Abstract
In a chemical mutagenesis screen we identified Szt2 (seizure threshold 2) as a gene that confers low seizure threshold to mice and may also enhance epileptogenesis. The semidominant phenotype was mapped to Chromosome 4 and narrowed further to a critical interval of approximately 650 kb. A novel large (>10 kb) transcript in the critical interval was found to have four-fold increased steady-state expression at the RNA level in Szt2 homozygous mutant brain. The corresponding 72 exon gene encodes a 378 kD protein with no significant or suggestive sequence similarities to any other protein. The mutant allele of Szt2 contains a splice donor mutation after exon 32, predicting transcriptional read-through, translational frameshift and premature stop. A second Szt2 allele, containing a gene-trap mutation in exon 21, also conferred a low seizure threshold and increased RNA expression, but unlike the ENU allele, some gene-trap homozygotes died embryonically. Szt2 is transcribed in many tissues, with the highest expression in brain, and it is also expressed during embryonic development. Szt2 is highly conserved in evolution, with a clear, single orthologue found in all land vertebrates and in many invertebrates. Interestingly, in mammals the Szt2 gene resides in a highly conserved head-to-head configuration with Med8 (which encodes a Mediator complex subunit), separated by only 91 nt. While the biological function of Szt2 remains unknown, its high conservation, unique structure and effect on seizure threshold suggest that it serves an important role in the central nervous system.
Keywords: epilepsy, seizure threshold, mutation, mutagenesis, feedback, mice
Introduction
Seizure threshold defines a theoretical “set-point” below which a stimulus may produce a seizure. Mouse mutants with spontaneous epilepsy also often have a low seizure threshold, for example, as assessed by sensitivity to pentylenetetrazole (PTZ), the non-competitive GABAA receptor antagonist – suggesting that seizure threshold is a natural component of genetic susceptibility to epilepsy. Still, until we learn more about the genetic variants that cause common epilepsies and study them in animal models, the distinction, if any, between seizure threshold and susceptibility per se is likely to remain blurry. There indeed may not be a unitary relationship between the two. For example, in a prior assessment of electroconvulsive threshold in various inbred mouse strains, it was determined that the threshold of the spontaneously epileptic SWXL4 strain is at the midpoint of the distribution, with a number of non-epileptic inbred strains having a lower seizure threshold than SWXL-4 (Frankel et al., 2001). In contrast, even single-gene mutants on a strain background with a high seizure threshold (e.g. C57BL/6J) can be spontaneously epileptic (Yang et al., 2007). Nevertheless, other epileptic strains such as PL/J do have very low seizure thresholds (Kitami et al., 2004). Together these results suggest that the relationship between threshold per se and epilepsy is not straightforward, and depends heavily upon the overall genetic context.
We previously examined the electroconvulsive thresholds of inbred strains as a framework for ENU mutagenesis forward genetics screens (Frankel et al., 2001). Three novel low seizure threshold mutations obtained from our screen occurred in the Kcnq2 gene (see Frankel, Kearney et al., 2006, Yang et al., 2003), whose human orthologue is mutated in Benign Neonatal Familial Convulsions, a form of human epilepsy (Singh et al., 1998). These mutations lowered the median electroconvulsive threshold by 1.5 mA, placing the Kcnq2 mutants approximately in the middle of the seizure threshold profile of inbred strains. Here we introduce Szt2 as a novel seizure threshold gene. The encoded protein is highly conserved in evolution – with clear orthologues in vertebrates and some invertebrates - but no structural similarities to known proteins. In addition to having a low acute seizure threshold, Szt2 mutant mice also kindle more readily than controls, making Szt2 a potentially interesting candidate gene for seizure susceptibility.
Methods
Mice
Mice carrying the Szt2m1Frk mutation were first detected in a mutagenesis screen for low electroconvulsive threshold in descendants of ENU-mutagenized C57BL/6J mice, as previously described (Yang et al., 2003). The Szt2 gene trap allele originated from feeder-independent ES cell line (XH662) derived from the 129/Ola mice that was obtained from BayGenomics (University of California, San Francisco). Microinjection of the ES cells into blastocysts was carried out by The Jackson Laboratory Microinjection Services. Chimeras were initially crossed to C57BL/6J mice to test germ-line transmission based on coat color. F1 progeny carrying the gene-trap allele were backcrossed to 129S1/SvImJ for 10 generations before analysis. All mice were housed in pathogen-free mouse facilities with 12 h light/dark cycle. Food and water were available ad libitum. All animal procedures was approved by The Jackson Laboratory Animal Care and Use Committee (ACUC).
Mutant allele detection
The Szt2 mutation creates a HphI restriction polymorphism, and was genotyped by restriction digestion following PCR amplification using the primers mutu: 5′-TGACCTGCCACCTCTCTTCT-3′ and mutd: 5′-GTCCGAGGCTGGAGGTAGTT-3′. The Szt2Gt(XH662)Byg (gene trap) mutation was genotyped using a three-primer PCR assay: TRAPD: 5′-GTTATCGATCTGCGATCTGCG-3′; Forward: 5′-CCCGTTCCACTTTGACCTACT-3′; S2e23R: 5′-TCGATCCTTAGCGACTGCATG-3′.
Seizure tests
Electroconvulsion
For electroconvulsive testing, we followed the procedures described previously (Frankel et al., 2001, Yang et al., 2003). Briefly, mice were restrained, a drop of anesthetic containing 0.5% tetracaine and 0.9% NaCl was placed onto each eye and a fixed electrical current was applied via silver transcorneal electrodes using a electroconvulsive stimulator (Ugo Basile model 7801). For acute electroconvulsion, the stimulator was set to produce rectangular wave pulses with the following parameters: 299 Hz, 0.2 s duration, 1.6 ms pulse width. For initial phenotypic deviant detection, and later for colony maintenance and for complementation testing, we challenged individual female and male mice with the CC3 (critical current level for a 3% response) for the minimal clonic seizure endpoint of wild-type C57BL/6J mice, previously determined for each sex separately, since for most strains and mutants, females have lower electroconvulsive thresholds than males (Frankel et al., 2001). These tests were done once a day – and twice total - with at least one of day rest between tests. For determining median acute electroconvulsive threshold (ECT), individual mice were tested once and analyzed using a modified staircase estimation procedure (Yang et al., 2007). For determining the latency to kindling, transcorneal electrodes were used with pulse parameters estimated to yield a similar integrated RMS as described previously by others using sinusoidal waveforms (Matagne & Klitgaard, 1998); 299 Hz, 3.0 s duration, 0.2 ms pulse width, 4.5 mA. Individual mice were challenged once daily until a partial seizure was observed, and group means were calculated to determine mean latency. For ECT median and kindling latency determinations, since female mice and male mice have different thresholds, only one sex was analyzed for convenience.
Pentylenetetrazole seizures (PTZ)
To determine susceptibility to chemically-induced tonic-clonic seizures, backcross Szt2m1Frk mice of each mutant genotype (heterozygous vs. homozygous) were injected subcutaneously with 50 mg/kg PTZ in fresh saline, placed onto clean bedding in a clear plastic box, and observed for at least 30 minutes. The incidence and latency to seizure endpoint standards (Racine, 1972) was recorded, and the average latency to tonic-clonic seizures was determined.
Real-Time PCR
Total RNA from adult male mouse brain (C57BL/6, Szt2 heterozygote, Szt2 homozygote from mapping cross and Szt2 homozygote from C57BL/6 strain) was prepared by using TRIzol (Invitrogen) and treated with DNase1 (Promega) using the manufacturers' suggested conditions. Two micrograms of RNA was transcribed with avian myeloblastosis virus reverse transcriptase (Promega). The cDNA was diluted 20-fold and added to Sybr Green PCR mix (Applied Biosystems) with primers. The PCR reactions were run in triplicate and analyzed on an ABI Prism 7700 sequence detector (Perkin Elmer). These studies used two biological replicates and three between-run experimental replicates, in addition to within-run triplicates.
Northern blot
Total RNA was prepared from mouse brains using TRIzol (Invitrogen), of which 25 μg was run on a 1.2% agarose/formaldehyde gel. After transfer, the blots were hybridized with a 32P-labeled probe containing Szt2 exons 1–3 in formamide/hybridization buffer at 45°C. Final wash after hybridization was 0.1× SSC, 0.1% SDS at 65°C. For the multiple tissue northern, two micrograms of poly(A)+ RNA from adult mouse tissue was prepared using the Oligotex mRNA kit (Qaigen) and run on a 1.2% agarose/formaldehyde gel. The blot was hybridized with a 685bp probe containing a portion of the Szt2 transcript from exons 69–72 in formamide/hybridization buffer at 42°C. Final wash after hybridization was 0.5× SSC, 0.1% SDS at RT. For all blots, a 1.8-kb mouse β actin probe was labeled and used as a control for any minor discrepancies in RNA loading.
Szt2-GFP fusion expression construct
First, KOD Hot Start DNA polymerase (Novagen) was used to amplify a 4.9kb fragment from reverse transcribed poly(A)+ RNA with primers Szt2Clau 5′ CCATCGATATGGCCTCGGAGCGCCCCGA 3′ (forward) and Szt2Xbad 5′ TCCCTCTAGACTGGAAAGCAC 3′ (reverse) encompassing the ORF in exons 1–32. The amplification product was gel purified, A-tailed using TAQ DNA polymerase and cloned into the TOPO cloning vector pCR2.1 (Invitrogen) for sequencing. A pCR2.1 plasmid with the correct sequence was restricted with ClaI and XbaI to release the 4.9kb fragment which was consequently ligated into the ClaI and XbaI sites of the pCMVGFP vector to create pCMVSzt2GFP5’. In the same way primers Szt2Xbau 5′ CAAGTGCTTTCCAGTCTAGAG 3′ (forward) and Szt2sped3 5′ CGACTAGTGAGAGAAGGCGGGTCCAGAG 3′ (reverse) encompassing the ORF in exons 32–72 were used to amplify a 5.5kb fragment that was cloned into pCR2.1. A pCR2.1 plasmid with the correct sequence was restricted with XbaI and SpeI to release the 5.5kb fragment which was consequently ligated into the XbaI digested and phosphatased pCMVSzt2GFP5’ plasmid to create pCMVSzt2GFP.
Immunoprecipitation
Briefly, COS-7 cells were transfected with pCMVSzt2GFP using the Lipofectamine kit, according to manufacturer’s instructions (Invitrogen, Inc). Two-to-three days following transfection, after observing fluorescence signal cells were harvested, lysed in buffer containing NP-40, centrifuged, incubated overnight at 4 C with rocking with 3 μL of mAb 3E6 anti-GFP from Invitrogen (A-11120), incubated for four hours with protein G-sepharose 4B at 4 C with rocking, washed twice in lysis buffer, resuspended in 1x Laemmli loading buffer, boiled, centrifuged and loaded onto SDS-PAGE (precast from Lonza Scientific), run for one hour at approximately 150 volts, stained with sypro-ruby and imaged.
Mass spectrometry
MALDI-TOF was used by The Jackson Laboratory’s Protein Chemistry Service to identify peptides corresponding to in vitro expressed SZT2-GFP. Briefly, protein bands were excised from the gel, equilibrated to remove residual acid and the cysteines were reduced and alykylated with iodoacetamide. The gel slice was then washed to remove salts and protein stains and dehydrated by the addition of acetonitrile. In-gel digestion of the proteins was performed by adding trypsin to the dehydrated gel slice and incubated over night at 37C. The solution was then removed from the gels slice into a clean test tube. The remaining peptides, that did not diffuse into the solution were extracted and pooled with the initial solution. The samples were then dried down by speed vac and brought up in 0.1% TFA followed by reverse phase chromatography using a C18 zip-tip (Millipore). The eluted peptides were combined with 10mg/ml alpha-Cyano-4 hydroxycinnaminic acid (HCCA) and spotted onto a stainless steel MALDI plate and dried. The samples were then subjected to MALDI-TOF MS in a Bruker Microflex mass spectrometer, and the identity of the protein was determined by peptide mass fingerprinting (PMF) using the Mascot software (Matrix Sciences, UK) and searching against an appropriate mouse protein database.
Accession numbers
The GenBank accession number for the C57BL/6J adult mouse brain cDNA sequence is FJ998170.
Results
Origin of Szt2 mutant mice
Szt2 mutant mice were identi ed originally in a small scale three-generation ENU mutagenesis screen where C57BL/6J mice were mutagenized by ENU, as described previously (Yang et al., 2003), and dominant or recessive seizure threshold were sought from subsequent generations. In this screen, 333 third generation (G3) mice from 19 independent G1 founder families were tested for their sensitivity to electrically-evoked minimal clonic seizures. In one family of 18 G3 mice (from three G2 females bred with a G1 male), two males had minimal forebrain clonic seizures at the 3% response level previously determined for wild-type C57BL/6J mice. In a mating of this male to C57BL/6J, 0/7 F1 progeny mice had a seizure but in further backcrosses to the G1 7/9 mice (2/3 females, 5/6 males) had a seizure, suggesting that the phenotype had a significant recessive component and was highly penetrant. Affected mice were viable, fertile and did not show spontaneous or handling provoked seizures at any age. Gross histological examination did not reveal abnormalities in brain, heart, liver, spleen, lung, kidney, gastric-intestine tract, muscle or gonads (data not shown). They also did not show any electroencephalographic, neurosensory or motor abnormalities (data not shown).
Szt2 confers low seizure threshold and is potentially epileptogenic
A B6-Szt2/Szt2 putative homozyous colony was obtained by mating affected mice inter se for several generations until greater than 90% of the mice tested had a seizure at the 3% response level of B6. Stimulus response curves were generated for putative homozygous Szt2 and wild-type control male mice, revealing a significantly decreased threshold to minimal forebrain clonic seizures (Figure 1A).
Figure 1. Szt2m1Frk homozygous mutants are susceptible to induced seizures.

A. Stimulus (in mA) response curves for B6-Szt2m1Frk homozygotes (“X’s”) and B6-+/+ control (triangles) male mice of approximately six to nine weeks of age (N=31; N=111, respectively). The dashed and solid lines represent the respective interpolated Probit curves for these responses. B. Mean latency (number of serial daily tests) to a partial seizure induced by corneal kindling in male B6-Szt2 homozygous vs. wild-type mice, approximately nine weeks of age. C. Mean latency (in minutes) to tonic-clonic seizures induced by acute pentylenetetrazole injection (PTZ, 40 mg/kg s.c.) in Szt2m1Frk homozygous vs. heterozygous male mice from the mapping cross (B6-Szt2 x BALB/cByJ)F1 X B6-Szt2, approximately eight weeks of age.
Although Szt2 mutants do not display spontaneous seizures, we tested the potential epileptogenicity of Szt2m1Frk homozygous males using a non-invasive kindling procedure originally described by Matagne and Klitgaard (Matagne & Klitgaard, 1998). For this experiment, males are tested daily with a tetanic, high-frequency electrical stimulus at a low amplitude which does not cause a seizure on the first few exposures, and their latency to the first seizure, usually a partial seizure, is determined. Interestingly, while wild-type C57BL/6J control mice did not have their first seizure until seven or eight tests, the latency for B6- Szt2m1Frk homozygotes was significantly earlier – four tests (Figure 1B). Unlike the study of Potschka and Loscher (Potschka & Loscher, 1999), we did not observe any lethality between tests and the kindled state also appeared to be stable, with four of four tested mice retaining the state four weeks after their last test. These results suggest that Szt2m1Frk mutants may be more epileptogenic than wild-type.
Szt2 maps to a 650 kb interval on Chr 4
To chromosomally map Szt2, affected, putative homozygous mice from the colony were mated to BALB/cByJ, which has a similar seizure threshold as C57BL/6J, and then crossed to the putative B6-Szt2/Szt2 homozygous mouse to collect backcross progeny. Backcross mice were tested twice for acute minimal forebrain clonic seizures at the 3% response level previously established for B6 and BALB (Yang et al., 2003). Interestingly, homozygotes that tested positive for seizures also had a shorter latency to pentylenetetrazole (PTZ) induced seizures (Figure 1C), suggesting that their seizure threshold phenotype was not specific to electrical stimuli.
For the genome scan, genomic DNA from 20 affected mice (minimal clonic seizure on both tests) and 20 unaffected mice (no seizure on either test) were pooled separately, and 34 markers across the genome were used to estimate marker allele intensity differences between the two pools and an F1 hybrid control. Two markers on Chr 4, D4Mit186 and D4Mit332 showed mostly the B6-derived allele in homozygous mutants compared to unaffected and wild-type control (data not shown). These markers and additional simple sequence length polymorphisms (SSLP) were used to genotype individual backcross progeny. A significant association was noted for markers on centromere-distal Chr 4, with the Szt2 mutation mapping to a 22 cM interval (Figure 2A). Once having mapped Szt2, further backcross progeny were genotyped and mice containing recombinations in the Szt2 interval (plus representative non-recombinant controls) were selectively phenotyped for minimal clonic seizures. After 380 backcross mice, mostly containing recombinants in the critical interval, and further SSLP markers were examined, Szt2 was narrowed to a 650 kb 95% confidence interval (Figure 2A).
Figure 2. Genomic map and candidate gene testing of Szt2 locus on Chr 4.

A. Interval map showing the respective LOD score curves based on the backcross population used initially to confirm linkage to Chr 4 (dashed curve) and that based on advanced backcross mice, mostly containing recombinations in the interval (solid curve). The relative marker distances are fixed to the physical map length, based on Ensembl build 36 of the C57BL/6J mouse genome sequence assembly. The stippled line shows the threshold for highly significant association, based on permutation tests. B. Genes corresponding to the indicated confidence interval (given as gene symbols or transcript or sequence identifiers from Ensembl build 36; number of exons and transcriptional orientation shown) were examined in homozygous Szt2m1Frk mice vs. wild-type controls for either expression differences by quantitative real-time PCR (qPCR), Northern blot, or the corresponding coding exons were examined for structural differences by single stranded confirmation assay (SSCP), or by DNA sequencing. As described in the text, BC05982 is the sequence identifier for the eventual Szt2 gene. Genes shown in parentheses are no longer models in the current genome assembly.
Szt2 encodes a large, highly-conserved protein of unknown function
To search for mutations, we began to examine candidate genes in the Szt2 critical interval. According to the Ensembl release available at the time (http://www.ensembl.org), eighteen transcripts resided in the interval but none were obvious candidates based on their identity or known expression patterns (Figure 2B). We began to examine the structure and expression of these genes in putative B6-Szt2/Szt2 homozygotes by various means (Figure 2B). Quantitative PCR was performed on 14 candidate transcripts in the Szt2 critical interval, three of which showed a difference between the wildtype B6 and Szt2 homozygotes ranging from 1.3 to 2.0 real-time PCR cycles fewer to amplify from homozygous mutant (Figure 2B; Figure 3A). By RT-PCR and analysis of cDNA, we then determined that these three transcripts were part of a larger transcription unit, together encompassing over 10 kb spanning approximately 47 kb of the genome. This gene was eventually identified as ENSMUSG00000033253 in Ensembl. A northern blot showed an approximately four-fold increase in mRNA in Szt2 mutant compared with wild-type, consistent with the quantitative PCR results (Figure 3B). To find a mutation, nine primer sets were used to amplify across the 10 kb cDNA in 1.2kb increments. One of the resultant fragments was larger in the Szt2 homozygote compared to wildtype. Sequencing the fragment revealed a mutation in the exon 32 splice site donor and the inclusion of intron 32 in the Szt2 mutant transcript. This mutation would result in a read-through of the splice site, creating an additional 32 amino acids prior to a premature stop codon (Figure 4).
Figure 3. Expression analysis of the Szt2 gene.
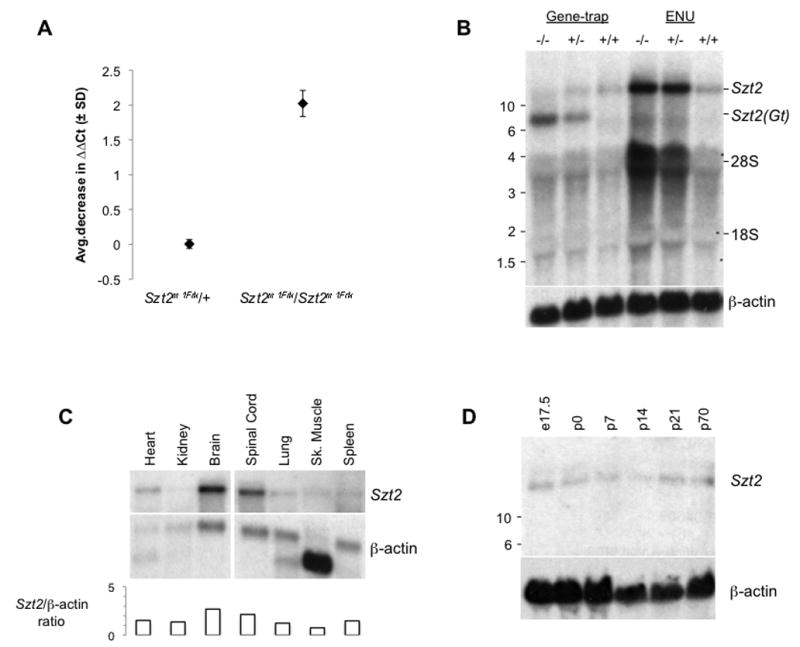
A. Analysis of real-time PCR results for one of the three primer sets listed in Figure 2, showing a significant decrease in the number of PCR cycles required to amplify Szt2 in homozygotes compared with wild-type. Two mutant replicates, and a minimum of six technical replicates (triplicate amplification, two separate runs) were used for these experiments. B Northern blot of whole brain total RNA from Szt2Gt(XH662)Byg (gene-trap) and Szt2m1Frk homozygotes, heterozygous and wild-type mice. The positions of the respective transcripts are indicated at the right, and the position of size markers on the left – hybridization control (β-actin) shown at the bottom. C. Northern blot of poly-A+ RNA from various tissues of adult C57BL/6J wild-type male mice, showing the most abundant expression to be in brain and spinal cord. The graph below is a visualization of the Szt2/β-actin ratio in each sample. D. Northern blot of total brain RNA showing the developmental expression of Szt2 from embryonic day 17.5 through postnatal development, plus an older adult sample.
Figure 4. The mouse Szt2 gene and its two germ-line mutations.
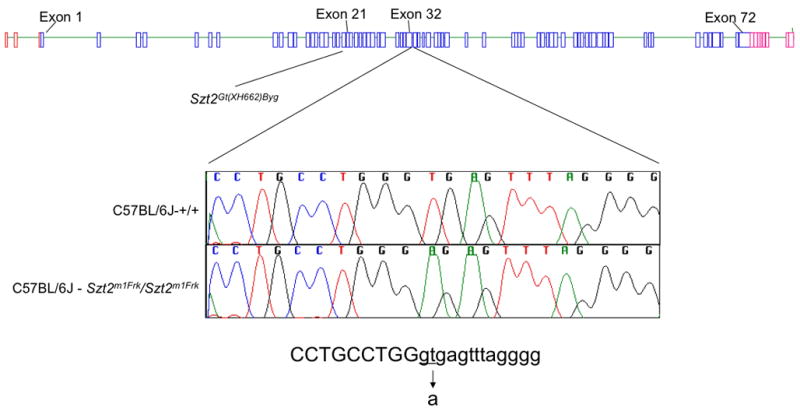
Shown is a graphic of the exons of Szt2 (blue boxes) and neighboring genes (red boxes), with the relative genomic positions based on Ensembl build 36 of the C57BL/6J mouse genome sequence assembly. Also indicated are the positions of the respective Szt2Gt(XH662)Byg (“gene trap”) and Szt2m1Frk (“ENU”) allelic mutations.
The Szt2 gene is expressed in many adult tissues, with its expression highest in brain and spinal cord (Figure 3C), although it is also expressed in the embryo (Figure 3D). An EST clone corresponding to a portion of Szt2 (BC059842) had been used as a probe by the Allen Brain Atlas (www.brain-map.org), and showed widespread but very low level of expression, with enrichment in hippocampus (pyramidal cell layer and dentate gyrus), cerebellar Purkinje cells, and olfactory lobe. Interestingly, the Szt2 gene resides in a head-to-head configuration with the Med8 gene, encoding a subunit of the Mediator transcription complex (Brower et al., 2002). In mammals, at least, the two first exons are 91 nt apart in the genome. Because the Allen Brain Atlas shows that Med8 has a very similar pattern of expression, we presume that they share a promoter. However, Med8 RNA is not upregulated in Szt2 mutant mice (Figure 2B and data not shown), suggesting that any self-regulation of Szt2 may be posttranscriptional in nature.
The putative 3431 amino acid protein encoded by murine Szt2 translates to a predicted mass of 378 kD (Supplemental Figure 1). We gained support for this prediction by expressing GFP-tagged full-length Szt2 cDNA in COS-7 cells and observing a high molecular weight band (Figure 5A). Mass spectrometry analysis confirmed that this band contained only peptides that correspond to the predicted SZT2 amino acid sequence, with a significant amount of coverage of the predicted protein (Figure 5B, Supplemental Figure 1).
Figure 5. Expression and evolutionary conservation of the SZT2 protein.

A. Sybro-ruby stained SDS-PAGE gel showing immunoprecipitation of pCMV-SZT2-GFP fusion protein (and CMV-GFP control) after transient transfection into COS-7 cells, indicating the position of SZT2 relative to size markers. The additional band represents co-precipitated immunoglobulins, present in both samples. B. MALDI-TOF profile of the SZT2 band, excised from the gel in panel A, with increasing mass (m/z) on the X-axis and arbitary units on the Y-axis. C. Conservation plot based on MAFFT protein sequence alignment of SZT2, in representative organisms from seven phyla, described in the text. The conservation score (Blosum 62 algorithm) for each residue was obtained using JalView Version 2 (Waterhouse et al., 2009). The highest scores represent the most conserved residues.
Protein database and peptide domain/motif searches (e.g. http://smart.embl-heidelberg.de/) revealed that SZT2 is unique – with no significant sequence similarities to any known protein. However, evolutionarily, Szt2 gene is very highly conserved. Based on primary sequence similarity and conserved synteny in vertebrates, mouse Szt2 appears to have a clear, single orthologue in all mammals, most vertebrates and at least some invertebrates (data not shown). We aligned murine SZT2 protein sequence with putative orthologues from organisms from seven divergent phyla, including mouse (Mus musculus) nematode (Caenorhabitis briggsae), red flour beetle (Tribolium castaneum), wasp (Nasonia vitripennis), amphioxus (Branchiostoma floridae), chicken (Gallus gallus) and sea squirt (Ciona intestinalis). Despite several phyla-specific gaps and insertions, a high degree of conservation was observed throughout the protein, with the first third of the N-terminus showing the highest degree of amino acid similarity (Figure 5B).
Second allele of Szt2
Because of the uniqueness of the SZT2 protein and uncertainty about the effect of the mutation – particularly given the increase in Szt2 transcript in Szt2m1Frk homozygotes - we obtained ES cells corresponding to a second allele – a gene-trap mutation – in exon 21 of Szt2 (Szt2Gt(XH662)Byg). Mice were obtained from these ES cells (129/Sv strain background), and chimeric mice were crossed to B6 or B6-Szt2/Szt2, to obtain compound and single heterozygotes for seizure analysis. The compound heterozygotes had a significantly higher incidence of seizures, when compared to either single heterozygote – supportive of allelism (Figure 6). Interestingly, in these analysis single heterozygotes from both alleles had a slightly higher incidence than wild-type, suggesting a small amount of dominance (also observed when progeny-testing Szt2m1Frk backcross mice carrying recombinant chromosomes – data not shown). We then backcrossed the Szt2Gt(XH662)Byg allele to 129S1/SvImJ, and analyzed the electroconvulsive threshold of individual males, which gave results consistent with the population median thresholds determined previously for the Szt2m1Frk allele (data not shown). However, the Szt2Gt(XH662)Byg homozygotes may be more severely affected than Szt2m1Frk, since several mice bypassed the minimal clonic seizure endpoint, proceeding directly to tonic hindlimb extension.
Figure 6. Genetic complementation analysis of the two Szt2 alleles.

Shown at the top is the incidence of electroconvulsion (minimal clonic seizure response) in (B6 × 129S1)F1 hybrids of Szt2Gt(XH662)Byg (gene-trap, or Gt) and Szt2m1Frk (ENU) male mice with compound and single heterozygous genotypes shown, at a stimulus level corresponding to the approximate 3% response level of wild-type F1 hybrid mice. The p-values from Fisher exact test are shown at the bottom.
It was also noted that approximately 90% of the Szt2Gt(XH662)Byg homozygotes died before birth on a congenic 129S1 or F1 hybrid background, and none survived on a congenic B6 background (Table 1). This is perhaps consistent with the expression in embryo. In addition, on the 129 background the surviving homozygotes had a slightly diluted coat color. However, strikingly, as with the ENU allele, the gene-trap allele also showed an increase of RNA expression in heterozygotes and homozygotes (Figure 3A, N.b. the smaller transcript size for mutant allele is presumably due to the gene trap chimeric transcript). Assuming that the lethality phenotype is caused by the gene-trap mutation, the gene-trap allele is more likely to represent a null allele, when compared to the ENU allele.
Table 1.
Yield of Szt2Gt(XH662)Byg homozygotes observed at weaning from intercross matings
| Strain background | # hom. | # total | # expected | % total |
|---|---|---|---|---|
| C57BL/6J (N10) | 0 | 173 | 43 | 0% |
| (B6 × 129S1)F1 | 4 | 46 | 12 | 8% |
| 129S1/SvImJ | 13 | 205 | 51 | 6% |
Discussion
Szt2 mutant mice were detected in a small-scale ENU mutagenesis screen in C57BL/6J mice for electroconvulsive threshold (ECT). The same type of screen led to the identification of three additional seizure threshold mutants, each of which were ultimately shown to be alleles of the gene encoding KCNQ2, one of two potassium channel defects that cause Benign Familial Neonatal Convulsions – a rare inherited form of epilepsy (Frankel, 2003, Kearney et al., 2006, Yang et al., 2003). Although it is likely that many genes are involved in setting seizure threshold, the overall yield of mutants from the mutagenesis screen was disappointingly low; that is, of thousands of potential mutations screened, despite the robustness of the ECT test only a handful of heritable, threshold-altering mutations were discovered – and three were in the same gene, Kcnq2. The low yield might be related to the fact that C57BL/6J has one of the highest seizure thresholds of all inbred mouse strains: there may not be too many single mutation events that can lower the threshold sufficiently to be detected in C57BL/6J in this type of screen – the well-established Kcnq2 being one of the few. In this context, Szt2 seems like a good candidate for having a very significant role in regulating seizure threshold and neuronal excitation.
The SZT2 protein is very highly conserved in evolution, as there appears to be a single orthologue encoded by every land vertebrate genome sequence represented in the Ensembl database, and in several invertebrates as well – suggesting its functional importance. The apparent nematode orthologue encoding the predicted protein F54B3.1, appears to be essential for viability in C. elegans, based on RNAi knockdown studies (www.wormbase.org). This is consistent with the apparent embryonic lethality of many mouse Szt2Gt(XH662)Byg homozygotes, although the fact that up to 10% can survive suggests that even the Szt2Gt(XH662)Byg allele might not be a functional null.
One potential clue to the function of Szt2 comes from its highly-conserved genomic proximity to Med8, encoding one of approximately 30 subunits of the Mediator of transcription complex. The two respective genes are very closely configured in vertebrate genomes and appear to share a promoter sequence. This, together with the observation that mutant Szt2 transcript is upregulated in Szt2 homozygous mice – not often observed in mutations with significant homozygous phenotypes - suggests a model whereby SZT2 protein is normally involved in the trans regulation of its own transcript, disrupted by loss-of-function in mutants. We did not, however, detect any direct protein interaction between the MED8 and SZT2 in co-immunoprecipitation experiments in transiently transfected COS-7 cells (unpublished results). Perhaps future exploratory proteomics studies will reveal SZT2 interaction partners. However, an alternate model is of a dose-dependent neomorphic function; without an antibody for SZT2 we cannot know formally whether truncated protein is made in the mutants. In the future, development of an antibody and exploratory proteomic analysis from mutant and wild-type brain may reveal SZT2 interaction partners and ultimately lead to an understanding of its molecular function.
Despite the fact that Szt2 mutants affect two aspects of epilepsy susceptibility – acute seizure threshold and shortened latency to kindling - its potential role in underlying human idiopathic epilepsy is currently difficult to predict. The approximately 20 idiopathic epilepsy genes that are known are thought to represent only a small fraction of genes that contribute to most idiopathic epilepsy – the rest remain to be discovered. While most of the phenotypic effect in Szt2 mutant mice is recessive, the fact that there is a detectable heterozygous effect leaves the possibility open that human SZT2 variants may play a role in common disease when expressed in the appropriate genetic context.
Supplementary Material
Supplemental Figure 1. Predicted sequence of Mus musculus (C57BL/6J) SZT2 protein and location of matching peptides from MALDI-TOF spectra. Probability-based Mowse score is − 10*Log(P), where P is the probability that the observed match is a random event. Protein scores greater than 60 are significant (p<0.05). The GenBank accession number for the Szt2 cDNA sequence is FJ998170.
Acknowledgments
We thank Greg Cox and Verity Letts for their feedback on this manuscript, Doug Hinerfeld and Rob Wilpan for their assistance with mass spectrometry, Ron Conaway for helpful discussions, Rod Bronson for reviewing our histological analysis, and Steve White for sharing with us his pilot studies on corneal kindling in mutants. This work was supported by NIH grant NS31348 (to WNF) NIH grant HD41066 (to TPO) and CA34196 (JAX Scientific Services).
References
- Brower CS, Sato S, Tomomori-Sato C, Kamura T, Pause A, Stearman R, Klausner RD, Malik S, Lane WS, Sorokina I, Roeder RG, Conaway JW, Conaway RC. Mammalian mediator subunit mMED8 is an Elongin BC-interacting protein that can assemble with Cul2 and Rbx1 to reconstitute a ubiquitin ligase. Proc Natl Acad Sci U S A. 2002;99:10353–10358. doi: 10.1073/pnas.162424199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel WN. nmf88, low threshold to electroconvulsive minimal clonic seizures. The Jackson Laboratory; 2003. [Google Scholar]
- Frankel WN, Taylor L, Beyer B, Tempel BL, White HS. Electroconvulsive thresholds of inbred mouse strains. Genomics. 2001;74:306–312. doi: 10.1006/geno.2001.6564. [DOI] [PubMed] [Google Scholar]
- Kearney JA, Yang Y, Beyer B, Bergren SK, Claes L, Dejonghe P, Frankel WN. Severe epilepsy resulting from genetic interaction between Scn2a and Kcnq2. Hum Mol Genet. 2006;15:1043–1048. doi: 10.1093/hmg/ddl019. [DOI] [PubMed] [Google Scholar]
- Kitami T, Ernest S, Gallaugher L, Friedman L, Frankel WN, Nadeau JH. Genetic and phenotypic analysis of seizure susceptibility in PL/J mice. Mamm Genome. 2004;15:698–703. doi: 10.1007/s00335-004-3007-7. [DOI] [PubMed] [Google Scholar]
- Matagne A, Klitgaard H. Validation of corneally kindled mice: a sensitive screening model for partial epilepsy in man. Epilepsy Res. 1998;31:59–71. doi: 10.1016/s0920-1211(98)00016-3. [DOI] [PubMed] [Google Scholar]
- Potschka H, Loscher W. Corneal kindling in mice: behavioral and pharmacological differences to conventional kindling. Epilepsy Res. 1999;37:109–120. doi: 10.1016/s0920-1211(99)00062-5. [DOI] [PubMed] [Google Scholar]
- Racine RJ. Modification of seizure activity by electrical stimulation. II. Motor seizure. Electroencephalogr Clin Neurophysiol. 1972;32:281–294. doi: 10.1016/0013-4694(72)90177-0. [DOI] [PubMed] [Google Scholar]
- Singh NA, Charlier C, Stauffer D, DuPont BR, Leach RJ, Melis R, Ronen GM, Bjerre I, Quattlebaum T, Murphy JV, McHarg ML, Gagnon D, Rosales TO, Peiffer A, VEA, Leppert M. A novel potassium channel gene, KCNQ2, is mutated in an inherited epilepsy of newborns. Nature Genet. 1998;18:25–29. doi: 10.1038/ng0198-25. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2 - a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Beyer BJ, Otto JF, O'Brien TP, Letts VA, White HS, Frankel WN. Spontaneous deletion of epilepsy gene orthologs in a mutant mouse with a low electroconvulsive threshold. Hum Mol Genet. 2003;12:975–984. doi: 10.1093/hmg/ddg118. [DOI] [PubMed] [Google Scholar]
- Yang Y, Mahaffey CL, Berube N, Maddatu TP, Cox GA, Frankel WN. Complex seizure disorder caused by Brunol4 deficiency in mice. PLoS Genet. 2007;3:e124. doi: 10.1371/journal.pgen.0030124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Predicted sequence of Mus musculus (C57BL/6J) SZT2 protein and location of matching peptides from MALDI-TOF spectra. Probability-based Mowse score is − 10*Log(P), where P is the probability that the observed match is a random event. Protein scores greater than 60 are significant (p<0.05). The GenBank accession number for the Szt2 cDNA sequence is FJ998170.


