Summary
The challenges associated with the structural characterization of disordered proteins have resulted in the application of a host of biophysical methods to such systems. NMR spectroscopy is perhaps the most readily suited technique for providing high-resolution structural information on disordered protein states in solution. Optical methods, solid state NMR, ESR and x-ray scattering can also provide valuable information regarding the ensemble of conformations sampled by disordered states. Finally, computational studies have begun to assume an increasingly important role in interpreting and extending the impact of experimental data obtained for such systems. This article discusses recent advances in the applications of these methods to intrinsically disordered proteins.
Introduction
A disordered protein is commonly defined as one that does not adopt a well-defined native structure when isolated in solution under near-physiological conditions. A disordered protein region can be defined as a region that both meets the above definition when excised from the full-length protein to which it belongs and retains its disordered character in the context of the complete protein. Interest in disordered proteins has swelled as a result of the realization that such proteins, instead of the rare exceptions they were once envisioned to be, are unexpectedly and perhaps even astonishingly common in human and other genomes [1–3]. Furthermore, disordered proteins do not appear to simply occur as ‘filler material’ amongst functional well-structured proteins, and instead are associated with a variety of biological functions, many of them intimately related to human disease [4–6]. Therefore, characterization of the structural properties of such proteins must be considered an important complement to more typical structural studies of well-ordered proteins.
The absence of a well-defined structure in disordered proteins complicates the approach that must be taken when considering structural studies, since the most common goal, the determination of a unique high-resolution structure, is not attainable for the isolated protein. Instead, the goal of such studies is usually to obtain experimental constraints on the ensemble of states that is sampled by the polypeptide in question, including the detection of residual secondary structure, transient long-range contacts, and regions of restricted or enhanced mobility, with the hope that such information may prove informative regarding the associated biological function [7–9]. It should be noted that many disordered proteins do adopt more highly ordered conformations upon interactions with other cellular components [10]. In some cases, these bound states are subject to the same methods used for solving the structures of well-structured protein complexes, permitting a high-resolution view of the bound protein in question. However, in at least some cases, the bound states of disordered proteins remain highly non-compact and retain substantial mobility, and a brief discussion of structural studies of such bound states is therefore included.
In many ways intrinsically disordered proteins resemble denatured states of well-structured proteins. The latter have been subjected to greatly increased scrutiny in the past few decades, largely in the context of attempts to better understand the process of protein folding. While the focus herein is primarily on studies of intrinsically disordered proteins rather than denatured states, many of the techniques that are applied to the former have been adapted directly from those used in studies of the latter, and a number of reviews of such studies are therefore directly relevant [11–14].
Residual Secondary Structure
Secondary structure in disordered proteins is typically transient and confined to short individual helical or extended segments with ensemble-averaged structured populations ranging from a few nearly 100 percent. Because these segments often comprise a small fraction of the total protein sequence, they are typically difficult to detect using techniques for measuring overall secondary structure content such as circular dichroism or infrared spectroscopies. Solution state NMR is probably the technique best suited for the detection and delineation of such segments. Chemical shifts, with their exquisite sensitivity to environment and structure, remain the most powerful tool for achieving this goal. Deviations of observed shifts from those estimated for an idealized unstructured state (so called random coil shifts) can be used to assess both the location and population of transient (or full formed) secondary structure [7] (Figure 1). In cases of very low populations (10% or lower) uncertainties in the random coil shifts can lead to ambiguities in this type of analysis, which can be lessened to some extent by looking at shifts from several different nuclei, linear combinations of shifts, or more sophisticated moving averages [15]. Short range NOEs can be used to corroborate secondary structure propensities detected through chemical shifts [16,17], although the effects of local dynamics on such an analysis must be considered [18]. NMR coupling constants, which can be directly related to bond torsion angles, can also be used to pick up local secondary structure preferences [19,20], although effects behind local variability in such measurements are not as well characterized as in the case of chemical shifts [21].
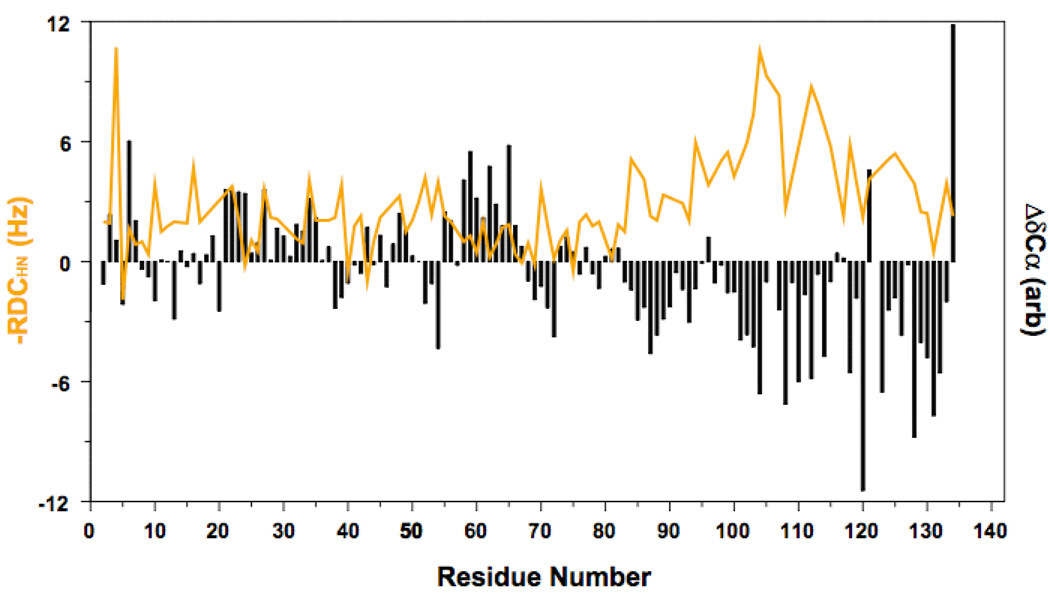
Figure 1. NMR α-carbon secondary chemical shifts and NH RDCs for β-synuclein.
Chemical shift deviations from random coil values indicate a preference for extended structure (negative values) in the C-terminal region and helical structure (positive values) in the N-terminal region. Large amplitude RDCs in the C-terminal region correlate qualitatively with negative shift deviations and likely reflect locally extended chain segments. Smaller amplitude RDCs in the N-terminal region occur largely in regions of positive shift deviations, although the RDCs do not change sign as would be expected for more highly populated helical structure. Adapted from [26].
Most recently, residual dipolar couplings (RDCs) have been shown to be sensitive to local secondary structure in disordered or unfolded proteins. In anisotropic media, polypeptide chain segments can align weakly along a preferred direction, leading to a weak recoupling of the spin-spin dipolar interaction and resulting in measurable NMR line splittings. Because bond vectors (and in particular NH bond vectors) can be orthogonally aligned in helical or extended structures, different local structure leads to opposite effects on the observed RDCs, leading to a characteristic signature for each type of secondary structure [22] (Figure 1). However, RDCs do not disappear for proteins even under strongly denaturing conditions expected to disrupt all secondary structure, possibly because they are dominated by contributions from highly extended conformations within the ensemble [23]. As a result, while robust helical structure is evidenced as RDCs of opposite sign [22,24], more transient helical structure may lead to only a decrease in RDC amplitude without a sign reversal [25,26] (Figure 1). A potential way to mitigate this difficulty may be to analyze RDC differences from those observed under strongly denaturing conditions [22,25]. At present, however, quantitative interpretation of disordered protein RDCs in terms of residual secondary structure awaits further work.
Many parameters that can be measured using solution state NMR can in principle be determined using solid state NMR as well. One advantage of solid state methods is that they may eliminate rapid interconversion of different populations within a single sample, facilitating the direct measurement of structural parameters such as chemical shifts for each sub-ensemble. A recent report demonstrates the use of solid state NMR to examine secondary structure preferences based on chemical shifts in a denatured villin headpiece subdomain [27], suggesting that this technique may also be applicable to the characterization of disordered proteins.
Long-range Contacts
Long-range contacts in disordered proteins are inherently transient and therefore difficult to detect. Furthermore, a clear definition for such contacts is important, since it is to be expected that different regions within a highly flexible disordered polypeptide chain will occasionally approach or contact each other in the absence of any preferred interactions. In general, this expectation has to be made explicit based on statistical or computational models of the distance distribution between two residues or locations within a polypeptide chain sampling an idealized random coil ensemble. Subsequently, such a model can be used to predict the corresponding experimental data in the absence of preferred contacts, which can then be compared with data obtained from actual measurements.
The overall dimensions of fully unfolded protein ensembles are well-predicted by statistical models [28] but are expected to decrease in the presence of long-range interactions. Measurements of parameters such as the hydrodynamic radius (Rh) or radius of gyration (Rg) of a protein can be used to detect such a decrease [29,30]. Techniques such as size-exclusion chromatography or analytical ultracentrifugation, which can easily differentiate between particles with dramatically different Rh have not typically been used for studies of disordered proteins, possibly because of relatively small anticipated changes. Instead, pulse-field gradient (PFG) NMR, small-angle x-ray scattering (SAXS) and fluorescence correlation spectroscopy (FCS) measurements have been more commonly used. PFG experiments enable measurements of Rh for a disordered protein [31–33], which can indicate the presence of long-range contacts within an ensemble, but they are not able to pinpoint specific regions or residues involved in such contacts. FCS measurements of fluorescently tagged disordered proteins can also provide diffusion coefficients [34,35]. In addition, however, FCS is sensitive to fluorophore quenching reactions that occur as a function of internal dynamics [12]. If specific quenching groups can be identified in the protein, some information regarding specific contacts can be obtained [36]. SAXS measurements principally provide a measurement of the radius of gyration (Rg) for a disordered protein ensemble [37]. However, for systems containing both fully ordered and disordered regions [24,38,39], or even less ordered and more ordered domains [40] pair distribution functions from SAXS data can provide powerful constraints on the relative topology of the different domains.
Experimental methods that report on distances are best suited for detecting specific long-range contacts. Fluorescence energy transfer, a sensitive measurement of specific distances, has proven useful in this context. Although various donor-acceptor pairs can be used, tryptophan is often convenient for disordered proteins as it rarely occurs within their native sequences. Acceptors used in recent reports include modified tyrosine residues (3-nitrotyrosine) [41] and 1,5-IAEDANS-labeled cysteine residues [42]. Dyes from the Alexa family (Invitrogen) have also been used effectively [36]. Measurements provide detailed information on the distance distribution between specific sites in a disordered protein, which can be used to constrain conformational ensembles. A shortcoming, however, is that multiple constructs with different donor-acceptor locations are necessary for a comprehensive characterization.
NMR long-range NOEs, traditionally used for obtaining topological distance constraints in well-structured proteins, have proven difficult to observe in disordered proteins. This likely results from two causes: first, the contacts in disordered proteins may be too short lived to allow for efficient buildup of NOE signals, and second, the very high degeneracy of side chain chemical shifts makes it very difficult to reliably assign long-range NOEs involving side chain nuclei. Nevertheless, successful detection of medium- and long-range NOEs in an unfolded state of a well-structured protein was reported in a study employing selective labeling strategies to greatly reduce the number of proton signals [43]. Thus, further advances may increase the applicability of NOE methods to intrinsically disordered systems.
Paramagnetic relaxation enhancement (PRE) has perhaps been most successful at detecting long-range contacts in disordered protein ensembles [44–47]. The method, which was again used first in studies of unfolded or partially folded proteins [48–50], relies on the dipolar interaction between nuclear spins and the spin of an unpaired electron, typically introduced in the form of a nitroxide compound, the so called spin label, conjugated to a cysteine residue. Alternately, chelating groups can also be used to introduce a paramagnetic metal [51,52]. Spins in the physical proximity of the spin label relax more efficiently, leading to broadened signals, an effect that is reliably detectable to approximately 25 ångströms [49]. Positions near the site of spin-label attachment in the polypeptide sequence are covalently constrained to be in the spatial vicinity of the spin label and always exhibit resonance broadening. For ideal random coils, however, residues increasingly distant in sequence from the labeled position spend relatively less time in the proximity of the label and should exhibit monotonically decreasing degrees of broadening. Deviations from this expected pattern are indicative of preferential long-range stabilizing interactions. Importantly, most reports to date measure PRE effects by comparing signal intensities in a single pair of spectra acquired in the presence and absence of a paramagnetic spin label. This approach is subject to systematic errors, and more accurate measurements are recommended to enable quantitative interpretation of PRE data [53,54].
In addition to measurements reflecting inter-spin distances, other NMR parameters have the potential to report on long-range contacts in disordered polypeptides. Chemical shifts are exquisitely sensitive to environmental perturbations, and might be expected to reflect any alteration of long-range interactions. Surprising, however, transient contacts that are observable using PRE methods do not appear to be robustly reflected in chemical shifts [45,55], possibly because of their highly fluctional nature. RDCs have also been proposed to be sensitive to long-range contacts, with reports attributing differences between predicted and observed RDCs to such interactions [56,57]. More recently, however, this interpretation has been brought into question by data suggesting that local transient secondary structure is responsible for the disagreement between the modeled and observed RDC data [26] (Figure 1 and Figure 2).
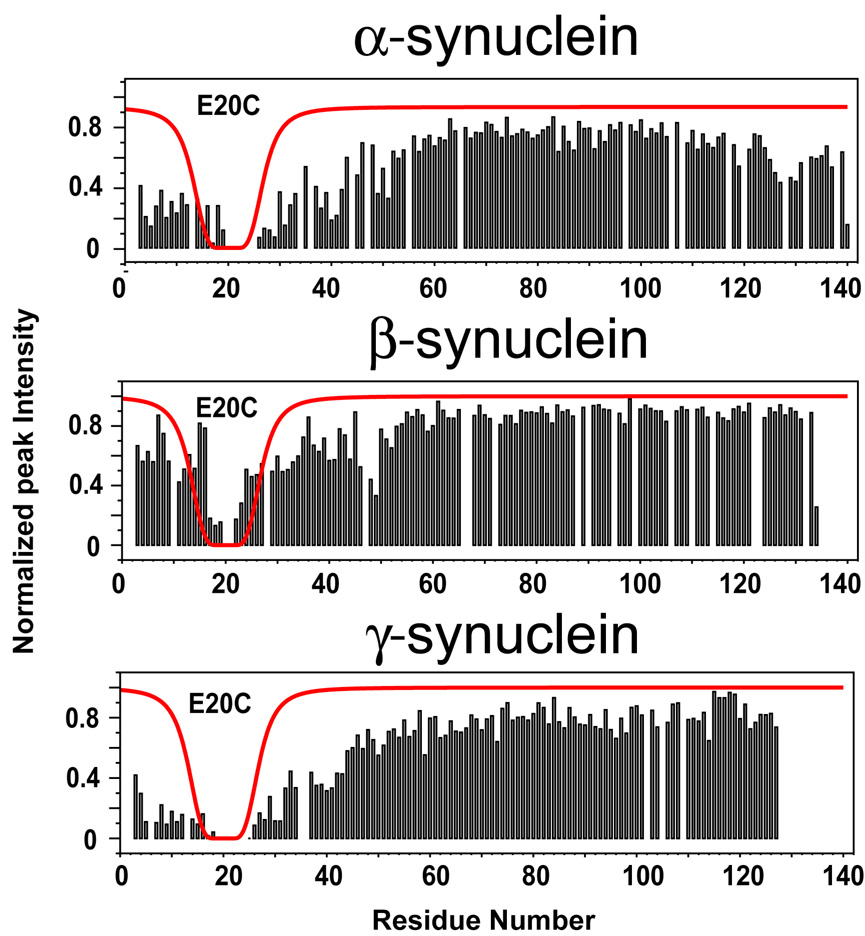
Figure 2. NMR PRE in α-, β- and γ-synuclein.
A paramagnetic spin label attached to a cysteine introduced at position 20 leads to clear broadening in the C-terminal tail of α-synuclein, but not of β- or γ-synuclein, suggesting long-range N- to C-terminal contacts are present in the former but not in the latter. The red lines represent the predicted broadening for an idealized random coil. Adapted from [26].
Dynamics
Disordered proteins are highly, but not uniformly, flexible. Different mobilities in different regions may be linked to function, making the characterization of dynamics in such proteins highly desirable. A number of techniques can provide dynamics information for specifically labeled sites. For example, FCS can report on the timescale of conformational dynamics associated with the quenching of specific intrinsic or conjugated fluorophores [36,58]. ESR can also be used to assess the flexibility of the polypeptide backbone at the location of spin-label attachment [42,59,60]. In contrast to such site-specific techniques, solution state NMR is capable of providing highly detailed dynamics information simultaneously at different sites throughout a protein, and has thus been the preferred tool for characterizing dynamics in disordered systems.
NMR measurements indicate that disordered proteins often exhibit rapid (on the NMR timescale) backbone motions throughout the polypeptide chain [16,61], in contrast to well structured systems where such motions are typically restricted to hinge or loop regions. When exceptions to this observation occur they are typically associated with secondary structure formation [17,62,63] or also local hydrophobic clusters [47,64], and are usually of interest from a functional perspective. Slower motions (on the NMR timescale), which have been attributed to conformational exchange arising from long-range contacts in unfolded proteins [65,66], have not thus far been commonly observed in disordered systems [67], although potential evidence for their presence exists [63]. However, such conformational exchange has been documented in folding-upon-binding events involving disordered proteins [68,69]. An accompanying article in this issue discusses recent developments in the study of such disorder-to-order transitions.
Computation
Despite the detailed information that can be obtained on residual secondary structure, transient long-range contacts, and dynamics in disordered proteins, describing the ensemble of conformations sampled by such systems remains a considerable challenge. Computational methods increasingly play an important role in helping to visualize the implications of experimentally observed constraints, as well as in generating de novo predictions and simulations. An early contribution was the development of the program AGADIR, which accurately predicts regions of significant helix propensity [70]. To date, no similarly successful predictor of strand propensity has been reported, possibly due to the more highly cooperative nature of helical structure. The reliability of residual secondary structure analysis based on NMR chemical shifts has been evaluated using molecular dynamics simulations [71] and improved statistical algorithms have been proposed [15].
Interpretation of RDCs has been facilitated by several approaches developed to better understand unfolded states of well-structured proteins. Calculations based on global alignments of large ensembles of conformations generated solely based on volume exclusion and backbone torsion angle parameters from loop regions of well-structured proteins [23,72] were found to reproduce experimental data for several systems. An approach using local alignment of chain segments in ensembles generated based on volume exclusion biased by experimentally determined secondary structure propensities gave similarly good agreement using considerably smaller ensembles [73]. Interestingly, a correlation between amino acid bulkiness and RDCs in disordered proteins has been observed and also interpreted to reflect locally extended conformations due to steric constraints [57]. The interpretation of SAXS data from disordered proteins has also been extended by computational generation and selection of consistent ensembles of polypeptide conformations [74,75].
Similarly, visualizing the implications of long-range contacts detected through PRE measurements has been facilitated by the development of computational analyses. PRE measurements have been used to constrain molecular dynamics simulations in order to generate an ensemble that is consistent with the experimental data. Comparison with an ensemble generated to resemble an excluded-volume random coil model allowed identification of regions involved in preferentially formed long-range contacts, as well as analysis of representative conformers [44]. An alternative approach uses PRE data to prune large ensembles of generated structures in order to enrich for conformations that are consistent with the data [52]. Either of these methods can also incorporate additional experimental constraints. More recently, models based on interactions of clusters of high AABUF (average area buried upon folding) value were found to successfully reproduce detailed features of PRE profiles [76].
Finally, de novo molecular dynamics simulations are also proving useful in providing general insights into the behavior of disordered proteins, especially for cases such as homopolymers that are particularly challenging experimentally [77–79].
Ordered States
Upon interacting with cellular components such as other proteins, membranes, or the cytoskeleton components, intrinsically disordered protein can fold into conformations that are more highly ordered. Folding can be restricted to short elements of secondary structure [80–82], or can encompass lengthy regions of the polypeptide chain [83]. If the resulting complex can be isolated in a well-structured form, standard methods can be applied to solve the structure of the ordered conformation. In some cases, however, it seems that the bound state retains a significant degree of mobility and in addition remains highly non-compact, precluding a typical structure determination. In such cases, the techniques described above can prove as useful in describing the bound state as they are in characterizing the free state, but additional become applicable as well. Solution state NMR relies largely on short inter-proton distances to constrain molecular structure at high resolution. For non-compact conformations, such constraints are rare or absent. In the absence of such constraints, a recent approach, molecular fragment replacement, has been developed which relies predominantly on RDCs, combined with the database of known structures, to assemble local trajectories of polypeptide chain segments into a complete structure [84]. This approach was combined with long-range PRE restraints to calculate a structure of the protein alpha-synuclein in its detergent micelle-bound state [51]. Longer-range distance constraints can also be obtained using pulsed ESR spectroscopy, as illustrated recently in studies of micelle-, bicelle- and vesicle-bound alpha-synuclein [85,86].
Conclusions
Detailed biophysical studies are likely to prove crucial in clarifying the relationship between biological function and underlying structure for intrinsically disordered proteins, much as they have for well-structured ones. Over the past several years, the application of increasingly sophisticated methods has revealed that disordered proteins are far from homogeneous statistical random coil polymers and instead exhibit a rich diversity of local and even long-range structural preferences, as well as dynamics, that are likely to be of functional import. The transient nature of such preferences makes them more challenging to detect and characterize than the stable structures of well-ordered proteins. Nevertheless, continuing technical improvements as well as a further increase in the amount and quality of available data, combined with improvements in computational methods, should improve our fundamental understanding of these systems and their increasingly recognized roles in biology.
Acknowledgements
This work was supported by NIH/NIA grants AG019391 and AG144051, the Irma T. Hirschl Foundation and a gift from Herbert and Ann Siegel.
Footnotes
Conflict of Interest
The author has no conflicts to declare.
References
- 1.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN, Dunker AK. Comparing and combining predictors of mostly disordered proteins. Biochemistry. 2005;44:1989–2000. doi: 10.1021/bi047993o. [DOI] [PubMed] [Google Scholar]
- 3.Tompa P, Dosztanyi Z, Simon I. Prevalent structural disorder in E. coli and S. cerevisiae proteomes. J Proteome Res. 2006;5:1996–2000. doi: 10.1021/pr0600881. [DOI] [PubMed] [Google Scholar]
- 4.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 5.Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18:1–9. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 7. Eliezer D. Characterizing residual structure in disordered protein States using nuclear magnetic resonance. Methods Mol Biol. 2007;350:49–67. doi: 10.1385/1-59745-189-4:49. Instructional review intended to enable non-experts to apply NMR to disordered proteins.
- 8.Receveur-Brechot V, Bourhis JM, Uversky VN, Canard B, Longhi S. Assessing protein disorder and induced folding. Proteins. 2006;62:24–45. doi: 10.1002/prot.20750. [DOI] [PubMed] [Google Scholar]
- 9.Mittag T, Forman-Kay JD. Atomic-level characterization of disordered protein ensembles. Curr Opin Struct Biol. 2007;17:3–14. doi: 10.1016/j.sbi.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 11.Dyson HJ, Wright PE. Unfolded proteins and protein folding studied by NMR. Chem Rev. 2004;104:3607–3622. doi: 10.1021/cr030403s. [DOI] [PubMed] [Google Scholar]
- 12.Chen H, Rhoades E. Fluorescence characterization of denatured proteins. Curr Opin Struct Biol. 2008;18:516–524. doi: 10.1016/j.sbi.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Millett IS, Doniach S, Plaxco KW. Toward a taxonomy of the denatured state: small angle scattering studies of unfolded proteins. Adv Protein Chem. 2002;62:241–262. doi: 10.1016/s0065-3233(02)62009-1. [DOI] [PubMed] [Google Scholar]
- 14.Schuler B, Eaton WA. Protein folding studied by single-molecule FRET. Curr Opin Struct Biol. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eliezer D, Barre P, Kobaslija M, Chan D, Li X, Heend L. Residual structure in the repeat domain of tau: echoes of microtubule binding and paired helical filament formation. Biochemistry. 2005;44:1026–1036. doi: 10.1021/bi048953n. [DOI] [PubMed] [Google Scholar]
- 17.Lee H, Mok KH, Muhandiram R, Park KH, Suk JE, Kim DH, Chang J, Sung YC, Choi KY, Han KH. Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J Biol Chem. 2000;275:29426–29432. doi: 10.1074/jbc.M003107200. [DOI] [PubMed] [Google Scholar]
- 18.Wong KB, Freund SM, Fersht AR. Cold denaturation of barstar: 1H, 15N and 13C NMR assignment and characterisation of residual structure. J Mol Biol. 1996;259:805–818. doi: 10.1006/jmbi.1996.0359. [DOI] [PubMed] [Google Scholar]
- 19.Smith LJ, Bolin KA, Schwalbe H, MacArthur MW, Thornton JM, Dobson CM. Analysis of main chain torsion angles in proteins: prediction of NMR coupling constants for native and random coil conformations. J Mol Biol. 1996;255:494–506. doi: 10.1006/jmbi.1996.0041. [DOI] [PubMed] [Google Scholar]
- 20.Plaxco KW, Morton CJ, Grimshaw SB, Jones JA, Pitkeathly M, Campbell ID, Dobson CM. The effects of guanidine hydrochloride on the 'random coil' conformations and NMR chemical shifts of the peptide series GGXGG. J Biomol NMR. 1997;10:221–230. doi: 10.1023/A:1018340217891. [DOI] [PubMed] [Google Scholar]
- 21.Penkett CJ, Redfield C, Dodd I, Hubbard J, McBay DL, Mossakowska DE, Smith RA, Dobson CM, Smith LJ. NMR analysis of main-chain conformational preferences in an unfolded fibronectin-binding protein. J Mol Biol. 1997;274:152–159. doi: 10.1006/jmbi.1997.1369. [DOI] [PubMed] [Google Scholar]
- 22.Mohana-Borges R, Goto NK, Kroon GJ, Dyson HJ, Wright PE. Structural characterization of unfolded states of apomyoglobin using residual dipolar couplings. J Mol Biol. 2004;340:1131–1142. doi: 10.1016/j.jmb.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 23.Jha AK, Colubri A, Freed KF, Sosnick TR. Statistical coil model of the unfolded state: resolving the reconciliation problem. Proc Natl Acad Sci U S A. 2005;102:13099–13104. doi: 10.1073/pnas.0506078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wells M, Tidow H, Rutherford TJ, Markwick P, Jensen MR, Mylonas E, Svergun DI, Blackledge M, Fersht AR. Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc Natl Acad Sci U S A. 2008;105:5762–5767. doi: 10.1073/pnas.0801353105. P53 contains well-structured DNA-binding and tetramerization domains as well as intervening disordered regions and a disordered N-terminal transactivation domain. This study uses RDCs to characterize helical and extended residual structure in the TAD as an isolated fragment and in the intact tetrameric protein. SAXS is also used to characterize the topology of the structured and disordered domains of the the tetrameric protein in the free and DNA-bound states.
- 25.Fieber W, Kristjansdottir S, Poulsen FM. Short-range, long-range and transition state interactions in the denatured state of ACBP from residual dipolar couplings. J Mol Biol. 2004;339:1191–1199. doi: 10.1016/j.jmb.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 26. Sung YH, Eliezer D. Residual Structure, Backbone Dynamics, and Interactions within the Synuclein Family. J Mol Biol. 2007;372:689–707. doi: 10.1016/j.jmb.2007.07.008. Comparison of residual secondary and long-range structure within the human synuclein family. Shows that large RDCs in the C-terminal tail of all three proteins are found to correlate with local secondary structure rather than long-range structure.
- 27.Havlin RH, Tycko R. Probing site-specific conformational distributions in protein folding with solid-state NMR. Proc Natl Acad Sci U S A. 2005;102:3284–3289. doi: 10.1073/pnas.0406130102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohn JE, Millett IS, Jacob J, Zagrovic B, Dillon TM, Cingel N, Dothager RS, Seifert S, Thiyagarajan P, Sosnick TR, et al. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc Natl Acad Sci U S A. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bilsel O, Matthews CR. Molecular dimensions and their distributions in early folding intermediates. Curr Opin Struct Biol. 2006;16:86–93. doi: 10.1016/j.sbi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Merchant KA, Best RB, Louis JM, Gopich IV, Eaton WA. Characterizing the unfolded states of proteins using single-molecule FRET spectroscopy and molecular simulations. Proc Natl Acad Sci U S A. 2007;104:1528–1533. doi: 10.1073/pnas.0607097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morar AS, Olteanu A, Young GB, Pielak GJ. Solvent-induced collapse of alphasynuclein and acid-denatured cytochrome c. Protein Sci. 2001;10:2195–2199. doi: 10.1110/ps.24301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Perugini MA, Yao S, Adda CG, Murphy VJ, Low A, Anders RF, Norton RS. Solution conformation, backbone dynamics and lipid interactions of the intrinsically unstructured malaria surface protein MSP2. J Mol Biol. 2008;379:105–121. doi: 10.1016/j.jmb.2008.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paleologou KE, Schmid AW, Rospigliosi CC, Kim HY, Lamberto GR, Fredenburg RA, Lansbury PT, Jr, Fernandez CO, Eliezer D, Zweckstetter M, et al. Phosphorylation at Ser-129 but not the phosphomimics S129E/D inhibits the fibrillation of alpha-synuclein. J Biol Chem. 2008;283:16895–16905. doi: 10.1074/jbc.M800747200. Phosphorylation is shown to disrupt long-range structure in alpha-synuclein, leading to a larger hydrodynamic radius in NMR PFG experiments.
- 34.Rhoades E, Ramlall TF, Webb WW, Eliezer D. Quantification of alpha-synuclein binding to lipid vesicles using fluorescence correlation spectroscopy. Biophys J. 2006;90:4692–4700. doi: 10.1529/biophysj.105.079251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crick SL, Jayaraman M, Frieden C, Wetzel R, Pappu RV. Fluorescence correlation spectroscopy shows that monomeric polyglutamine molecules form collapsed structures in aqueous solutions. Proc Natl Acad Sci U S A. 2006;103:16764–16769. doi: 10.1073/pnas.0608175103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc Natl Acad Sci U S A. 2007;104:2649–2654. doi: 10.1073/pnas.0611503104. Single molecule FRET and FCS study of the yeast prion protein SUP35 revealed a collapsed state with rapid internal fluctuations
- 37.Uversky VN, Li J, Fink AL. Evidence for a partially folded intermediate in alpha-synuclein fibril formation. J Biol Chem. 2001;276:10737–10744. doi: 10.1074/jbc.M010907200. [DOI] [PubMed] [Google Scholar]
- 38. Galea CA, Nourse A, Wang Y, Sivakolundu SG, Heller WT, Kriwacki RW. Role of intrinsic flexibility in signal transduction mediated by the cell cycle regulator, p27 Kip1. J Mol Biol. 2008;376:827–838. doi: 10.1016/j.jmb.2007.12.016. The C-terminal regulatory domain of the CDK inhibitor p27 is shown by NMR to remain disordered in the p27/CDK2/Cyclin A ternary complex. SAXS data was used to define a molecular envelope for the ternary complex which was in qualitative agreement with MD simulations.
- 39.Violot S, Aghajari N, Czjzek M, Feller G, Sonan GK, Gouet P, Gerday C, Haser R, Receveur-Brechot V. Structure of a full length psychrophilic cellulase from Pseudoalteromonas haloplanktis revealed by X-ray diffraction and small angle X-ray scattering. J Mol Biol. 2005;348:1211–1224. doi: 10.1016/j.jmb.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 40.Mylonas E, Hascher A, Bernado P, Blackledge M, Mandelkow E, Svergun DI. Domain conformation of tau protein studied by solution small-angle X-ray scattering. Biochemistry. 2008;47:10345–10353. doi: 10.1021/bi800900d. [DOI] [PubMed] [Google Scholar]
- 41.Lee JC, Langen R, Hummel PA, Gray HB, Winkler JR. Alpha-synuclein structures from fluorescence energy-transfer kinetics: implications for the role of the protein in Parkinson's disease. Proc Natl Acad Sci U S A. 2004;101:16466–16471. doi: 10.1073/pnas.0407307101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeganathan S, von Bergen M, Brutlach H, Steinhoff HJ, Mandelkow E. Global hairpin folding of tau in solution. Biochemistry. 2006;45:2283–2293. doi: 10.1021/bi0521543. [DOI] [PubMed] [Google Scholar]
- 43.Crowhurst KA, Forman-Kay JD. Aromatic and methyl NOEs highlight hydrophobic clustering in the unfolded state of an SH3 domain. Biochemistry. 2003;42:8687–8695. doi: 10.1021/bi034601p. [DOI] [PubMed] [Google Scholar]
- 44.Dedmon MM, Christodoulou J, Wilson MR, Dobson CM. Heat shock protein 70 inhibits alpha-synuclein fibril formation via preferential binding to prefibrillar species. J Biol Chem. 2005;280:14733–14740. doi: 10.1074/jbc.M413024200. [DOI] [PubMed] [Google Scholar]
- 45.Bertoncini CW, Jung YS, Fernandez CO, Hoyer W, Griesinger C, Jovin TM, Zweckstetter M. Release of long-range tertiary interactions potentiates aggregation of natively unstructured alpha-synuclein. Proc Natl Acad Sci U S A. 2005;102:1430–1435. doi: 10.1073/pnas.0407146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vise P, Baral B, Stancik A, Lowry DF, Daughdrill GW. Identifying long-range structure in the intrinsically unstructured transactivation domain of p53. Proteins. 2007;67:526–530. doi: 10.1002/prot.21364. [DOI] [PubMed] [Google Scholar]
- 47. Dancheck B, Nairn AC, Peti W. Detailed Structural Characterization of Unbound Protein Phosphatase 1 Inhibitors. Biochemistry. 2008 doi: 10.1021/bi801308y. Two protein phosphatase 1 inhibitors, DARPP-32 and I-2 are shown to be intrinsically disordered, yet to posess significant residual secondary and long-range structure, which is proposed to be functionally relevant. AABUF is shown to correlate with restricted motions in regions without residual secondary structure.
- 48.Gillespie JR, Shortle D. Characterization of long-range structure in the denatured state of staphylococcal nuclease. I. Paramagnetic relaxation enhancement by nitroxide spin labels. J Mol Biol. 1997;268:158–169. doi: 10.1006/jmbi.1997.0954. [DOI] [PubMed] [Google Scholar]
- 49.Gillespie JR, Shortle D. Characterization of long-range structure in the denatured state of staphylococcal nuclease. II. Distance restraints from paramagnetic relaxation and calculation of an ensemble of structures. J Mol Biol. 1997;268:170–184. doi: 10.1006/jmbi.1997.0953. [DOI] [PubMed] [Google Scholar]
- 50.Lietzow MA, Jamin M, Jane Dyson HJ, Wright PE. Mapping long-range contacts in a highly unfolded protein. J Mol Biol. 2002;322:655–662. doi: 10.1016/s0022-2836(02)00847-1. [DOI] [PubMed] [Google Scholar]
- 51.Ulmer TS, Bax A, Cole NB, Nussbaum RL. Structure and dynamics of micelle-bound human alpha-synuclein. J Biol Chem. 2005;280:9595–9603. doi: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- 52. Marsh JA, Neale C, Jack FE, Choy WY, Lee AY, Crowhurst KA, Forman-Kay JD. Improved structural characterizations of the drkN SH3 domain unfolded state suggest a compact ensemble with native-like and non-native structure. J Mol Biol. 2007;367:1494–1510. doi: 10.1016/j.jmb.2007.01.038. Describes improvements in a flexible algorithm that accepts large initial ensembles generated by a variety of methods and selects out families of conformations that are consistent with a wide spectrum of experimental constraints, including PRE data.
- 53. Clore GM, Tang C, Iwahara J. Elucidating transient macromolecular interactions using paramagnetic relaxation enhancement. Curr Opin Struct Biol. 2007;17:603–616. doi: 10.1016/j.sbi.2007.08.013. Reviews a number of considerations required for accurate measurement and interpretation of PRE data.
- 54.Iwahara J, Tang C, Marius Clore G. Practical aspects of (1)H transverse paramagnetic relaxation enhancement measurements on macromolecules. J Magn Reson. 2007;184:185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernandez CO, Hoyer W, Zweckstetter M, Jares-Erijman EA, Subramaniam V, Griesinger C, Jovin TM. NMR of alpha-synuclein-polyamine complexes elucidates the mechanism and kinetics of induced aggregation. Embo J. 2004;23:2039–2046. doi: 10.1038/sj.emboj.7600211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bernado P, Bertoncini CW, Griesinger C, Zweckstetter M, Blackledge M. Defining long-range order and local disorder in native alpha-synuclein using residual dipolar couplings. J Am Chem Soc. 2005;127:17968–17969. doi: 10.1021/ja055538p. [DOI] [PubMed] [Google Scholar]
- 57. Cho MK, Kim HY, Bernado P, Fernandez CO, Blackledge M, Zweckstetter M. Amino acid bulkiness defines the local conformations and dynamics of natively unfolded alpha-synuclein and tau. J Am Chem Soc. 2007;129:3032–3033. doi: 10.1021/ja067482k. Notes a correlation between amino acid bulkiness and RDCs that may arise from locally extended conformations adopted to minimize steric overlap. Posits that deviations from this correlation may reflect long-range contacts.
- 58.Chen H, Rhoades E, Butler JS, Loh SN, Webb WW. Dynamics of equilibrium structural fluctuations of apomyoglobin measured by fluorescence correlation spectroscopy. Proc Natl Acad Sci U S A. 2007;104:10459–10464. doi: 10.1073/pnas.0704073104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen M, Margittai M, Chen J, Langen R. Investigation of alpha-synuclein fibril structure by site-directed spin labeling. J Biol Chem. 2007;282:24970–24979. doi: 10.1074/jbc.M700368200. [DOI] [PubMed] [Google Scholar]
- 60. Belle V, Rouger S, Costanzo S, Liquiere E, Strancar J, Guigliarelli B, Fournel A, Longhi S. Mapping alpha-helical induced folding within the intrinsically disordered C-terminal domain of the measles virus nucleoprotein by site-directed spin-labeling EPR spectroscopy. Proteins. 2008;73:973–988. doi: 10.1002/prot.22125. ESR data show decreased flexibility in a region of residual helical structure in the disordered C-terminal domain of the measles virus nucleoprotein, and demonstrate the further ordering of this region upon interaction with a binding partner.
- 61.Bussell R, Jr, Eliezer D. Residual structure and dynamics in Parkinson's disease-associated mutants of alpha-synuclein. J Biol Chem. 2001;276:45996–46003. doi: 10.1074/jbc.M106777200. [DOI] [PubMed] [Google Scholar]
- 62.Lacy ER, Filippov I, Lewis WS, Otieno S, Xiao L, Weiss S, Hengst L, Kriwacki RW. p27 binds cyclin-CDK complexes through a sequential mechanism involving binding-induced protein folding. Nat Struct Mol Biol. 2004;11:358–364. doi: 10.1038/nsmb746. [DOI] [PubMed] [Google Scholar]
- 63. Baker JM, Hudson RP, Kanelis V, Choy WY, Thibodeau PH, Thomas PJ, Forman-Kay JD. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat Struct Mol Biol. 2007;14:738–745. doi: 10.1038/nsmb1278. Documents residual helical structure in the regulatory region of CFTR using NMR and shows that such structure likely mediates dynamic regulatory interactions with possibly overlapping sites on a nucleotide binding domain of CFTR and that phosphorylation may influence these interactions by modulating helical propensity. Posits that long-range contacts within the R region may also contribute to observed slower time scale dynamics.
- 64.Yao J, Chung J, Eliezer D, Wright PE, Dyson HJ. NMR structural and dynamic characterization of the acid-unfolded state of apomyoglobin provides insights into the early events in protein folding. Biochemistry. 2001;40:3561–3571. doi: 10.1021/bi002776i. [DOI] [PubMed] [Google Scholar]
- 65.Klein-Seetharaman J, Oikawa M, Grimshaw SB, Wirmer J, Duchardt E, Ueda T, Imoto T, Smith LJ, Dobson CM, Schwalbe H. Long-range interactions within a nonnative protein. Science. 2002;295:1719–1722. doi: 10.1126/science.1067680. [DOI] [PubMed] [Google Scholar]
- 66.Wirmer J, Schlorb C, Klein-Seetharaman J, Hirano R, Ueda T, Imoto T, Schwalbe H. Modulation of compactness and long-range interactions of unfolded lysozyme by single point mutations. Angew Chem Int Ed Engl. 2004;43:5780–5785. doi: 10.1002/anie.200460907. [DOI] [PubMed] [Google Scholar]
- 67. Wu KP, Kim S, Fela DA, Baum J. Characterization of conformational and dynamic properties of natively unfolded human and mouse alpha-synuclein ensembles by NMR: implication for aggregation. J Mol Biol. 2008;378:1104–1115. doi: 10.1016/j.jmb.2008.03.017. A comparison of human and mouse alpha-synuclein at low temperatures shows that anomalous NMR R2 values in both proteins do not reflect conformational exchange.
- 68.Eliezer D, Palmer AG., 3rd Biophysics: proteins hunt and gather. Nature. 2007;447:920–921. doi: 10.1038/447920a. [DOI] [PubMed] [Google Scholar]
- 69. Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. Employs NMR methods for detecting conformational exchange to characterize intermediates associated with the binding and folding of a disordered protein to its binding partner.
- 70.Lacroix E, Viguera AR, Serrano L. Elucidating the folding problem of alpha-helices: local motifs, long-range electrostatics, ionic-strength dependence and prediction of NMR parameters. J Mol Biol. 1998;284:173–191. doi: 10.1006/jmbi.1998.2145. [DOI] [PubMed] [Google Scholar]
- 71. Weinstock DS, Narayanan C, Baum J, Levy RM. Correlation between 13Calpha chemical shifts and helix content of peptide ensembles. Protein Sci. 2008;17:950–954. doi: 10.1110/ps.073365408. Uses MD simulations to demonstrate that C〈 secondary chemical shifts accurately reflect percent helicity, but that some underprediction may occur due to averaging over ensembles including extended conformations.
- 72.Bernado P, Blanchard L, Timmins P, Marion D, Ruigrok RW, Blackledge M. A structural model for unfolded proteins from residual dipolar couplings and small-angle x-ray scattering. Proc Natl Acad Sci U S A. 2005;102:17002–17007. doi: 10.1073/pnas.0506202102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Marsh JA, Baker JM, Tollinger M, Forman-Kay JD. Calculation of residual dipolar couplings from disordered state ensembles using local alignment. J Am Chem Soc. 2008;130:7804–7805. doi: 10.1021/ja802220c. RDCs in good agreement with experiment are calculated using local alignment of structures in ensembles generated based on excluded volume biased by residual secondary structure. Biasing ensembles by tertiary contacts was not necessary, despite evidence for transient long-range structure in the proteins studied. The ensemble size necessary for convergence was greatly reduced when using local vs. global alignment.
- 74. Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. Uses a genetic algorithm to select a subset of conformations that together reproduce SAXS data for disordered systems from larger random ensembles based on loop database derived torsion angles and volume exclusion.
- 75.Hammel M, Fierobe HP, Czjzek M, Kurkal V, Smith JC, Bayer EA, Finet S, Receveur-Brechot V. Structural basis of cellulosome efficiency explored by small angle X-ray scattering. J Biol Chem. 2005;280:38562–38568. doi: 10.1074/jbc.M503168200. [DOI] [PubMed] [Google Scholar]
- 76. Felitsky DJ, Lietzow MA, Dyson HJ, Wright PE. Modeling transient collapsed states of an unfolded protein to provide insights into early folding events. Proc Natl Acad Sci U S A. 2008;105:6278–6283. doi: 10.1073/pnas.0710641105. Transient interactions between clusters of residues with high AABUF, modeled according to contact energies and configurational entropies of corresponding loop closure events, accurately reproduce detailed features of PRE profiles in acid-denatured apomyoglobin.
- 77.Armen RS, Bernard BM, Day R, Alonso DO, Daggett V. Characterization of a possible amyloidogenic precursor in glutamine-repeat neurodegenerative diseases. Proc Natl Acad Sci U S A. 2005;102:13433–13438. doi: 10.1073/pnas.0502068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Vitalis A, Wang X, Pappu RV. Quantitative characterization of intrinsic disorder in polyglutamine: insights from analysis based on polymer theories. Biophys J. 2007;93:1923–1937. doi: 10.1529/biophysj.107.110080. Together with the subsequent reference, analyzes MD simulations of polyglycine and polyglutamine sequences in water and/or chemical denaturants and concludes that compactness in disordered proteins results at least in part from water being a poor solvent for flexible polypeptide backbones.
- 79.Tran HT, Mao A, Pappu RV. Role of backbone-solvent interactions in determining conformational equilibria of intrinsically disordered proteins. J Am Chem Soc. 2008;130:7380–7392. doi: 10.1021/ja710446s. [DOI] [PubMed] [Google Scholar]
- 80.Fuxreiter M, Simon I, Friedrich P, Tompa P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J Mol Biol. 2004;338:1015–1026. doi: 10.1016/j.jmb.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 81.Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. Analysis of molecular recognition features (MoRFs) J Mol Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 82.Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005;44:12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- 83.Dames SA, Martinez-Yamout M, De Guzman RN, Dyson HJ, Wright PE. Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response. Proc Natl Acad Sci U S A. 2002;99:5271–5276. doi: 10.1073/pnas.082121399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Delaglio F, Kontaxis G, Bax A. Protein structure determination using molecular fragment replacement and NMR dipolar couplings. J Am Chem Soc. 2000;122:2142–2143. [Google Scholar]
- 85.Borbat P, Ramlall TF, Freed JH, Eliezer D. Inter-helix distances in lysophospholipid micelle-bound alpha-synuclein from pulsed ESR measurements. J Am Chem Soc. 2006;128:10004–10005. doi: 10.1021/ja063122l. [DOI] [PubMed] [Google Scholar]
- 86. Georgieva ER, Ramlall TF, Borbat PP, Freed JH, Eliezer D. Membrane-bound alpha-synuclein forms an extended helix: long-distance pulsed ESR measurements using vesicles, bicelles, and rodlike micelles. J Am Chem Soc. 2008;130:12856–12857. doi: 10.1021/ja804517m. Pulsed ESR measurements of distances up to 70 Å are used to constrain the topology of lipid vesicle and bicelle bound alpha-synuclein.