, Braig HR, Andreadis TG, Tesh RB. In vitro cultivation of Wolbachia pipientis in an Aedes albopictus cell line. Insect Mol Biol. 1997;6:33–39. doi: 10.1046/j.1365-2583.1997.00157.x. [DOI] [PubMed] [Google Scholar]
- Rasgon JL, Gamston CE, Ren X. Survival of Wolbachia pipientis in cell-free medium. Appl Env Microbiol. 2006;72:6934–6937. doi: 10.1128/AEM.01673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruang-areerate T, Kittayapong P, McGraw EA, Baimai V, O’Neill SL. Wolbachia replication and host cell division in Aedes albopictus. Curr Microbiol. 2004;49:10–12. doi: 10.1007/s00284-003-4245-8. [DOI] [PubMed] [Google Scholar]
- Saito K, Mattheakis LC, Nomura M. Post-transcriptional regulation of the str operon in Escherichia coliRibosomal protein S7 inhibits coupled translation of S7 but not its independent translation. J Mol Biol. 1994;235:111–124. doi: 10.1016/s0022-2836(05)80020-8. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Braig HR, O’Neill SL. Analysis of Wolbachia protein synthesis in Drosophila in vivo. Insect Mol Biol. 1998;7:101–105. doi: 10.1046/j.1365-2583.1998.72057.x. [DOI] [PubMed] [Google Scholar]
- Schneider I. Differentiation of larval Drosophila eye-antennal discs in vitro. J Exp Zool. 1964;156:91–104. doi: 10.1002/jez.1401560107. [DOI] [PubMed] [Google Scholar]
- Shih KM, Gerenday A, Fallon AM. Culture of mosquito cells in Eagle’s medium. In Vitro Cell Dev Biol, Anim. 1998;34:629–630. doi: 10.1007/s11626-996-0010-1. [DOI] [PubMed] [Google Scholar]
- Sinkins SP. Wolbachia and cytoplasmic incompatibility in mosquitoes. Insect Biochem Mol Biol. 2004;34:723–729. doi: 10.1016/j.ibmb.2004.03.025. [DOI] [PubMed] [Google Scholar]
- Sinkins SP, Gould F. Gene drive systems for insect disease vectors. Nat Rev, Genet. 2006;7:427–435. doi: 10.1038/nrg1870. [DOI] [PubMed] [Google Scholar]
- Varma MGR, Pudney M. The growth and serial passage of cell lines from Aedes aegypti (L.) larvae in different media. J Med Entomol. 1969;6:432–439. doi: 10.1093/jmedent/6.4.432. [DOI] [PubMed] [Google Scholar]
- Wu M, Sun LV, Vamathevan J, Riegler M, Deboy R, Brownlie JC, et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: A streamlined genome overrun by mobile genetic elements. PloS Biol. 2004;2:E69. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen JH, Barr AR. New hypothesis of the cause of cytoplasmic incompatibility in Culex pipiensL . Nature. 1971;232:657–658. doi: 10.1038/232657a0. [DOI] [PubMed] [Google Scholar]


